Human macrophage inflammatory protein-3α (MIP-3α) has been crystallized in a new space group I4. Crystals diffract to 1.81 Å.
Keywords: macrophage inflammatory protein-3α
Abstract
The structure of the human macrophage inflammatory protein-3α (MIP-3α) has been determined at 1.81 Å resolution by X-ray crystallography. The dimer crystallized in the tetragonal space group I4, with unit-cell parameters a = b = 83.99, c = 57.20 Å. The crystals exhibit two molecules in the asymmetric unit. The structure was solved by the molecular-replacement method and the model was refined to a conventional R value of 20.6% (R free = 25.7%). MIP-3α possesses the same monomeric structure as previously described for other chemokines. However, in addition to limited structural changes in the β1–β2 hairpin of monomer B, the electron density is fully defined for a few extra residues at the N- and C-termini of monomer A and the C-terminus of monomer B compared with MIP-3α in space group P61. As the N-terminal and loop regions have been shown to be critical for receptor binding and signaling, this additional structural information may help in determining the basis of the CCR6 selectivity of MIP-3α.
1. Introduction
Leukocyte trafficking is mediated by a group of structurally related proteins known as chemokines (Rodig et al., 2002 ▶). The chemokines are a group of small (8–14 kDa) basic soluble proteins that are produced and released by a wide variety of cell types during the initial phase of host response to injury, allergens, antigens or invading microorganisms (Zlotnik & Yoshie, 2000 ▶). To date, 50 chemokines and 20 receptors have been identified in humans (Zlotnik & Yoshie, 2000 ▶; Horuk, 2001 ▶). Chemokines have been identified as chemotactic cytokines and can be divided into C, CC, CXC and CX3C subgroups based upon the position of cysteine residues located near the N-terminus. Although the primary function of chemokines is the regulation of leukocyte trafficking, chemokines also play an important role in angiogenesis, tumor metastasis, hematopoiesis and human immunodeficiency virus infection (Zlotnik & Yoshie, 2000 ▶; Rollins, 1997 ▶; Baggiolini, 1998 ▶; Rossi & Zlotnik, 2000 ▶; Müller et al., 2001 ▶). The chemokine macrophage inflammatory protein-3α (MIP-3α) has been noted as being expressed in human liver, lung, appendix, tonsillar crypts and skin epidermal cells (Hieshima et al., 1997 ▶; Hromas et al., 1997 ▶; Tanaka et al., 1999 ▶; Charbonnier et al., 1999 ▶). A member of the CC chemokines, MIP-3α is chemotactic for memory T cells and immature dendritic cells (Hieshima et al., 1997 ▶; Varona et al., 1998 ▶; Liao et al., 1999 ▶; Dieu-Nosjean et al., 2000 ▶; Krzysiek et al., 2000 ▶), while MIP-3β is chemotactic for activated T lymphocytes and mature dendritic cells (Dieu-Nosjean et al., 1999 ▶). MIP-3α and MIP-3β bind and activate CCR6 and CCR7 receptors, respectively (Yang et al., 1999 ▶).
In order to identify the motifs that are key to the specificity of receptor binding and activation, various structural studies of chemokines have been made (Baldwin et al., 1991 ▶; Schraufstätter et al., 1993 ▶; Perez-Canadilla et al., 2001 ▶; Hoover et al., 2002 ▶; Luz et al., 2005 ▶) by X-ray crystallography and/or NMR techniques. In this report, we present the structure of MIP-3α in the tetragonal space group I4 (MIPI4) at 1.81 Å resolution. Structural analysis of MIPI4 reveals subtle structural differences between the published crystal structure of human MIP-3α in space group P61 (MIPP61) at resolution 1.7 Å (Hoover et al., 2002 ▶) and the current structure in space group I4.
2. Experimental procedures
2.1. Crystallization
The chemokine macrophage inflammatory protein-3α (MIPI4) was expressed, purified and refolded as described in a previously published protocol (Schibli et al., 2002 ▶).
Crystals of MIPI4 were grown by the hanging-drop vapor-diffusion method by mixing equal volumes (2 µl each) of protein (10 mg ml−1 in 0.1 M Tris–HCl pH 8.0) and reservoir solution. Two diffraction-quality crystal forms, tetragonal and hexagonal, grew together in solution No. 23 of Crystal Screen 2 (Cudney et al., 1994 ▶) containing 1.6 M ammonium sulfate, 10%(v/v) dioxane and 0.1 M MES pH 6.5. Small crystals appeared within 24 h, continued to grow slowly for 10–15 d at 277 K and reached maximum dimensions in 18 d. These crystals were flash-frozen with a cryoprotectant solution [10% PEG 400, 1.6 M ammonium sulfate, 10%(v/v) dioxane and 0.1 M MES pH 6.5] for low-temperature data collection. The two crystal forms (tetragonal and hexagonal) differ only in their packing. Here, we present and discuss the structure of the tetragonal form of the crystal.
2.2. Data collection
Initial testing of the crystals for X-ray diffraction was performed on a rotating-anode RU-H3R X-ray generator equipped with state-of-the-art focusing mirrors (from Osmic) and a Rigaku R-AXIS IV++ image-plate detector. The diffraction data were collected (from flash-cooled crystals at 100 K) at the SBC-CAT beamline 17-ID at the Advanced Photon Source, Argonne National Laboratory. A complete data set from the tetragonal crystal form was collected by exposing a crystal to the X-ray beam for 1 s for each 0.5° oscillation. A total of 180 images were collected at 150 mm crystal-to-detector distance.
Data were processed using the program d*TREK (Pflugrath, 1999 ▶). The tetragonal form of the crystal belongs to space group I4 and diffracted to 1.81 Å. The data-collection statistics are shown in Table ▶1. The scaled data were truncated to obtain structure-factor amplitudes using the CCP4 (Collaborative Computational Project, Number 4, 1994 ▶) program TRUNCATE (French & Wilson, 1978 ▶).
Table 1. Data-measurement and refinement statistics for the structure of MIP-3α (1.81 Å).
Values in parentheses are for the outermost resolution shell (1.99–1.81 Å).
| Data collection | |
| Crystal system | Tetragonal |
| Unit-cell parameters (Å) | a = b = 83.99, c = 57.20 |
| Space group | I4 |
| Resolution (Å) | 21.57–1.81 (1.99–1.81) |
| Measured reflections | 62834 |
| Unique reflections | 17465 |
| Completeness (%) | 100 (100) |
| I/σ(I) | 15.0 (2.4) |
| Rsym† (%) | 6.2 (30.8) |
| Solvent content (%) | 61.55 |
| No. of molecules per ASU | 2 |
| Refinement | |
| Resolution range (Å) | 21.57–1.81 |
| R factor (%) | 20.6 |
| Rfree (%) | 25.7 |
| R.m.s. deviation in bond distances (Å) | 0.019 |
| R.m.s. deviation in bond angles (°) | 1.7 |
| Water molecules | 128 |
R
sym = 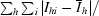
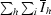 , where I
hi is the observed intensity of the ith measurement of reflection h and
, where I
hi is the observed intensity of the ith measurement of reflection h and 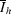 is the mean intensity of reflection h.
is the mean intensity of reflection h.
2.3. Structure determination and refinement
The structure was solved by molecular replacement using AMoRe from the CCP4 program suite (Navaza, 1994 ▶) using the human MIP-3α dimer as a search model (PDB code 1m8a; Hoover et al., 2002 ▶). A solution with a correlation coefficient of 75% and an R factor of 40% was obtained. Based on the calculated molecular volume (Matthews, 1968 ▶), the solvent content suggested there to be a dimer in the asymmetric unit. After an initial run of rigid-body refinement, the structure was refined using the maximum-likelihood program REFMAC (Murshudov et al., 1997 ▶) in combination with ARP (Lamzin & Wilson, 1993 ▶). After a single round of refinement, the R factor fell to 29% with a free R factor of 32% (Brünger, 1992 ▶). Model building was performed using the program O (Jones et al., 1991 ▶). All residues were shown to lie within the allowed regions of the Ramachandran plot. The final refinement statistics are given in Table 1 ▶. Validation of the structure was performed using the program WHATIF (Vriend, 1990 ▶) using the validation server at EMBL, Heidelberg, Germany.
3. Results and discussion
The structure of human MIP-3α has been determined by X-ray crystallography in space group I4. The final model of MIP-3α at 1.81 Å resolution consists of 66 residues in monomer A and 63 in monomer B, representing a total of 129 protein residues. No electron density can be seen for the first residue (Ala1) or for the side chains of the last two residues (Lys68, Asn69) of monomer A. In monomer B, the first four (Ala1–Phe4) residues and the side chains of the last two residues (Lys68, Asn69) also showed no electron density. The final crystallographic R factor and free R factor were 20.6 and 25.7%, respectively, for all reflections (21.57–1.8 Å). The r.m.s. deviations in bond lengths and bond angles are 0.019 and 1.7 Å, respectively. The Ramachandran plot calculated for the final MIP-3α model with the program PROCHECK (Laskowski et al., 1993 ▶) shows that 95.0% of the residues are in the most favored regions and 5.0% of residues are in the additionally allowed regions.
3.1. Description of the tertiary structure of the monomer
MIPI4 possesses the same monomeric structure previously described for other chemokines. The N-terminal region is attached to the β1–β2 loop (Leu27–Asn35) and the β3 strand (Ser46–Ala49) by the disulfide bonds 6–32 and 7–48. Other interactions between the N-terminus and the core of the protein include the main-chain N and O atoms of Cys7 with Gln26 of the β1 strand. The main-chain N atom of Thr11 also forms a hydrogen bond with the main-chain O atom of Cys48 of the β3 strand.
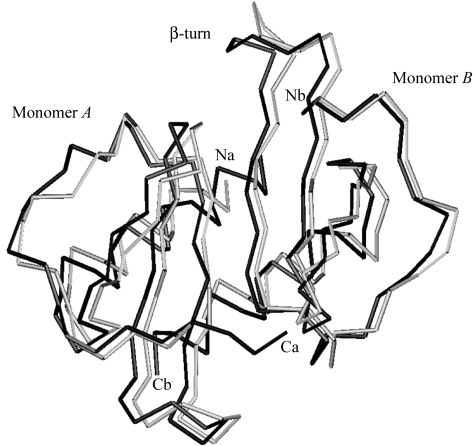
Superposition of MIPI4 on to the structure of MIPP61 shows limited but significant structural differences at the N-terminus of monomer A and in the β-turn loop between the β1 and β2 strands of monomer B (Fig. 1 ▶). The primary differences between MIPP61 and MIPPI4 lie in the electron density for previously undefined residues at the N-terminus of monomer A and the C-terminal helices in both monomers A and B of MIPI4. The conformation of the Asn27–Cys33 loop between the β1 and β2 strands is slightly different in monomer B of MIPI4, where Arg25 and Glu30 brace each other by forming a hydrogen bond between the side-chain OE1 and OE2 atoms of Glu30 and the NH1 and NH2 of Arg25 of the same monomer B. This interaction may play some role in the bend of the β-turn towards the core of the protein. Glu30 OE1 forms another hydrogen bond with water in the same monomer B, while Glu30 OE2 may stabilize the dimer by forming a hydrogen bond with the side chain of Lys42 in an adjacent monomer A. In MIPP61 the side chain of Glu30 of monomer B is exposed to water, while Arg25 is oriented inward, forming a hydrogen bond with a water molecule and main-chain O atom of Leu63 of the C-terminal α-helix in monomer A. The hydrophobic interaction of the side chains of the C-terminal helix with the β-sheet is similar to those in MIPP61 except that Leu15 is facing Phe19 and Ile20. These differences may arise as a consequence of the different packing of the molecules in the crystal.
Figure 1.
Superimposed Cα backbone structures of human MIPP61 in grey and MIPPI4 in black showing the long N- and C-termini of monomer A. The alternative orientation of the β-turn can also be seen in monomer B. The r.m.s. difference in Asp5–Lys65 Cα atomic positions, calculated with the LSQMAN option in O, is 0.85 Å (Jones et al., 1991 ▶). This figure was created using the program MOLMOL (Koradi et al., 1996 ▶).
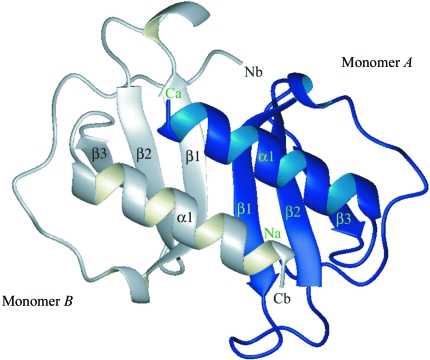
3.2. Description of dimerization in the crystal
The dimerization in the MIP-3α crystal structure is similar to that seen in several CXC chemokines (Baldwin et al., 1991 ▶). The dimer is primarily stabilized by hydrogen bonds between the β1 strands of the two monomers as well as by additional side-chain interactions. Thus, the dimeric molecule consists of a six-stranded sheet and two antiparallel helices inclined at an angle of ∼60° to the β-strands (Fig. 2 ▶). Several hydrogen-bond interactions between monomers A and B are similar to those seen in MIPP61.
Figure 2.
Ribbon diagram of the overall three-dimensional structure of the dimer of human MIP-3α. The dimer is composed of a six-stranded β-sheet and two parallel α-helices. β-Strands are shown as arrows and α-helices as coils. This figure was generated with the program MOLMOL (Koradi et al., 1996 ▶).
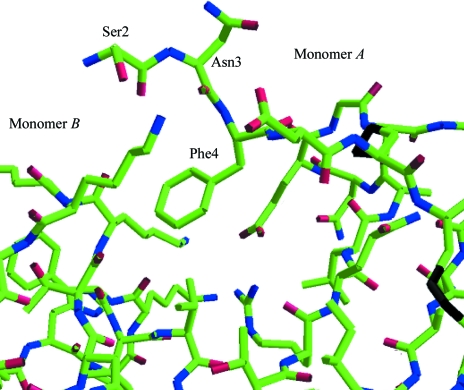
Several studies have shown interaction between the amino-terminus of chemokines (MIP-1α and RANTES) and the transmembrane helix bundle of the cellular receptors, such as CCR5 (Murphy et al., 2000 ▶; Rollins, 1997 ▶; Zlotnik & Yoshie, 2000 ▶; Blanpain et al., 2003 ▶). Amino-acid residues preceding the first cysteine and the N-loop (second receptor-binding site, residues 10–20) of chemokines are important for binding and receptor activation (Clark-Lewis et al., 1995 ▶; Lowman et al., 1996 ▶). We found fully defined electron density for the N-terminal Ser2, Asn3 and Phe4 of monomer A that was not visible in MIPP61. These well defined residues of monomer A may play a role in stabilization of the dimer, as the side-chain O atom of Ser2 and the main-chain O atom of Asn3 of monomer A form two hydrogen bonds with the side chain of Lys44 of monomer B (Fig. 3 ▶). Residues 41–44 of the loop between strands β2 and β3 of monomer B are also shifted, perhaps as a consequnce of the presence of Phe4 in monomer A. This probably results in restricting the freedom of this loop in monomer B. The dimer in MIPI4 is further stabilized by the weak hydrogen bond between the side chain of Glu30 of monomer A and Lys42 of monomer B (Fig. 3 ▶).
Figure 3.
Stick representation of the well defined N-terminal residues Ser2, Asn3 and Phe4 of monomer A that may play a role in the stabilization of the dimer.
Studies on IL-8, which is structurally related to MIP-3α, suggest that the C-terminus also plays an important role in binding to receptors (Schraufstätter et al., 1993 ▶), indicating that the C-terminus could be important for the function of the chemokine. The orientation of the C-terminal helix to the triple-stranded β-sheet within each monomer is stabilized by a number of hydrophobic interactions. There are hydrophobic contacts between Val67 of the C-terminus of monomer A and the methyl groups of Val60 and Ile37 from the β2 strand of monomer B. There is also a hydrogen bond between Arg61 of monomer A and Ser64 of monomer B, while Arg61 of monomer B makes a van der Waals interaction with Ser64 of monomer A. These interactions may serve to maintain the orientation of the C-terminal helices with respect both to each other and to the β-sheet below. In MIPI4 the long C-terminal helix forms two more hydrogen bonds between the side-chain N atom of Lys57 of monomer A and the main-chain O atom of Val67 of monomer B. Likewise, the main-chain O atom of Val67 of monomer A forms a hydrogen bond with the side chain of Lys57 of monomer B. These side-chain interactions are different from those seen in MIPP61 (Hoover et al., 2002 ▶).
Structural analysis of chemokines has revealed that the α/β-fold is highly conserved between both the CXC and CC chemokine classes (Schraufstätter et al., 1993 ▶). The electron-density map was ambiguous for N- and C-terminus residues in MIPP61, but is significantly clearer in the MIPI4 structure, especially at the amino-terminus, which comprises one of the receptor-recognition and binding motifs. We have also identified two residues at the C-terminus of both monomers that were disordered and could not be interpreted in MIPP61.
These new experimentally observed side-chain interactions, especially the significant differences at the N-terminus and β-turn of monomer B, coupled with the C-terminal hydrophobic interactions may help to increase our understanding of the interaction between chemokines and their receptors. These results will also aid the rational design of variants to modulate or interfere with its important biological functions.
Supplementary Material
PDB reference: human MIP-3α chemokine, 2hci, r2hcisf
Acknowledgments
We thank the protein crystallography facility and Dr S. Ramaswamy at the Department of Biochemistry, University of Iowa for help during data collection. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. W-31-109-Eng-38.
References
- Baggiolini, M. (1998). Nature (London), 392, 565–568. [DOI] [PubMed] [Google Scholar]
- Baldwin, E. T., Weber, I. T., St Charles, R., Xuan, J. C., Appella, E., Yamada, M., Matsushima, K., Edwards, B. F., Clore, G. M. & Gronenborn, A. M. (1991). Proc. Natl Acad. Sci. USA, 88, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain, C., Doranz, J. B., Bondue, A., Govaerts, C., De Leener, A., Vassart, G., Doms, W. R., Proudfoot, A. & Parmentier, M. (2003). J. Mol. Biol.278, 5179–5187. [DOI] [PubMed]
- Brünger, A. T. (1992). Nature (London), 355, 472–474. [DOI] [PubMed]
- Charbonnier, A. S., Kohrgruber, N., Kriehuber, E., Stingl, G., Rot, A. & Maurer, D. (1999). J. Exp. Med.190, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Lewis, I., Kim, K. S., Rajarathnam, K., Gong, J. H., Dewald, B., Moser, B., Baggiolini, M. & Sykes, B. D. (1995). J. Leukoc. Biol.57, 703–711. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Cudney, R., Patel, S., Weisgraber, K., Newhouse, Y. & McPherson, A. (1994). Acta Cryst. D50, 414–423. [DOI] [PubMed] [Google Scholar]
- Dieu-Nosjean, M. C., Massacrier, C., Homey, B., Vanbervliet, B., Pin, J. J., Vicari, A., Lebecque, S., Dezutter-Dambuyant, C., Schmitt, D., Zlotnik, A. & Caux, C. (2000). J. Exp. Med.192, 705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieu-Nosjean, M. C., Vicari, A., Lebecque, S. & Caux, C. (1999). J. Leukoc. Biol.66, 252–262. [DOI] [PubMed] [Google Scholar]
- French, G. S. & Wilson, K. S. (1978). Acta Cryst. A34, 517–525. [Google Scholar]
- Hieshima, K., Imai, T., Opdenakker, G., Van Damme, J., Kusuda, J., Tei, H., Takasuki, K., Miura, R., Yoshie, O. & Nomiyama, H. (1997). J. Biol. Chem.272, 5846–5853. [DOI] [PubMed] [Google Scholar]
- Hoover, D. M., Boulègue, C., Yang, D., Oppenheim, J. J., Tucker, K., Lu, W. & Lubkowski, J. (2002). J. Biol. Chem.277, 37647–37654. [DOI] [PubMed] [Google Scholar]
- Horuk, R. (2001). Cytokines Growth Factors Rev.12, 313–335.
- Hromas, R., Gray, P. W., Chantry, D., Godiska, R., Krathwohl, M., Fife, K., Bell, I., Takeda, J., Aronica, S., Gordon, M., Cooper, S., Broxmeyer, H. E. & Klemsz, M. J. (1997). Blood, 89, 3315–3322. [PubMed] [Google Scholar]
- Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M. & Wüthrich, K. (1996). J. Mol. Graph.14, 51–55. [DOI] [PubMed] [Google Scholar]
- Krzysiek, R., Lefevre, E. A., Bernard, J., Foussat, A., Galanaud, P., Louache, F. & Richard, Y. (2000). Blood, 96, 2338–2345. [PubMed] [Google Scholar]
- Lamzin, V. & Wilson, K. S. (1993). Acta Cryst. D49, 129–147. [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst.26, 283–290. [Google Scholar]
- Liao, F., Rabin, R. L., Smith, C. S., Sharma, G., Nutman, T. B. & Farber, J. M. (1999). J. Immunol.162, 186–194. [PubMed] [Google Scholar]
- Lowman, H. B., Stagele, P. H., Deforgo, L. E., Wirth, C. M., Gillece-Castro, B. L., Bourell, J. H. & Fairbrother, W. J. (1996). J. Biol. Chem.271, 14344–14352. [DOI] [PubMed] [Google Scholar]
- Luz, J. G., Yu, M., Su, Y., Wu, Z., Zhou, Z., Sun, R. & Wilson, I. A. (2005). J. Mol. Biol.352, 1019–1028. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Müller, A., Homey, B., Soto, H., Ge, N., Catron, D., Buchanan, M. E., McClanahan, T., Murphy, E., Yuan, W., Wagner, S. N., Barrera, J. L., Mohar, A., Verástegui, E. & Zlotnik, A. (2001). Nature (London), 410, 50–56. [DOI] [PubMed] [Google Scholar]
- Murphy, P. M., Baggiolini, M., Charo, I. F., Hebert, C. A., Horuk, R., Matsushima, K., Miller, L. H., Oppenheim, J. J. & Power, C. A. (2000). Pharmacol. Rev.52, 145–176. [PubMed] [Google Scholar]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Perez-Canadilla, J. M., Zaballaos, A., Gutierrez, J., Varona, R., Roncal, F., Albar, J. P., Marquez, G. & Bruix, M. (2001). J. Biol. Chem.276, 28372–28379. [DOI] [PubMed] [Google Scholar]
- Pflugrath, J. W. (1999). Acta Cryst. D55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- Rodig, S. J., Jones, D., Shahsafaei, A. & Dorfman, D. M. (2002). Hum. Pathol.33, 1227–1233. [DOI] [PubMed] [Google Scholar]
- Rollins, B. J. (1997). Blood, 90, 909–928. [PubMed] [Google Scholar]
- Rossi, D. & Zlotnik, A. (2000). Annu. Rev. Immunol.18, 217–242. [DOI] [PubMed] [Google Scholar]
- Schibli, J. D., Hunter, H. N., Aseyev, V., Starner, T. D., Wiencek, J. M., McCray, P. B. Jr, Tack, B. F. & Vogel, H. J. (2002). J. Mol. Biol.277, 8279–8289. [DOI] [PubMed]
- Schraufstätter, I. U., Barritt, D. S., Ma, M., Oades, Z. G. & Cochrane, C. G. (1993). J. Immunol.151, 6418–6428. [PubMed] [Google Scholar]
- Tanaka, Y., Imai, T., Baba, M., Ishikawa, I., Uehira, M., Nomiyama, H. & Yoshie, O. (1999). Eur. J. Immunol.29, 633–642. [DOI] [PubMed] [Google Scholar]
- Varona, R., Zaballos, A., Gutierrèz, J., Martin, P., Roncal, F., Albar, J. C., Ardavin, C. & Màrquez, G. (1998). FEBS Lett.440, 188–194. [DOI] [PubMed] [Google Scholar]
- Vriend, G. (1990). J. Mol. Graph.8, 52–56. [DOI] [PubMed] [Google Scholar]
- Yang, D., Chertov, O., Bykovskaia, S. N., Chen, Q., Buffo, M. J., Shogan, J., Anderson, M., Schöder, J. M., Wang, J. M., Howard, O. M. Z. & Oppenheim, J. J. (1999). Science, 286, 525–528. [DOI] [PubMed] [Google Scholar]
- Zlotnik, A. & Yoshie, O. (2000). Immunity, 12, 121–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human MIP-3α chemokine, 2hci, r2hcisf