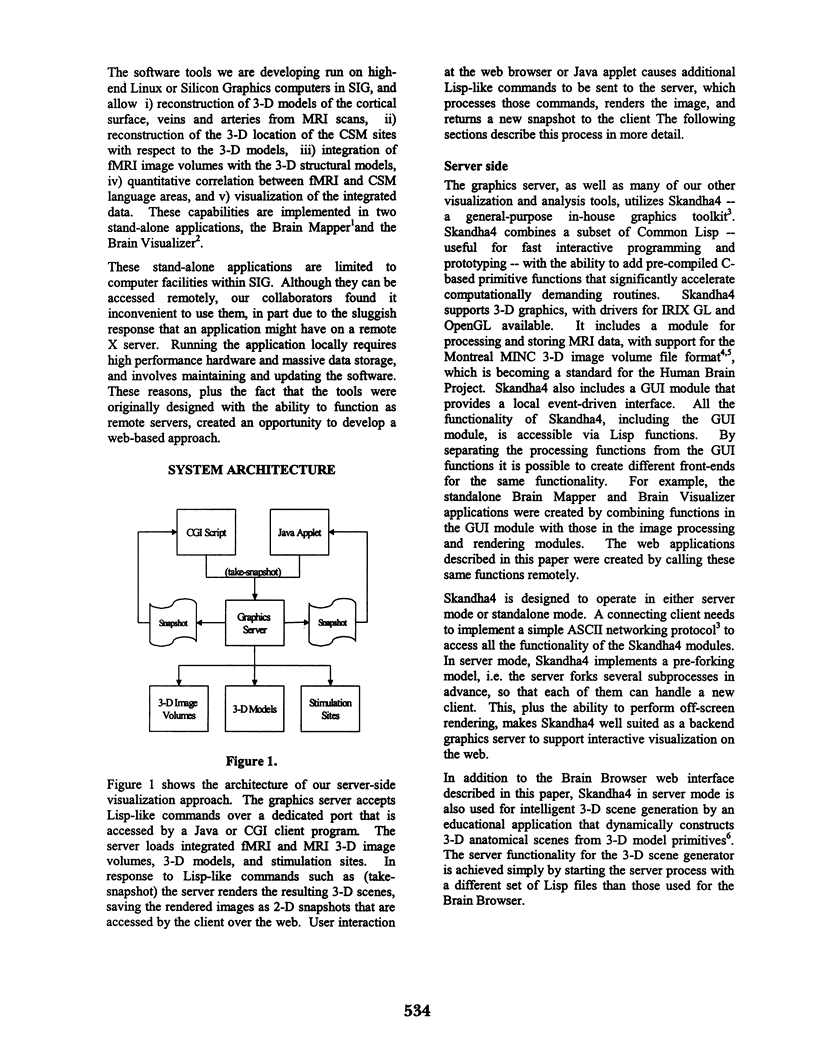
Abstract
Although computer processing power and network bandwidth are rapidly increasing, the average desktop is still not able to rapidly process large datasets such as 3-D medical image volumes. We have therefore developed a server side approach to this problem, in which a high performance graphics server accepts commands from web clients to load, process and render 3-D image volumes and models. The renderings are saved as 2-D snapshots on the server, where they are uploaded and displayed on the client. User interactions with the graphic interface on the client side are translated into additional commands to manipulate the 3-D scene, after which the server re-renders the scene and sends a new image to the client. Example forms-based and Java-based clients are described for a brain mapping application, but the techniques should be applicable to multiple domains where 3-D medical image visualization is of interest.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Collins D. L., Neelin P., Peters T. M., Evans A. C. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr. 1994 Mar-Apr;18(2):192–205. [PubMed] [Google Scholar]
- Hinshaw K. P., Brinkley J. F. Incorporating constraint-based shape models into an interactive system for functional brain mapping. Proc AMIA Symp. 1998:921–925. [PMC free article] [PubMed] [Google Scholar]
- Tarczy-Hornoch P., Kwan-Gett T. S., Fouche L., Hoath J., Fuller S., Ibrahim K. N., Ketchell D. S., LoGerfo J. P., Goldberg H. I. Meeting clinician information needs by integrating access to the medical record and knowledge resources via the Web. Proc AMIA Annu Fall Symp. 1997:809–813. [PMC free article] [PubMed] [Google Scholar]


