The metallo-glycerophosphodiesterase from E. aerogenes (GpdQ) has been cloned, expressed in E. coli and purified. Initial screening of crystallization conditions for this enzyme resulted in the identification of needles from one condition in a sodium malonate grid screen. Removal of the metals from the enzyme and subsequent optimization of these conditions led to crystals.
Keywords: glycerophosphodiesterases, Enterobacter aerogenes, GpdQ
Abstract
The metallo-glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) has been cloned, expressed in Escherichia coli and purified. Initial screening of crystallization conditions for this enzyme resulted in the identification of needles from one condition in a sodium malonate grid screen. Removal of the metals from the enzyme and subsequent optimization of these conditions led to crystals that diffracted to 2.9 Å and belonged to space group P213, with unit-cell parameter a = 164.1 Å. Self-rotation function analysis and V M calculations indicated that the asymmetric unit contains two copies of the monomeric enzyme, corresponding to a solvent content of 79%. It is intended to determine the structure of this protein utilizing SAD phasing from transition metals or molecular replacement.
1. Introduction
Glycerophosphodiesterases (GDPDs; EC 3.1.4.46) belong to a relatively small family of enzymes that function by catalyzing the hydrolysis of the 3′-5′ phosphodiester bond of glycerophosphodiesters; for instance, hydrolyzing glycerophosphocholine to sn-glycerol-3-phosphate (G3P) and choline (Larson et al., 1983 ▶). Maintenance of G3P concentration within bacterial cells is essential to enable phospholipid synthesis (Raetz, 1986 ▶). This is achieved in a number of ways in bacterial cells: in Escherichia coli the Glp and Ugp pathways are used to transport glycerophosphodiesters into the cell, with hydrolysis occurring during and after transport, respectively (Brzoska & Boos, 1988 ▶; Tommassen et al., 1991 ▶). In addition to maintaining G3P levels for phospholipid synthesis, Haemophilus influenzae has been shown to use the other hydrolytic product, transferring choline from the host-organism cells onto the bacterial cell surface. This is believed to enable the bacteria to evade the immune response, implicating the GDPD from H. influenzae in pathogenesis (Janson et al., 1992 ▶).
An operon has been cloned from Enterobacter aerogenes that is homologous to the Ugp pathway from E. coli. With the exception of the GDPD, GpdQ, every gene shares significant sequence homology to the corresponding genes in E. coli. Indeed, sequence analysis indicates that GpdQ may be a structurally unique member of the GDPD family (McLoughlin et al., 2004 ▶). Secondary-structure prediction and identification of conserved sequence motifs suggest that GpdQ is likely to be structurally similar to the large and functionally diverse α/β-sandwich family of metallo-dependent phosphoesterases that includes purple acid phosphatase (Klabunde et al., 1996 ▶), 5′-nucleotidase (Knofel & Strater, 1999 ▶), Mre11 nuclease (Hopfner et al., 2001 ▶) and the Ser/Thr protein phosphatases (Rusnak & Mertz, 2000 ▶). These enzymes are typically monomeric or dimeric, with active sites consisting of two transition metals. There is still debate over some features of the catalytic mechanism of these enzymes. GpdQ has been shown to have very broad substrate specificity, catalysing the hydrolysis of phosphomonoesters, diesters and triesters in addition to phosphothiolates. Understanding the structure–function relationship underlying this substrate promiscuity may advance our general understanding of the binuclear metallo-phosphatase mechanism. Our interest in the structural determination of GpdQ is therefore twofold: to further investigate the catalytic mechanism of the α/β-sandwich metallo-phosphatases and to improve our understanding of bacterial GDPDs.
2. Experimental methods
2.1. Protein expression and purification
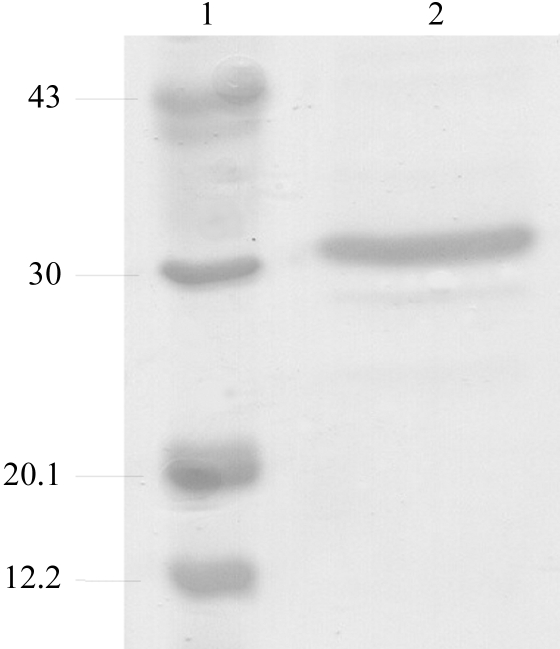
Recombinant E. aerogenes GpdQ was expressed and purified as described elsewhere (McLoughlin et al., 2004 ▶), with a few minor differences. Briefly, E. coli DH5α cells expressing GpdQ were lysed in a French press and purified using DEAE Fractogel (0–1 M NaCl) and phenyl Sepharose [1–0 M (NH4)2SO4] columns (Amersham Pharmacia). After purification, the protein was dialyzed against the storage buffer (50 mM HEPES pH 7.0; buffer A). To remove the metal from the active site, GpdQ was dialysed against a solution of 50 mM MES pH 5.0, 1 mM EDTA for several days before dialysis against buffer A. GpdQ and apo-GpdQ were concentrated to 12 and 10 mg ml−1, respectively, using 30 kDa molecular-weight cutoff membranes (Centricon). The concentration of the recombinant protein was determined by measuring the absorbance at 280 nm. The purity of GpdQ was assessed by 20% SDS–PAGE (Fig. 1 ▶). Two additional bands were present at approximately 29 and 25 kDa which could not be removed by further purification.
Figure 1.
Assessment of the purity of GpdQ by SDS–PAGE (20% gel) analysis. Lane 1 shows molecular-weight markers (kDa) and lane 2 purified GpdQ.
2.2. Crystallization
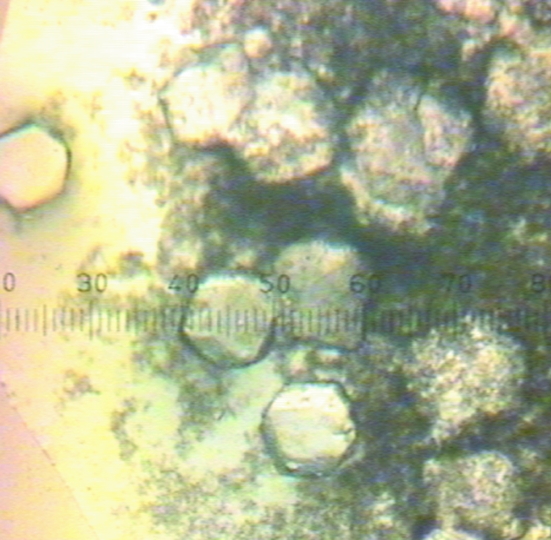
Initial crystallization conditions were screened using the sparse-matrix approach as described by Jancarik & Kim (1991 ▶). Crystallization trials were set up in 96-well plates (Hampton Research) using the sitting-drop vapour-diffusion method at 277 K with Hampton Research Crystal Screens I and II, Index Screen and PEG/Ion Screen. Each sitting drop consisted of 1 µl reservoir solution and 1 µl protein solution (12 mg ml−1 GpdQ in buffer A). Grid-screening trials were then set up in VDX plates (Hampton Research) using the hanging-drop vapour-diffusion method at 277 K with Hampton Research Grid Screens (Sodium Malonate, Ammonium Phosphate, PEG/LiCl and Quick Screens). Each hanging drop consisted of 3 µl reservoir solution and 3 µl protein solution (12 mg ml−1 GpdQ in buffer A). The reservoir volume was 1 ml. We obtained small crystals (Fig. 2 ▶) in 1.9 M sodium malonate without added buffer. Removing the metals from the active site of GpdQ and increasing the crystallization temperature to 291 K optimized the crystallization (Fig. 3 ▶).
Figure 2.
Needles of GpdQ in 1.9 M sodium malonate pH 7.0. The scale is 0.025 mm per gradation.
Figure 3.
Crystals of apo-GpdQ in 1.9 M sodium malonate pH 7.0. The scale is 0.025 mm per gradation.
2.3. Data collection and processing
Sodium malonate was used as the cryoprotectant (Holyoak et al., 2003 ▶); before cryocooling, crystals of the apoenzyme were immersed stepwise for 15 min per step in increasing concentrations of sodium malonate pH 7.0 to a final concentration of 4 M. The final 4 M cryoprotectant solution contained either 500 µM ZnCl2 or 500 µM CoCl2. All crystals were flash-cooled by immersion in liquid nitrogen. All diffraction data were collected under cryogenic conditions (100 K) at the Structural Biology Consortium Collaborative Access Team (SBC-CAT) facilities at the Advanced Photon Source (Argonne National Laboratory, IL, USA). Data were integrated and scaled using the programs DENZO and SCALEPACK (Otwinowski & Minor, 1977 ▶). Data sets were collected for crystals of apo-GpdQ and GpdQ with CoCl2 in the cryobuffer at 0.97929 Å and a data set for a GpdQ crystal with ZnCl2 in the cryobuffer was collected at the Zn K edge at 1.28269 Å.
3. Results and discussion
The expressed GpdQ construct encodes the mature GpdQ sequence composed of 274 amino acids with a calculated molecular weight of 30 839.93 Da and a theoretical pI of 5.45 (EMBL accession No. AAO83402; McLoughlin et al., 2004 ▶). The yield of GpdQ was ∼40 mg from 1 l LB medium. When analysed with SDS–PAGE, the major band is visible at ∼31 kDa, which is in accordance with the calculated molecular-weight analysis of concentrated GpdQ and indicates high purity (Fig. 1 ▶).
No suitably diffracting crystals were obtained from any of the sparse-matrix screens. Small needles appeared after two months in 1.9 M sodium malonate pH 7.0 from the Sodium Malonate Grid Screen (Fig. 2 ▶). A dramatic improvement in the size of the crystals was observed when the experiments were set up with apo-GpdQ (∼10 mg ml−1 in buffer A) at 291 K. These crystals had a cubic morphology, were not birefringent and grew to dimensions of ∼0.25 × 0.25 × 0.25 mm within two weeks (Fig. 3 ▶).
In an effort to obtain a physiologically relevant data set of GpdQ with bound active-site metals and to assist in phase determination, two apo-GpdQ crystals were soaked in cryobuffer containing transition-state metals (ZnCl2 and CoCl2). A crystal of Zn-GpdQ diffracted to 2.9 Å and crystals of apo-GpdQ and Co-GpdQ diffracted to 3.0 Å using synchrotron radiation. The data-collection statistics are listed in Table 1 ▶ and indicate that the metals used in the cryobuffer did not adversely affect the diffraction of the crystals. All crystals were cubic and were indexed in point group P23. Systematic absences indicate that the correct space group is P213. The molecular weight of the enzyme is 30 800 Da and the volume of the asymmetric unit is 388 819.6 Å3; the distribution range of the volume-to-weight ratio (V M) values (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶) indicated that the most likely number of monomers in the asymmetric unit was four, with a Matthews coefficient of 2.9 Å3 Da−1 and a solvent content of 57.7%. However, other numbers of monomers are also possible. To resolve this ambiguity, a self-rotation map was calculated using MOLREP (Vagin & Teplyakov, 1997 ▶). A non-crystallographic twofold rotation axis was indicated at (θ, ϕ) = (141, −17°). No fourfold non-crystallographic axes were indicated. The asymmetric unit therefore probably contains two monomers (corresponding to a V M of 5.9 Å3 Da−1 and a solvent content of 78.9%). Although this is a high value of V M, it is not unprecedented (Blamey et al., 2005 ▶). Thus, it appears that under crystallization conditions GpdQ tends to form a dimer. Whether this aggregation state reflects the actual biological assembly of GpdQ remains to be determined.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Crystal | Apo-GpdQ | Zn-GpdQ | Co-GpdQ |
|---|---|---|---|
| Space group | P213 | P213 | P213 |
| Unit-cell parameter a (Å) | 164.13 | 164.26 | 164.46 |
| Temperature (K) | 100 | 100 | 100 |
| Wavelength (Å) | 0.97929 | 1.28269 | 0.97929 |
| Resolution (Å) | 50–3.0 (3.19–3.0) | 50–2.9 (3.08–2.9) | 50–3.0 (3.19–3.0) |
| Unique reflections | 29709 | 32293 | 29959 |
| Redundancy | 6.8 (6.4) | 12.5 (11.2) | 14.0 (12.3) |
| 〈I/σ(I)〉 | 19.7 (2.8) | 32.2 (4.6) | 34.3 (4.6) |
| Completeness | 99.9 (99.9) | 100 (100) | 100 (100) |
| Rsym (%) | 11.2 (50.7) | 7.7 (43.6) | 7.3 (42.4) |
Acknowledgments
We thank the staff of the SBC-CAT beamline for their technical support during data collection. Use of the SBC-CAT beamline was supported by a grant from the Australian Synchrotron Radiation Project (ASRP). This research was supported by an ARC grant to DO.
References
- Blamey, C. J., Ceccarelli, C., Naik, U. P. & Bahnson, B. J. (2005). Protein Sci.14, 1214–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzoska, P. & Boos, W. (1988). J. Bacteriol.170, 4125–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoak, T., Fenn, T. D., Wilson, M. A., Moulin, A. G., Ringe, D. & Petsko, G. A. (2003). Acta Cryst. D59, 2356–2358. [DOI] [PubMed] [Google Scholar]
- Hopfner, K. P., Karcher, A., Craig, L., Woo, T. T., Carney, J. P. & Tainer, J. A. (2001). Cell, 105, 473–485. [DOI] [PubMed] [Google Scholar]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Janson, H., Heden, L. O. & Forsgren, A. (1992). Infect. Immun.60, 1336–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde, T., Strater, N., Frohlich, R., Witzel, H. & Krebs, B. (1996). J. Mol. Biol.259, 737–748. [DOI] [PubMed] [Google Scholar]
- Knofel, T. & Strater, N. (1999). Nature Struct. Biol.6, 448–453. [DOI] [PubMed] [Google Scholar]
- Larson, T. J., Ehrmann, M. & Boos, W. (1983). J. Biol. Chem.258, 5428–5432. [PubMed] [Google Scholar]
- McLoughlin, S. Y., Jackson, C., Liu, J. W. & Ollis, D. L. (2004). Appl. Environ. Microbiol.70, 404–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1977). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Raetz, C. R. (1986). Annu. Rev. Genet.20, 253–295. [DOI] [PubMed] [Google Scholar]
- Rusnak, F. & Mertz, P. (2000). Physiol. Rev.80, 1483–1521. [DOI] [PubMed] [Google Scholar]
- Tommassen, J., Eiglmeier, K., Cole, S. T., Overduin, P., Larson, T. J. & Boos, W. (1991). Mol. Gen. Genet.226, 321–327. [DOI] [PubMed] [Google Scholar]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025. [Google Scholar]