Abstract
A series of oligopeptides with β-forming and adhesive properties, were synthesized and analyzed for adhesion shear strength, secondary structure, and association properties. The sequences contained related hydrophobic core segments varying in length from 5 to 12 residues and flanked by di- or tri-lysine segments. Three remarkable peptides consisting of just 11 residues with hydrophobic core sequences of FLIVI, IGSII, and IVIGS flanked by three lysine residues gave the highest dry adhesion shear strength and displayed unusual biophysical properties in the presence and absence of water. KKKFLIVIKKK had its highest adhesion strength at 2% (w/v) at pH 12.0 and showed the highest adhesion strength after exposure to water (water resistance). Both KKKIGSIIKKK and KKKIVIGSKKK, at 4% (w/v) at pH 12.0, displayed nearly identical dry shear strength values to that with the FLIVI core sequence. The peptide with IGSII core, however, displayed a lower water resistance and the latter, IVIGS, showed no water resistance, completely delaminating upon soaking in water. These are the smallest peptides with adhesive properties reported to date and show remarkable adhesion strength even at lower concentrations of 0.2% (w/v), which corresponds to 1.6 mM. The FLIVI containing peptide adopted a β-sheet secondary structure in water while the IGSII- and IVIGS-containing sequences folded similarly only in the absence of water. Analytical ultracentrifugation studies showed that when the FLIVI sequence adopts β-structure in aqueous solution, it associates into a large molecular weight assembly. The random coils of IGSII and IVIGS showed no tendency to associate at any pH.
Introduction
Biological protein-based adhesives have attracted considerable attention in recent years. These protein adhesion molecules are classified based on three functional structures:
-
1.
Chemical modification of amino acids such as the enzymatic conversion of L-tyrosine to 3,4-dihydroxy-L-phenylalanine (L-DOPA) that can then form cross-links or chelate metals (1);
-
2.
Covalent bonds involving inter- and intra-S-S bonding (2); and
-
3.
Noncovalent bonds utilizing just hydrogen bonds and van der Waals interactions (3,4).
Most of the reported protein-based adhesives originate from anchoring proteins secreted by marine organisms, such as mussels, oysters, and barnacles (5). These bioadhesives are strong, durable, and water-resistant (6) and have been suggested for use in medical and dental applications (7–9). It was found that marine adhesive proteins have similar repetitive polypeptides with basic isoelectric points and contain high levels of L-DOPA, which has been identified as one of the key components for molecular cross-linking during formation of the water-resistant adhesive (10). Unfortunately, extraction of precursor adhesive proteins from marine organisms is expensive and gives low yields. Their larger size and heterogeneity makes structural studies on the aggregated adhesive state impossible to do with current methodologies.
Another major source of protein-based adhesives is from plants such as the soybean. They have several advantages over traditional petroleum-based adhesives including being environmentally friendly and renewable. Soy proteins are considered potential alternatives to petroleum-based adhesives in applications such as particleboard, fiberboard, wood panel, and construction (11–14). Soy protein-based adhesives demonstrate dry adhesion strength similar to petroleum-based urea formaldehyde in particleboard (15–17). However, they display reduced water resistance. By isolating and testing the constituent subunit components from soybean storage proteins, net charge and hydrophobicity play an important role in protein adhesion (18).
Recently we adopted a synthetic strategy that provides a reductionist model for studying protein adhesion with regard to preferred sequence, secondary structure, and mean hydrophobicity (4). A series of peptides were designed and synthesized with a common nine-amino-acid residue hydrophobic core (FLIVIGSII, abbreviated as h9) and flanked by clustered positively or negatively charged lysine, arginine, histidine, or glutamate residues. Among K3h9K3, E3h9E3, K3h9E3, E3h9K3, R3h9R3, and H3h9H3 peptides, K3h9K3 was found to have the highest adhesion strength using a simple glued wood assay, especially at pH 12, with an adhesion strength value of 3.7 MPa when cured at 170°C. Hydrophobicity, secondary structure, and curing temperatures appeared to be the most important factors influencing peptide adhesion.
While the adhesion strength of this sequence is considerably less than that observed with commercial petroleum-based products, this adhesive works in the absence of covalent bonds. These peptide sequences can be modified easily to include other functionalities that form a variety of tunable covalent interactions that will greatly increase adhesive strength. Toward this objective, this study defines a minimal core peptide length that still functions in the absence of covalent bonds. Synthetic peptides with varying hydrophobic core lengths and flanking tri-lysine clusters were synthesized and tested. For peptides with hydrophobic core segments of five amino acids, the contribution of amino-acid composition and the number of lysines in the flanking clusters were further studied. Identifying a small adhesive peptide would further secondary structural and modeling studies aimed at identifying and improving the physical properties of biorenewable adhesives.
Materials and methods
Materials
Dichloromethane, dimethylformamide, diethyl ether, and n-methylpyrrolidone were purchased from Fisher Biotech (Fair Lawn, NJ); 1,2-ethanedithiol, n, n-diisopropylethylamine, piperidine, and trifluoroacetic acid were purchased from Aldrich (Milwaukee, WI); 2-(1H-benzotriazol-1-yl) 1,1,3,3-tetramethyluronium hexafluorophosphate and 1-hydroxybenzotriazole were purchased from Q-Biogene (Carlsbad, CA); all protected amino acids including Nɛ-formyl-lysine were purchased from Anaspec (San Jose, CA). CLEAR-amide resin was purchased from Peptide International (Louisville, KY). All reagents were ACS certified unless specified otherwise. Cherry wood samples were purchased from the Veneer One (Oceanside, NY).
Peptide synthesis
All peptides were synthesized by an automated 9-fluorenylmethoxycarbonyl (Fmoc) strategy using Fmoc-protected amino acids and CLEAR-amide resin on a model No. 431 peptide synthesizer (Applied Biosystems, Foster City, CA). The C-termini were blocked as the carboxyamide. The peptides were cleaved from the resin with simultaneous deprotection by treatment with water in 95% trifluoroacetic acid for 2 h at room temperature. The cleaved peptides were washed three times with diethyl ether and dissolved in 20% acetonitrile in water, then lyophilized. All syntheses were characterized by matrix-assisted, laser desorption-ionization time-of-flight (MALDI-TOF) mass spectroscopy on an Ultraflex II instrument (Bruker Daltronics, Billerica, MA).
Hydrophobicity calculations
Mean residue hydrophobicity values are reported as ΔGavg in kcal/mol. To obtain ΔGave, the ΔGresidue contribution for each amino acid were summed and then divided by the number of residues. Values were taken from the Octanol-Interface Scale published on Professor Stephen White's (University of California, Irvine) website: http://blanco.biomol.uci.edu/. Depending on whether you have two or three lysines at each end, pI values for the peptides change. Applying the Henderson-Hasselbach equation at pH 12.0 and using a pKa value of 10.5 for the bis-K2 adducted peptide and 10.7 for the bis-K3 adducted peptide, the percentages of charged lysine residues were 3.2% and 5.0%, respectively. Since there are no published values for the hydrophobicity of lysine at pH 12.0, mixed hydrophobicity values of −0.61 and −0.57 kcal/mol were calculated based on the values of leucine (−0.69 kcal/mole) (96.8%) and lysine (+1.81 kcal/mol) (when present at either 3.2% or 5%).
Mean residue β-sheet propensity calculations
Mean residue β-sheet propensity values for the peptides in the adhesive state (pH 12.0) were calculated using two β-sheet propensity scales, from Chou and Fasman (19) and Koehl and Levitt (20). Using both scales, the propensity value for each amino acid in the sequence was summed and then divided by the number of residues in the sequence to obtain the mean. For lysine at pH 12.0, mixed propensity values for the two scales were calculated using leucine and lysine based on the mole percents of lysine in its charged state (as stated in the preceding paragraph). Values of −0.14 and 0.37 were calculated for the Chou and Fasman and Koehl and Levitt scales, respectively.
Sample preparation
Adhesive solutions at various peptide concentrations were prepared in water and stirred for 1 h, subsequently the pH of each solution was adjusted using either 1.0 N sodium hydroxide or 1.0 N hydrochloric acid. The adhesive solution (360 μL) was brushed onto one side of a wood sample with a marked area of 8 cm × 20 cm. The treated wood pieces were allowed to rest at room temperature for 15 min. Two coated wood surfaces were assembled and then pressed by using a Hot Press (model No. 3890 Auto “M”; Carver, Wabash, IN) at pressure of 1.4 MPa cm−2 at 130°C for 5 min.
Adhesion strength measurements
The wood specimens for shear strength testing were prepared according to the ASTM D2339-98 (21). The peptide-glued cherry wood test pieces were conditioned at 23°C and 50% relative humidity for three days, cut into three strips with a glue area of 20 mm × 20 mm. The samples were subsequently conditioned (as described above) for an additional four days before testing for shear strength. An Instron testing machine (model No. 4465; Canton, MA) with a crosshead speed of 1.6 mm/min was used. Stress at maximum load was recorded. Wood failure was estimated according to Standard Practice for estimating the percentage of wood failure in adhesive bonded joints ASTM D5266-99 (22). The reported results were an average of six replicates.
Water resistance
Water resistance was measured following Standard Test Methods for Resistance of Adhesives to Cyclic Laboratory Aging Conditions (23) and Standard Test Methods for Effect of Moisture and Temperature on Adhesive Bonds (24). The fully cured glued wood pieces were allowed to soak in tap water at 23°C for 48 h. The wet strength was obtained by testing immediately after soaking. The shear strength was tested as described above.
Glass slide preparation
Polished glass microscope slides were initially washed with a sponge using a laboratory detergent (cat. No. 04-320-4; Fisher Scientific, Waltham, MA). Washed slides were rinsed with tap water and blown dry using N2 gas. All the following wash steps were carried out in a laminar flow hood. Individual slides were then sonicated for 5 min sequentially in acetone, ethanol, and toluene. Between each solvent wash, the slides were dried using a stream of N2. The cleaned slides were placed in washed petri dishes and transported to the plasma cleaner. The slides were plasma-cleaned for 5 min using a model No. PDC-32G plasma cleaner (Harrick Scientific, Ossining, NY). Room atmosphere air was used as the plasma gas and introduced after the onset of plasma formation. The process of plasma-cleaning serves to hydroxylate the surface and ensures that the surface is hydrophilic before coating.
Sialyation of glass slides
The trichlorododecylsilane and trichlorohexadecylsilane, purchased from Fluka (Buchs, Switzerland) and the (3-cyanopropyl) trichlorosilane, obtained from Fisher Scientific were applied to the glass surface using vapor deposition. Several drops of the individual silanes were placed in a closed clean container (6” × 4.5” × 1.75”) made of polyethylene terephthalate and with three glass slides. The deposition time for the reaction was monitored by recording the contact angle measure with a drop of water. After 10 min, no further increase in contact angle was observed. The coated slides were stored in a rack in a sealed plastic container until used for the adhesion shear strength tests. Peptide was applied to the surface and subsequently tested for adhesion strength as previously described for the wood samples.
Circular dichroism spectra
The circular dichroic (CD) spectra were recorded on a model No. J-720 (Jasco, Tokyo, Japan) spectropolarimeter with a model No. RTE-111M circulator bath (Neslab Instruments, Portsmouth, NH) using a 1.0 mm quartz cuvette from 190 to 260 nm at 25°C. The spectra were an average of five scans recorded at a scan rate of 20 nm/min with a 0.2-nm step interval. For CD measurements, 1 mL samples of synthetic peptides were prepared with different pH adjusted solutions. The following solutions were used: water, pH 5.5; and 10 mM NaOH, pH 12.0. The final peptide concentrations ranged from 45 to 178 μM. All spectra were corrected by subtracting the baseline of the solutions recorded under the same condition. The CD absorbances were expressed as the mean residue ellipticity in units of degrees cm2 dmol−1. Peptide concentrations were determined with a BCA assay (Pierce, Rockford, IL) using a small purified synthetic peptide containing a tryptophan as the peptide standard. The concentration of the standard was determined by measuring the absorbance of the stock solution at 280 nm.
Infrared spectroscopy
A 0.5% w/v K3h5mIK3 solution was prepared by dissolving the peptide in 10 mM NaOH, pH 12.0. The solution was then applied to the ends of cleaned glass slides, such that when placed face to face a partial overlap of 400 mm2 (20 × 20 mm) was created. The paired slides were then hot-pressed as before (4) at 130°C. For IR measurements, five pairs of glued glass slides were pulled apart and the dried peptide film collected by scraping with a razor. Approximately 0.5 mg of dried adhesive was ground in a mortar and pestle with 5 mg predried KBr in a glove box. This protocol was followed to minimize any contributions of water to the IR bands centered at ∼1635–1640 cm−1, since such stretches can obscure or be mistaken for the Amide I of β-sheets. The finely ground mixture was pressed to 5000–6000 lbs for 4 min in a model B Laboratory Press (Carver) to produce a translucent pellet. The IR spectra were recorded on a Nexus 670 (Nicolet Biomedical, Madison, WI) Fourier-transform infrared spectroscopy (FTIR) ESP. The reported spectrum is an average of 32 scans at 2 cm−1 resolution. All spectra were corrected by subtracting the background. The instrument was purged every 5 min with nitrogen. Sample spectra were recorded immediately after the instrument was purged.
Analytical ultracentrifugation
Sedimentation velocity experiments were conducted using an Optima XL-I ultracentrifuge (Beckman Coulter, Fullerton, CA) with an An-60 Ti rotor. Sedimentation was monitored by absorbance at 260 nm with a 5-min interval for 120 min and thereafter by interference optics with a 30-s interval. Sample (400 μL) was loaded into double sector cells and sedimentation was done at 45,000 rpm at 20°C. Sedimentation data were analyzed using DCDT+ software, Ver. 1.16 (http://www.jphilo.mailway.com). Most of the buffer density and viscosity measurements were calculated by Sednterp, Ver. 1.08 (http://www.jphilo.mailway.com). The viscosity of peptide in 10 mM NaOH was measured using an Ostwald viscometer. The partial specific volumes of peptides were calculated from their amino acids’ composition using Sednterp, Ver. 1.08.
Results and discussion
Peptides design and hydrophobicity
As previously reported (4), peptides containing the nine-amino-acid residue-hydrophobic core (FLIVIGSII) termed h9 and flanked by different charged amino-acid residues, were analyzed for adhesion strength and secondary structure at acidic, neutral, and basic pH. The 15-amino-acid peptide (K3h9K3) was found to have the highest adhesion strength (3.0 MPa) at pH 12.0 using wood strips as the substrate. When standard glass microscope slides (silica) were used as the substrate, the adhesion strength dropped dramatically (0.32 MPa), suggesting that adhesion was occurring through a mechanical process, with the cohesive forces between the peptides providing the strength. Here K3h9K3 was used as a reference point to evaluate the effects of length and amino-acid composition of the hydrophobic core on adhesion strength. Peptides with hydrophobic core lengths of 5-, 7-, 11-, and 12-amino-acid residues were prepared (Table 1). To increase the length of the hydrophobic core, additional hydrophobic amino acids were incorporated from the parent IVS3 sequence (4) into the nine-amino-acid residue hydrophobic core FLIVIGSII to yield the peptides designated as K3h11K3 and K3h12K3. For hydrophobic core sequences with reduced lengths, the nine-amino-acid residue hydrophobic core h9 was truncated by removing the two C-terminal isoleucine residues, yielding a peptide denoted as K3h7K3. The five-amino-acid residue segments, from either the N- or C-terminus or middle of h9, were synthesized and denoted as K3h5nK3, K3h5cK3, and K3h5mK3, respectively. Peptides with reduced numbers of lysines, K2h5nK2 and K2h5cK2, were also prepared. Two five-residue peptides were prepared to test for the effect of packing interactions on adhesion strength with the replacement of the central amino-acid isoleucine of K3h5mK3 with leucine (K3h5mLK3) and alanine (K3h5mAK3). Finally, KAE16-IV (25), a peptide containing alternating polar and nonpolar residues and known to adopt β-sheet structure at both neutral and high pH, was used as a negative control. The sequences of peptides along with their masses, calculated pI values, mean residue hydrophobicity for both the hydrophobic core residues and for the lysine adducted sequences, and β-sheet propensities (using two different scales) are presented in Table 1.
Table 1.
Amino-acid sequences with calculated mean residue hydrophobicity of synthesized peptides and mean residue β-sheet propensity
| ΔGavg (kcal/mol) |
Mean residue β-sheet propensities, pH 12 |
|||||||
|---|---|---|---|---|---|---|---|---|
| Peptides | Sequence | MW | pI | hx | pH 7.0 | pH 12.0 | C-F | K-L |
| K3h9K3 | KKK-FLIVIGSII-KKK | 1742.3 | 10.7 | −0.39 | 0.49 | −0.46 | 0.15 | 0.23 |
| K3h11K3 | KKK-VFFLIVIGSII-KKK | 1988.6 | 10.7 | −0.43 | 0.36 | −0.48 | 0.14 | 0.21 |
| K3h12K3 | KKK-FLIVIGSIIVIL-KKK | 2067.8 | 10.7 | −0.47 | 0.29 | −0.50 | 0.16 | 0.23 |
| K3h7K3 | KKK-FLIVIGS-KKK | 1516.0 | 10.7 | −0.29 | 0.68 | −0.42 | 0.14 | 0.24 |
| K3h5cK3 | KKK-IGSII-KKK | 1269.7 | 10.7 | −0.20 | 0.90 | −0.40 | 0.013 | 0.25 |
| K3h5nK3 | KKK-FLIVI-KKK | 1371.8 | 10.7 | −0.69 | 0.67 | −0.62 | 0.15 | 0.26 |
| K2h5cK2 | KK-IGSII-KK | 1013.3 | 10.5 | −0.20 | 0.69 | −0.31 | 0.14 | 0.26 |
| K2h5nK2 | KK-FLIVI-KK | 1115.5 | 10.5 | −0.6 | 0.42 | −0.53 | 0.16 | 0.28 |
| K3h5mIK3 | KKK-IVIGS-KKK | 1255.6 | 10.7 | −0.149 | 0.92 | −0.37 | 0.10 | 0.21 |
| K3h5mLK3 | KKK-IVLGS-KKK | 1255.6 | 10.7 | −0.11 | 0.98 | −0.36 | 0.12 | 0.23 |
| K3h5mAK3 | KKK-IVAGS-KKK | 1213.6 | 10.7 | 0.08 | 1.02 | −0.35 | 0.06 | 0.17 |
| KAE16-IV | (KA)4(EA)4-CO-NH2 | 1614.8 | 6.3 | — | 1.22 | 0.62 | −0.07 | 0.01 |
MW, molecular weight; pI, isoelectric point; ΔGavg, calculated mean residue hydrophobicity for entire sequence; and hx, calculated mean residue hydrophobicity for hydrophobic core segment. The β-sheet propensities were calculated using the Chou and Fasman (19) (C-F) and Koehl and Levitt (20) (K-L) scales.
The hydrophobic core sequences showed a range of mean residue hydrophobicities (ΔGave) from +0.08 to −0.69 kcal/mol. The h5x sequences span the full range of mean residue hydrophobicities. Since no ionizable amino acids were present within the core sequences, hydrophobicities were not affected by alterations in pH. Incorporating ionizable lysines, the KxhxKx peptides displayed dramatic increases in hydrophobicity in moving from neutral pH to high pH. The presence of the lysine groups showed calculated mean residue hydrophobicities at pH 7.0 that were positive and ranged from 0.29 to 1.02 kcal/mol, explaining high aqueous solubility of the peptides at neutral pH. At pH 12.0, the KxhxKx peptides showed significant increases in hydrophobicity with the mean residue hydrophobicity values ranging from −0.62 to −0.30 kcal/mol. At pH 12.0, the N-terminal α-amino groups as well as the lysine ɛ-amino groups become mostly deprotonated, thereby increasing the lysine's hydrophobicity to a value that we have assigned as equivalent to leucine. As stated in Materials and Methods, 3.2% of the lysine ɛ-amino groups retain a positive charge. The control sequence KAE16-IV had mean residue hydrophobicities that remained positive at both pH 7.0 and 12.0 with values of 1.22 and 0.62 kcal/mol, respectively. This sequence, while a strong β-former, has the least hydrophobic character of any sequence tested for adhesive properties.
Adhesion strength
The adhesion strengths for many of the peptides in Table 1 are shown in Fig. 1. At 4% (w/v), a concentration employed previously (4), all KxhxKx peptides demonstrated higher adhesion strength at pH 12.0 compared to values obtained at pH 7.0. Adhesives prepared at pH 12.0 displayed wood failure (the substrate fails before the glue) in 15% of the tests, whereas those at pH 7.0 had a value closer to 5%, suggesting that adhesives made from peptides at pH 12.0 displayed stronger protein molecular cohesion. All of the KxhxKx sequences shown in Fig. 1 had secondary structures (as determined by circular dichroism) that changed from random coils at neutral pH to β-sheet structures at pH 12.0. The KAE16-IV sequence was β-sheet-structured at both pH values, yet had only minimal adhesion strengths. Adhesion values for KAE16-IV were equivalent to those recorded for the unstructured KxhxKx sequences in Fig. 1 at pH 7.0. This suggests that β-sheet structure alone is not sufficient to convey adhesive behavior. The β-peptides that displayed adhesive properties possessed mean hydrophobicity values of < −0.30 kcal/mol.
Figure 1.

Shear strength of adhesives prepared from peptides with differing hydrophobic core lengths. Adhesives were prepared at either pH 7.0 (open bar) or 12.0 (solid bar) and pressed at 130°C, 1.4 MPa for 5 min.
The sequence length of a peptide hydrophobic core was tested as a potential factor in determining adhesion strength. Compared to the nine-amino-acid residue hydrophobic core at pH 12.0, adhesion strength increased to a value of 2.9 ± 0.2 MPa as the core length increased to 11-amino-acid residues. The 12-residue sequence showed a value of just 2.24 ± 0.1 MPa. This result is somewhat paradoxical because the h12 core is slightly more hydrophobic than the h11, −0.52 vs. −0.49 kcal/mol, respectively. This result suggests that both amino-acid composition and positional effects can alter adhesion strength.
For the shorter hydrophobic cores, the peptide with a seven-amino-acid residue showed similar adhesion strength to the nine-residue core. However, further reduction of the core length to five-amino-acid residue segments derived from the N- and C-termini, and middle segments of the h9 peptide produced surprisingly higher adhesion strengths (Fig. 2). Adhesives from the peptide with five-amino-acid residue core K3h5nK3, K3h5mIK3, and K3h5cK3, had higher adhesion strengths of 3.27 ± 0.25, 3.24 ± 0.13, and 3.44 ± 0.23 MPa, respectively, at 4% (w/v). The hydrophobic core sequences of the β-forming peptides presented in this article share compositional similarities with regard to mean residue hydrophobicity at pH 12.0 with the putative nucleation sites of the Aβ (1–42) peptide found in senile fibril plaques in Alzheimer's patients and the islet amyloid polypeptides. These short segments within the longer peptides have been proposed as the sites of nucleation for the β-strand aggregates. Within the Aβ sequence there are two sites, residues 16–20 (KLVFF) and 30–35 (AIIGLM) (26) and one in the islet amyloid peptide, residues 23–28 (IFGAIL) (27). In the Aβ study (26), enhanced aggregation of the peptides occurred only when residues 17–20 or 30–35 were present. Enhanced fibril formation was seen using atomic force microscopy when peptides containing these two sites were added to Aβ (1–40). The Aβ 16–20 (KLVFF) sequence is similar to our peptides containing KFLIVI while the Aβ AIIGLM 30–35) sequence is similar to our sequences that contain IGSII. Our different five-residue core sequences contain one of these two motifs and indicate that these core sequences contain unique aggregating properties that lead to the formation of protofibrils. The fact that some of the sequences contain analogs for both of the Aβ peptide nucleation sites suggests we could potentially produce fibrils that propagate in two separate planes.
Figure 2.
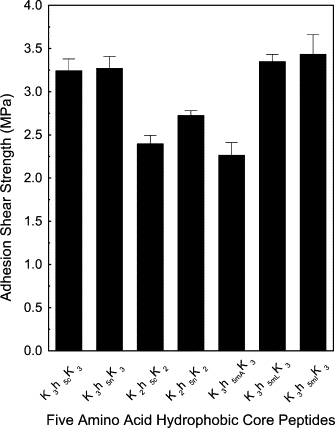
Shear strength for the different Knh5Kn adhesives peptides prepared at pH 12.0 and pressed at 130°C, 1.4 MPa for 5 min.
To statistically ascertain the contribution of peptide hydrophobicity to adhesion strength, linear regression was used to analyze the adhesion strength data at both pH 7.0 and pH 12.0 against their corresponding mean residue hydrophobicities. A significant (P < 0.0001 and R2 = 0.713) negative linear correlation was found (Fig. 3), which, at first glance, supports a view that increased hydrophobicity is a contributing factor in the adhesion strength. On closer inspection, however, it should be noted that the data points are clustered about hydrophobicity values observed at neutral and elevated pH. Within each cluster, the data points are randomly distributed, yielding no discernable correlation within each pH grouping. In raising the pH, the net charge on the lysine residues was virtually eliminated, thereby reducing electrostatic repulsion. In the absence of charge, the core sequences can then come together. This interpretation is supported by earlier work (4) where the six lysines were replaced by the smaller lysine analog, diaminopropionic acid, with a shorter side chain. No difference in adhesion strength was observed when the potentially more hydrophobic lysines were replaced with this less hydrophobic analog.
Figure 3.
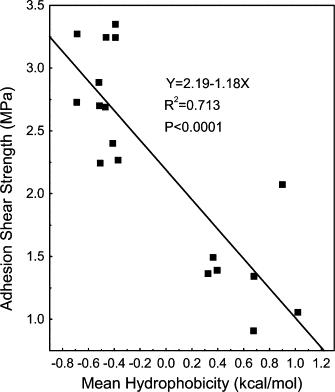
Correlation of adhesion shear strengths of peptides with the calculated mean residue hydrophobicity values (ΔGave) at pH 12.0. These values shown were taken from those presented in Table 1 and Figs. 1 and 2.
Somewhat contradictory results were observed for peptides flanked with just two lysine residues. Specifically, adhesion strengths for 4% (w/v) solutions of K2h5nK2 and K2h5cK2 at pH 12.0 were 2.73 ± 0.06 and 2.40 ± 0.1 MPa, respectively. These values were substantially lower than that measured for the tri-lysine flanking derivatives. The calculated mean hydrophobicity values for ΔGavg at pH 12.0 were −0.42 and −0.31 kcal/mol for the K3h5cK3 and K2h5cK2 pair, with −0.65 and −0.53 kcal/mol for K3h5nK3 and K2h5nK2 pair. While the hydrophobicity values are higher at elevated pH for the K3-containing peptides, they are substantially lower at pH 7.0. The overall net charge is higher at both pH 7.0 and 12.0 for the K3-containing sequences. Since our protocol has the peptides dissolved initially at neutral pH where they are unstructured and then raised to pH 12.0, peptide solubility will have a large influence on adhesion strength. Insoluble peptides show no adhesion properties (unpublished observations). Having three versus two lysines on either side of the hydrophobic core while slightly increasing hydrophobicity at pH 12.0 also increases the net percent of charged lysines at pH 12.0 to 5% vs. 3.2%, thereby influencing solubility at both pH values. In a previous study (28), we observed a direct positive correlation between the number of adducted lysines at either terminus of a hydrophobic peptide and solubility.
Another statistical analysis was performed looking at the mean residue β-sheet propensities for the different sequences shown in Table 1. Linear regression was used yielding P < 0.01 and R2 values of 0.50 and 0.49 for the Chou and Fasman (19) and Koehl and Levitt (20) scales, respectively (figure not shown). These calculated values indicate a modest correlation between the adhesion shear strength and β-sheet propensity.
Comparing the five-residue core sequences K3h5mIK3 and K3h5cK3, these peptides are made up of a mixture of three residues with larger molal volumes, Ile = 101.9 and Val = 85.2, and two smaller ones, Ser = 54.9 and Gly = 36.5 mL/mol (29). For IVIGS (h5mI), the large and small residues are side by side, whereas, in the case of K3h5cK3 (IGSII), the small residues are flanked by larger ones. The third sequence, K3h5nK3 (FLIVI), contains only five larger amino acids—Phe = 113.3, Leu = 101.9, Ile = 101.9, and Val = 85.2 mL/mol—and gave a similar adhesion strength. The adhesion strength differences between the mixed size cores and the all-large sized residue core suggests no difference between mixture of all large versus large and small residues for dry adhesion strength. To test whether introduction of a third smaller residue increases or decreases adhesion strength, Ile-6 in K3h5mIK3 was substituted with either Leu (101.9 mL/mol) or Ala (52.6 mL/mol). As shown in Fig. 2, addition of a different large hydrophobic residue, Leu, resulted in an equivalent adhesive strength of 3.35 ± 0.08 MPa, with replacement of the smaller Ala residue decreasing adhesion strength to a value of 2.67 ± 0.14 MPa. In the case of the K3h5mK3 sequence, introducing a smaller residue that decreases both the hydrophobicity and molal volume of the sequence weakens the adhesion strength.
In our previous study (4), the sequence K3h9K3 displayed maximal strength at 4% (w/v), which corresponds to a concentration of 23 mM. Both K3h5n K3 and K3h5mIK3, on the other hand, showed concentration dependences that were significantly left-shifted (Fig. 4). Maximal adhesion strength was observed at 0.5% (3.2 mM) for K3h5mIK3, with significant adhesion strength observed at 0.1% (0.6 mM). The sequence K3h5nK3 showed considerable strength starting at 0.1%, followed by a more linear concentration-dependence profile with maximal strength of 3.9 ± 0.2 MPa observed at 2%. At 4%, there was a significant drop-off due to decreased solubility for the more hydrophobic sequence. These are remarkable values relative to commercial protein-based adhesives such as those made from fragmented soy protein fractions, where typical concentrations ranging from 8 to 12% are required for maximal adhesion strength (30).
Figure 4.
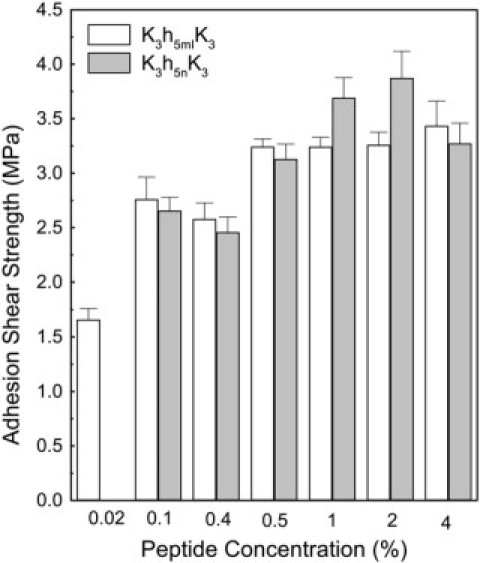
Concentration dependence of adhesion strength for K3h5nK3 and K3h5mIK3 prepared at pH 12.0 and pressed at 130°C, 1.4 MPa for 5 min.
The adhesion strength of the K3h5n K3 peptide (2%) at pH 12.0 was tested on cleaned glass slides that had been precoated with alkyl silanes by vapor deposition. This treatment eliminates the net negative charge of the glass as well as fills all of the deformations in the surface, yielding a smooth hydrophobic surface. The peptide exhibited a shear strength of 0.23 MPa for the plasma-cleaned slides but was unable to glue any of the trichlorododecyl-silane, trichlorohexadecylsilane or (3-cyanopropyl) trichlorosilane-treated slides (data not shown). These data showed that the hydrophobic amino acids of the adhesive peptide did not interact significantly with the hydrophobic glass surface and suggests that the peptide's adhesive strength comes from a mechanical mechanism where cohesive forces between aggregated peptides trapped inside surface deformities prevail.
Another important property for adhesives is resistance in the presence of water. This value is termed “wet” shear strength and is a predictor of how glued wood products would behave after being exposed to environmental elements. A comparison of dry and wet strengths for K3h5nK3, K3h5cK3, and K3h5mIK3 is shown in Table 2. Using a hot-press temperature of 130°C for 5 min followed by soaking in water yielded residual strength percentages of 34.6, 20.4, and 0, for K3h5nK3, K3h5cK3, and K3h5mIK3, respectively. The residual wet-strength value of 34.6% for K3h5nK3 far surpasses the value of 22.0% obtained for K3h9K3, our best previously-reported sequence (4). In the absence of covalent bonds, water is able to penetrate the adhesive substrate and interfere with H-bonding networks.
Table 2.
Comparison of dry and wet adhesion strengths for three of the five-residue hydrophobic core sequences
| Sequence | Dry strength (MPa) | Wet strength (MPa) | % residual strength |
|---|---|---|---|
| K3h5nK3 | 3.27 ± 0.19 | 1.13 ± 0.25 | 34.6 |
| K3h5cK3 | 3.24 ± 0.13 | 0.66 ± 0.20 | 20.4 |
| K3h5mIK3 | 3.44 ± 0.23 | Delaminated | 0 |
| K3h9K3* | 3.05 ± 0.23 | 0.67 ± 0.08 | 22.0 |
All samples were used at 4% w/v and hot-pressed at 130°C. Glue samples were soaked in water for 48 h before strength test.
These values were reported previously in Shen et al. (17) and are shown here for comparison purposes.
The calculated mean residue hydrophobicity for K3h5nK3 (pH 12.0) was −0.69 kcal/mol and most likely accounts for the improved water resistance over K3h5cK3 and K3h5mIK3. These two sequences have similar hydrophobicities, −0.33 and −0.34 kcal/mol (Table 1); however, their behavior after water exposure differed substantially. This minimal difference in free energy is unlikely to account for the observed water susceptibilities. Rather, we propose a proximity effect is responsible for the difference. For the core sequences of K3h5cK3 (IGSII) and K3h5mIK3 (IVIGS), the more water-resistant IGSII sequence has hydrophobic residues flanking the more hydrophilic glycine and serine, whereas for IVIGS, the hydrophilic residues are closer to the peptide's terminus. Having the more hydrophilic residues closer to the end of the peptide could allow for more rapid solvation by water molecules.
Structural properties
With regard to structure, the sequence K3h5nK3 readily formed β-structure in 10 mM NaOH as measured by CD, while the sequences K3h5cK3 and K3h5mIK3 did not (Fig. 5). All of the sequences were unstructured under pH conditions where the lysine residues were nearly fully protonated. The transition for K3h5nK3 from unstructured at neutral pH to structured at pH 12.0 is in agreement with our previous study using K3h9K3 (4). The lack of structure for the other h5c and h5mI sequences is a new observation, and suggests that they remain highly solvated at pH 12.0. The water is able to compete for the intermolecular hydrogen-bonding sites necessary for the formation and stabilization of β-structure. There appears to be a direct correlation between the inability of the K3h5cK3 and K3h5mIK3 sequences to adopt β-structure in water as measured by CD and their reduced water resistance as adhesives. The adhesive matrix formed by these peptides may be hygroscopic, and, once present, the water competes for and displaces the intermolecular hydrogen bonds. The CD measurements were carried out over a range of peptide concentrations that was somewhat lower than the concentrations that exhibited maximal adhesion strength. Clearly secondary structure is present in the μM range of concentrations. The FTIR spectra obtained from dried adhesive material collected off cured glass slides confirms that β-structure is preserved at high concentrations.
Figure 5.
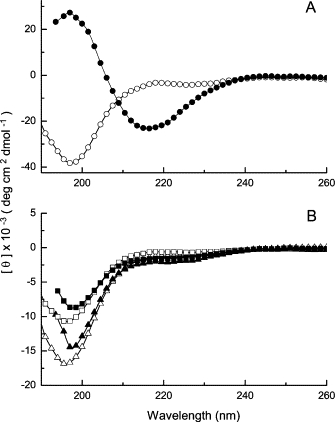
Circular dichroic spectra for K3h5K3 adhesive peptides. (A) K3h5nK3 in water (open circle) and in 10 mM NaOH (solid circle); (B) K3h5mIK3 in water (open triangle) and in 10 mM NaOH (solid triangle) and K3h5cK3 in water (open square) and in 10 mM NaOH (solid square).
Viscosity and mass analyses of aqueous K3h5nK3 and K3h5mIK3 peptides solutions (5 mM) were performed to see what sized assemblies were present at different pH values. Time-dependent viscosity changes for both peptides were studied at pH values of 6.8, 9.6, and 12.0. At all pH values K3h5mIK3 showed no changes in viscosity. At pH 6.8, K3h5nK3 showed no change in viscosity. At pH 12.0, K3h5nK3 immediately adopted a gel-like state. At pH 9.6, the viscosity increased steadily following second-order kinetics, reaching a plateau at 2 h (data not shown). The viscosity values calculated in this experiment were used in the analytical ultracentrifugation experiments (Fig. 6).
Figure 6.
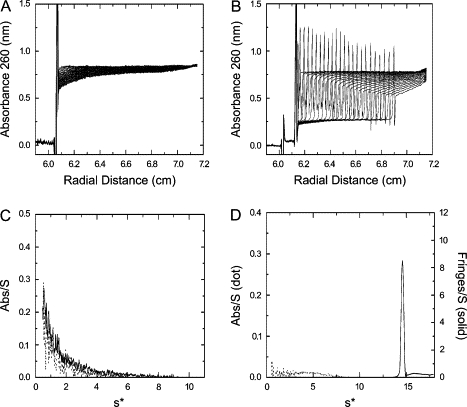
Sedimentation velocity profiles for K3h5nK3 and K3h5mIK3 at pH 6.8 and pH 9.6. The sedimentation velocity scans at pH 9.6 are shown for K3h5mIK3 (A) and K3h5nK3 (B). Bottom panels show g(s*) profiles at pH 6.8 (dotted line) and pH 9.6 (solid line) from sedimentation velocity profiles of K3h5mIK3 (C) and K3h5nK3 (D), respectively. Sedimentation conditions were as described in Materials and Methods.
The raw analytical ultracentrifugation data for the two peptides are shown (Fig. 6 A for pH 6.8 and Fig. 6 B for pH 9.6). The apparent masses of K3h5nK3 and K3h5mIK3 (5 mM) were analyzed at pH 6.8 and 9.6 after an incubation period of 120 min (Fig. 6, C and D, respectively). Both peptides at neutral pH showed no tendency to associate. The calculated sedimentation coefficients were <1.0, indicating that both peptides were in a low molecular weight form (possibly monomer). The K3h5mIK3 peptide at pH 9.6 behaved similarly to the neutral pH sample. Fig. 6 B shows the time-dependent sedimentation boundaries with absorption optics of K3h5nK3 at pH 9.6. Sharp spike peaks were seen at the sedimentation boundary, which might be the refraction of incident light due to the steep peptide concentration gradient. Dogic et al. (31) showed a similar sedimentation phenomenon using a rodlike colloid macromolecule assembly. They suggested that steep gradient was due to the pronounced self-sharpening effect leading to a hyper-sharp phenomenon. They proposed that these effects occur with elongated colloids, possibly protofibrils, rather than globular ones.
Fig. 6 D shows the S distributions at higher pH. The sedimentation coefficient is 14.6 S and the distribution is a single sharp peak, which is an indicator of the homogeneity of peptide assemblies and lower diffusion coefficient. Using the g(s*) fitting function in DCDT+ software, the molecular mass was estimated in excess of 106. These results show that, in the case of K3h5nK3, the peptide starts to associate in solution before becoming an adhesive. In the case of the K3h5mIK3 sequence, no such association takes place before the removal of the water. It appears that assembly in solution is not a prerequisite for adhesive formation.
FTIR (Fig. 7) was performed on dried K3h5mIK3 in a KBr pellet where the peptide was collected from glass slides that had been previously pressed at 130°C for 5 min and cured at an elevated relative humidity as described in Materials and Methods. This spectrum shows the characteristic amide I (1685 cm−1) CO stretching vibrations, the amide II (1556 cm−1) C-N stretching vibration, and N-H deformation vibration stretches, suggesting the presence of an antiparallel β-sheet conformation. The amide-I peak was split in two (common to β-sheet peptides) and the peaks appear at 1685 and 1631 cm−1. Likewise, the amide-II peak was split in two, showing stretches at 1556 and 1538 cm−1. No amide-III peak was present. This result indicated that what had been random-coil for K3h5mIK3 in solution at pH 12.0 became β-like upon the removal of the water. Folding and association of this peptide required elevated temperatures and desiccation, conditions that would usually denature most peptides. As part of this study, we located several glass slides that had been glued together at pH 12.0 more than four years earlier to assess the long-term stability of the peptide K3h9K3. Excess peptide that remained outside the fused joint as well as peptide under the joint were recovered and analyzed by matrix-assisted, laser desorption-ionization time-of-flight (MALDI-TOF) mass spectroscopy. Both the air-exposed material and the peptide that had formed the adhesive layer were not degraded and no cross-linking was detected (spectra not shown). The peptide was stable.
Figure 7.
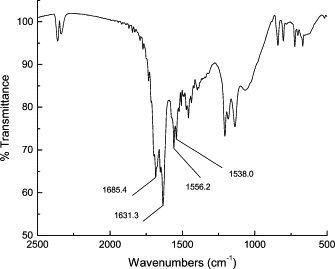
FTIR spectra of dried K3h5mIK3 in KBr after being harvested from glued glass slides prepared as described in Materials and Methods.
To assess the contributions of the remaining partial charge on the lysines at pH 12.0, K3h5nK3 and K3h5mIK3 were resynthesized using Nα- Fmoc, Nɛ-formyl-lysine at all six positions. Formylation eliminated all charge on the molecule at any pH, theoretically allowing it to be used at lower pH values. Due to aqueous insolubilities, the formylated K3h5nK3 and K3h5mIK3 were dissolved in ethylene glycol (HO-CH2-CH2-OH). Both peptides displayed β-structure in ethylene glycol (Fig. 8); however, the nonformylated forms of K3h5nK3 and K3h5mIK3 remained as random coil structures (data not shown). Elimination or reduction of the charge contributions of lysine by formylation or deprotonation at pH 12.0 greatly enhances β-structure formation due, most likely, to decreased repulsive Coulombic interactions.
Figure 8.
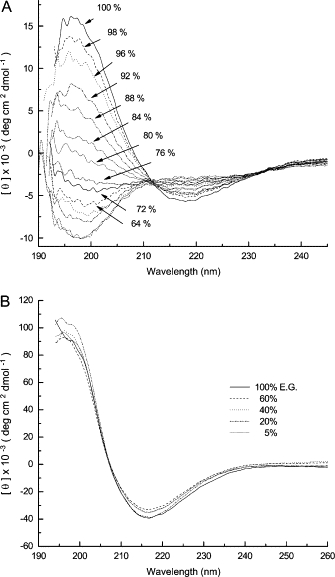
Circular dichroic spectra of peptides dissolved in mixtures of ethylene glycol and water. (A) The spectra of K3h5mIK3 (140 μM) are generated in the indicated percentages of ethylene glycol. (B) The spectra of K3h5nK3 (44 μM) are generated in the indicated percentages of ethylene glycol. Spectra were recorded at room temperature.
The stability of β-structured formylated peptides in the presence of increasing concentrations of water in ethylene glycol was examined by CD (Fig. 8). β-Structure was reduced by half in formylated K3h5mIK3 in the presence of 16% water (84% ethylene glycol). The formylated K3h5nK3 retained its β-structure in 90% water (10% ethylene glycol) (Fig. 8 B). Water disrupts more easily the hydrogen-bonding network in the β-structured formylated K3h5mIK3. Formylated K3h5nK3 proved much more resistant to the effects of water, suggesting that the accessibility of water to a K3h5nK3 hydrogen-bonding network is reduced. The adhesion strength for the two formylated sequences dissolved in ethylene glycol was not measured due to the high boiling temperature of ethylene glycol (196°C), higher than the 130°C curing temperature. These data suggest that limiting water penetration into the H-bonded β-structure is an important factor in maintaining adhesion strength in aqueous environments.
From this and previous work (4), the adhesive form of the desiccated nonformylated peptide at pH 12.0 is a β-sheet, but for the peptide to act as a strong adhesive, the folded peptide must undergo supramolecular assembly during desiccation to form a three-dimensional structure that can lead to entanglements.
Summary
In our earlier study (4) of examining the adhesive properties of K3h9K3 we hypothesized that two physiochemical properties were contributing to maximum adhesion strength: hydrophobicity and the ability of the sequence to adopt a stable β-sheet structure before the hot pressing, all in the absence of covalent bonding. In this study, three peptides, with five-residue hydrophobic cores analogous to protofibril nucleating segments in amyloid peptides, were identified. They had similar high dry adhesion strength, with K3h5nK3 as the most hydrophobic (at pH 12.0); adopting a β-sheet structure in water at pH 12.0, it displayed the best adhesion strength at 2% (w/v) and with substantial water resistance. The adhesion strength measured for this sequence is the strongest observed to date for any of the small adhesive peptides we have studied. The results herein also suggested that the length of the hydrophobic core, the hydrophobicity, the amino-acid order, and the chemical composition are important factors affecting adhesion. The bonding surface should be rough and possibly porous. The other two peptides, K3h5cK3 and K3h5mIK3, had no β-structure in aqueous solution, with only low or no water resistance, respectively. Only at extremely low water concentrations was β-structure observed. The work presented here suggests that the internal hydrophobic sequences are quite unique and that what may govern adhesion strength is the ability of these hydrophobic sequences to facilitate nucleated peptide associations into ordered higher structures. The flanking lysine tri-peptides aid solubility and act as a switch at elevated pH, allowing the peptide-peptide interactions to occur as the electrostatic barrier is reduced.
It also appears that having a β-sheet secondary structure before application to the wood surface is not needed for dry adhesion, but it is required for water resistance. We propose that hydrophobic core composition and arrangement along with β-structure in the presence of water, are crucially important for the water resistance adhesion. While the K3h5mIK3 peptide may not be particularly useful as a long-lasting adhesive where water is present, it may find application in switching devices where the adhesive bond degrades as a function of relative humidity or as a strong adhesive in environments where water is not present, such as in vacuums or in outer space.
Having peptides that spontaneously form noncovalent adhesives is a good starting point for engineering new sequences with other DNA-encoding amino acids that could introduce covalent bonds, thereby increasing both dry and wet adhesive strength. The small size of these sequences make them ideal for constructing plasmids or mini-genes that could express high-molecular-weight tandem repeats of this sequence that could be used in commercial applications.
Acknowledgments
We thank Elizabeth Blaesi for the viscosity measurements and Sean P. McBride for preparing the silane-treated glass slides.
This article is contribution No. 06-167-J from the Kansas Agricultural Experimental Station, Manhattan, Kansas. The study was supported in part by the United States Department of Agriculture, The Kansas Agricultural Research Station, and a Kansas State University Targeted Excellence Award.
Footnotes
Editor: Heinrich Roder.
References
- 1.Sagert J., Sun C., Waite J.H. Chemical subtleties of mussel and polychaete holdfasts. In: Smith A.M., Callow J.A., editors. Biological Adhesives. Springer-Verlag; Berlin, Heidelberg: 2006. [Google Scholar]
- 2.Humphrey A.J., Finlay J.A., Pettitt M.E., Stanley M.S., Callow J.A. Effect of Ellman's reagent and dithiothreitol on the curing of the spore adhesive glycoprotein of the green alga. Ulva. J. Adhesion. 2005;81:791–803. [Google Scholar]
- 3.Kamino K. Barnacle underwater attachment. In: Smith A.M., Callow J.A., editors. Biological Adhesives. Springer-Verlag; Berlin, Heidelberg: 2006. [Google Scholar]
- 4.Shen X.C., Mo X.Q., Moore R., Frazier S.J., Iwamoto T., Tomich J.M., Sun X.Z. Novel pH-dependent adhesive peptides. J. Nanosci. Nanotechnol. 2006;6:837–844. doi: 10.1166/jnn.2006.126. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto H., Sakai Y., Ohkawa K. Synthesis and wettability characteristics of model adhesive protein sequences inspired by a marine mussel. Biomacromolecules. 2000;1:543–551. doi: 10.1021/bm000061p. [DOI] [PubMed] [Google Scholar]
- 6.Papov V.V., Diamond T.V., Biemann K., Waite J.H. Hydroxyarginine-containing polyphenolic proteins in the adhesive plaques of the marine mussel. Mytilus edulis. J. Biol. Chem. 1995;34:20183–20192. doi: 10.1074/jbc.270.34.20183. [DOI] [PubMed] [Google Scholar]
- 7.Waite J.H. Nature's underwater adhesive specialist. Int. J. Adhes. 1987;7:9–14. [Google Scholar]
- 8.Benedict C.V., Picciano P.T. In: Adhesives from Renewable Resource. Hemingway R.W., Connor A.H., Branham S.J, editors. American Chemical Society; Washington, DC: 1989. [Google Scholar]
- 9.Strausberg R.L., Link R.P. Protein-based medical adhesives. Trends Biotechnol. 1990;8:53–57. doi: 10.1016/0167-7799(90)90134-j. [DOI] [PubMed] [Google Scholar]
- 10.Yu M., Deming T.J. Synthetic polypeptide mimics of marine adhesives. Macromolecules. 1998;31:4739–4745. doi: 10.1021/ma980268z. [DOI] [PubMed] [Google Scholar]
- 11.Clay J.D., Vijayendran B., Moon J. 57th Annual Technical Conference, Society of Plastics Engineers 1. 1999. Rheological study of soy protein-based wood adhesives; pp. 1298–1301. [Google Scholar]
- 12.Kumar R., Choudhary V., Mishra S., Varma I.K., Mattiason B. Adhesives and plastics based on soy protein products. Ind. Crops Pro. 2002;16:155–172. [Google Scholar]
- 13.Kuo M., Adams D., Myers D., Curry D., Heemstra H., Smith J.L., Bian Y. Properties of wood/agricultural fiberboard bonded with soybean-based adhesives. Forest Prod. J. 1998;48:71–75. [Google Scholar]
- 14.Thames S.F., Cook R.C., Wang Q., Mendon S.K. U.S. Pat. Publ. US; 2004. Particleboard and method for forming particleboard using soy protein binder. #173306. [Google Scholar]
- 15.Mo X.Q., Hu J., Sun X.S., Ratto J.A. Compression and tensile strength of low-density straw-protein particleboard. Ind. Crops Prod. 2001;14:1–9. [Google Scholar]
- 16.Li K., Peshkova S., Geng X. Investigation of soy protein-Kymene® adhesive systems for wood composites. J. Am. Oil Chem. Soc. 2004;81:487–491. [Google Scholar]
- 17.Cheng E., Sun X.Z., Karr G.S. Adhesive properties of modified soybean flour in wheat straw particleboard. Composites Part A: Appl. Sci. Mfg. 2004;35A:297–302. [Google Scholar]
- 18.Mo X.Q., Zhong Z.K., Wang D.H., Sun X.Z. Soybean glycinin subunits: characterization of physicochemical and adhesion properties. J. Agric. Food Chem. 2006;54:7589–7593. doi: 10.1021/jf060780g. [DOI] [PubMed] [Google Scholar]
- 19.Chou P.Y., Fasman G.D. Empirical predictions of protein conformation. Annu. Rev. Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 20.Koehl P., Levitt M. Structure-based conformational preferences of amino acids. Proc. Natl. Acad. Sci. USA. 1999;96:12524–12529. doi: 10.1073/pnas.96.22.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.American Society for Testing and Materials . Annual Book of ASTM Standards. Vol. 15. ASTM; Philadelphia, PA: 2002. Standard test method for strength properties of adhesive in two-ply wood construction in shear by tension loading. D2339-98. [Google Scholar]
- 22.American Society for Testing and Materials . Annual Book of ASTM Standards. Vol. 15. ASTM; Philadelphia, PA: 2002. Standard practice for estimating the percentage of wood failure in adhesive bonded joints. D5266-99. [Google Scholar]
- 23.American Society for Testing and Materials . Annual Book of ASTM Standards. Vol. 15. ASTM; Philadelphia, PA: 2002. Standard test methods for resistance of adhesives to cyclic laboratory aging conditions. D1183-96. [Google Scholar]
- 24.American Society for Testing and Materials . Annual Book of ASTM Standards. Vol. 15. ASTM; Philadelphia, PA: 2002. Standard test methods for effect of moisture and temperature on adhesive bonds. D1151-00. [Google Scholar]
- 25.Zhang S., Rich A. Direct conversion of an oligopeptide from a β-sheet to an α-helix: a model for amyloid formation. Proc. Natl. Acad. Sci. USA. 1997;94:23–28. doi: 10.1073/pnas.94.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu R., McAllister C., Lyubchenko Y., Sierks M.R. Residues 17–20 and 30–35 of β-amyloid play critical roles in aggregation. J. Neurosci. Res. 2004;75:162–171. doi: 10.1002/jnr.10859. [DOI] [PubMed] [Google Scholar]
- 27.Azriel R., Gazit E. Analysis of the minimal amyloid-forming fragment of the islet amyloid polypeptide. An experimental support for the key role of the phenylalanine residue in amyloid formation. J. Biol. Chem. 2001;276:34156–34161. doi: 10.1074/jbc.M102883200. [DOI] [PubMed] [Google Scholar]
- 28.Tomich J.M., Wallace D.P., Henderson K., Brandt R., Ambler C.A., Scott A.J., Mitchell K.E., Radke G., Grantham J.J., Sullivan L.P., Iwamoto T. Aqueous solubilization of transmembrane peptide sequences with retention of membrane insertion and function. Biophys. J. 1998;74:256–267. doi: 10.1016/S0006-3495(98)77784-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gekko K., Hasegawa Y. Compressibility-structure relationship of globular proteins. Biochemistry. 1986;25:6563–6571. doi: 10.1021/bi00369a034. [DOI] [PubMed] [Google Scholar]
- 30.Zhong Z.K., Sun X.S., Fang X.H., Ratto J.A. Adhesive properties of soy protein with fiber cardboard. J. Am. Oil Chem. Soc. 2001;78:37–41. [Google Scholar]
- 31.Dogic Z., Philipse A.P., Fraden S., Dhont J.K.G. Concentration-dependent sedimentation of colloidal rods. J. Chem. Phys. 2000;113:8368–8380. [Google Scholar]


