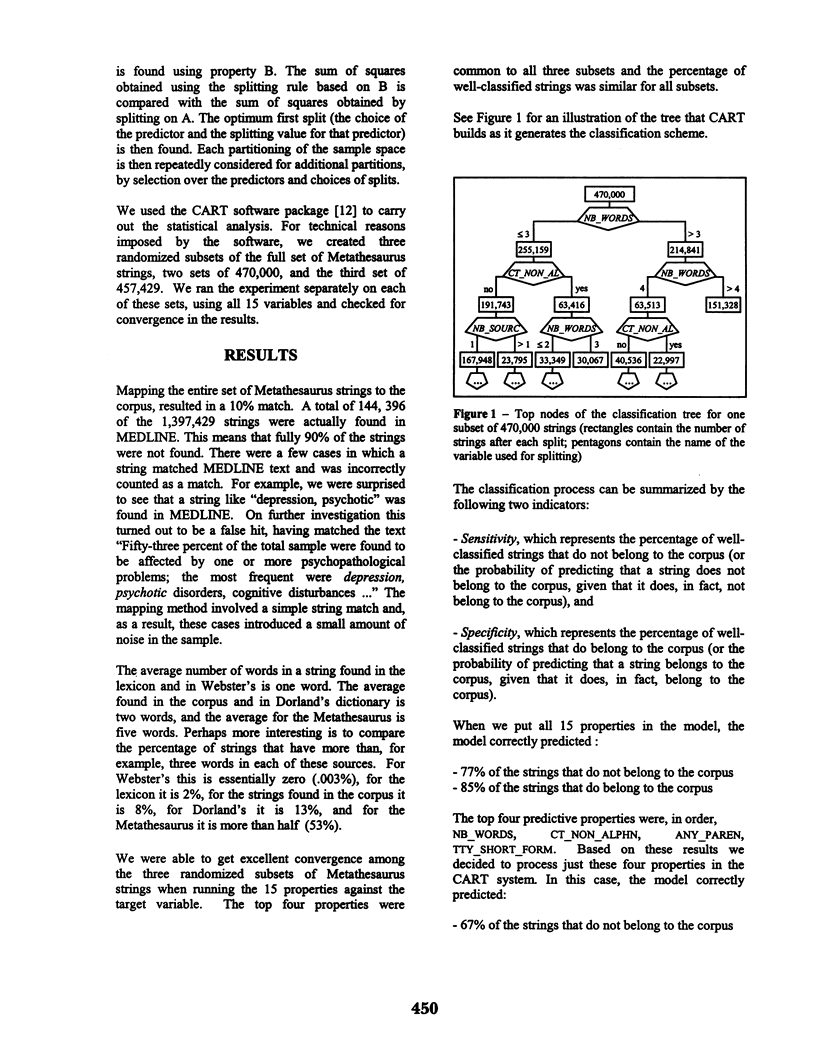
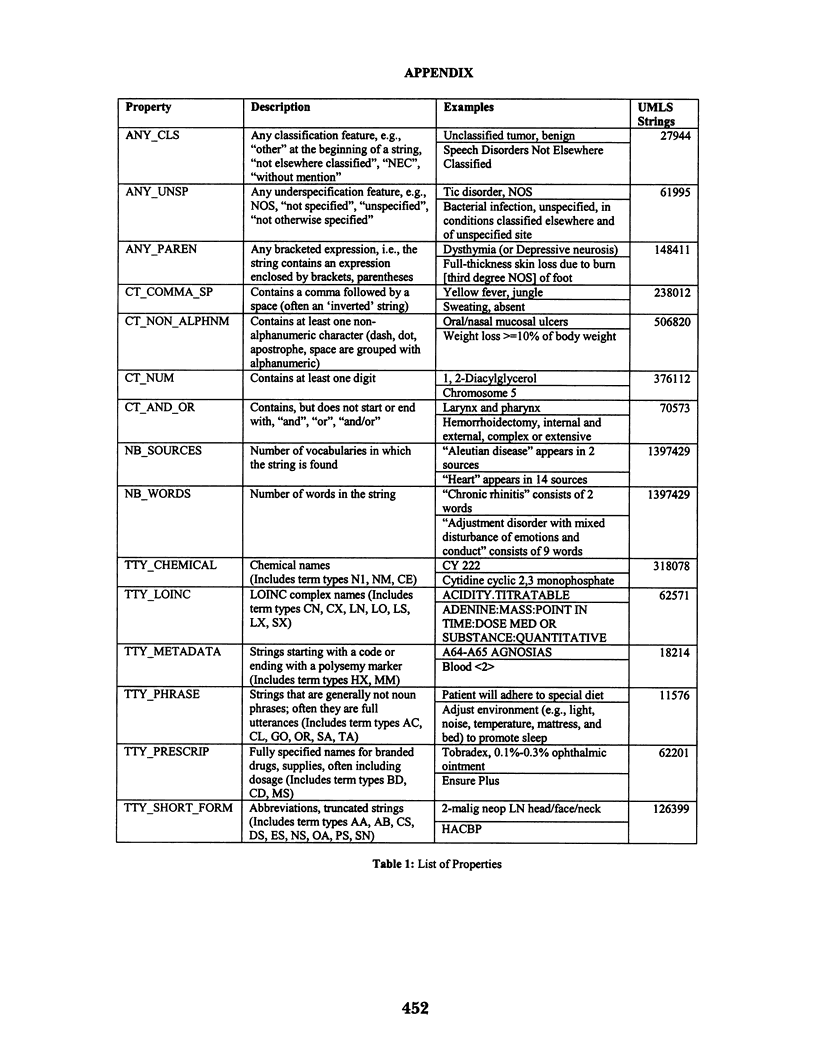
Abstract
The National Library of Medicine's Unified Medical Language System (UMLS) is a rich source of knowledge in the biomedical domain. The UMLS is used for research and development in a range of different applications, including natural language processing (NLP). In this paper we investigate the nature of the strings found in the UMLS Metathesaurus and evaluate them for their usefulness in NLP. We begin by identifying a number of properties that might allow us to predict the likelihood of a given string being found or not found in a corpus. We use a statistical model to test these predictors against our corpus, which is derived from the MEDLINE database. For one set of properties the model correctly predicted 77% of the strings that do not belong to the corpus, and 85% of the strings that do belong to the corpus. For another set of properties the model correctly predicted 96% of the strings that do not belong to the corpus and 29% of the strings that do belong to the corpus.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aronson A. R. The effect of textual variation on concept based information retrieval. Proc AMIA Annu Fall Symp. 1996:373–377. [PMC free article] [PubMed] [Google Scholar]
- Baclawski K., Cigna J., Kokar M. M., Mager P., Indurkhya B. Knowledge representation and indexing using the unified medical language system. Pac Symp Biocomput. 2000:493–504. doi: 10.1142/9789814447331_0047. [DOI] [PubMed] [Google Scholar]
- Friedman C., Hripcsak G. Natural language processing and its future in medicine. Acad Med. 1999 Aug;74(8):890–895. doi: 10.1097/00001888-199908000-00012. [DOI] [PubMed] [Google Scholar]
- Hahn U., Romacker M., Schulz S. How knowledge drives understanding--matching medical ontologies with the needs of medical language processing. Artif Intell Med. 1999 Jan;15(1):25–51. doi: 10.1016/s0933-3657(98)00044-x. [DOI] [PubMed] [Google Scholar]
- Johnson S. B. A semantic lexicon for medical language processing. J Am Med Inform Assoc. 1999 May-Jun;6(3):205–218. doi: 10.1136/jamia.1999.0060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray A. T. The nature of lexical knowledge. Methods Inf Med. 1998 Nov;37(4-5):353–360. [PubMed] [Google Scholar]
- Spyns P. Natural language processing in medicine: an overview. Methods Inf Med. 1996 Dec;35(4-5):285–301. [PubMed] [Google Scholar]


