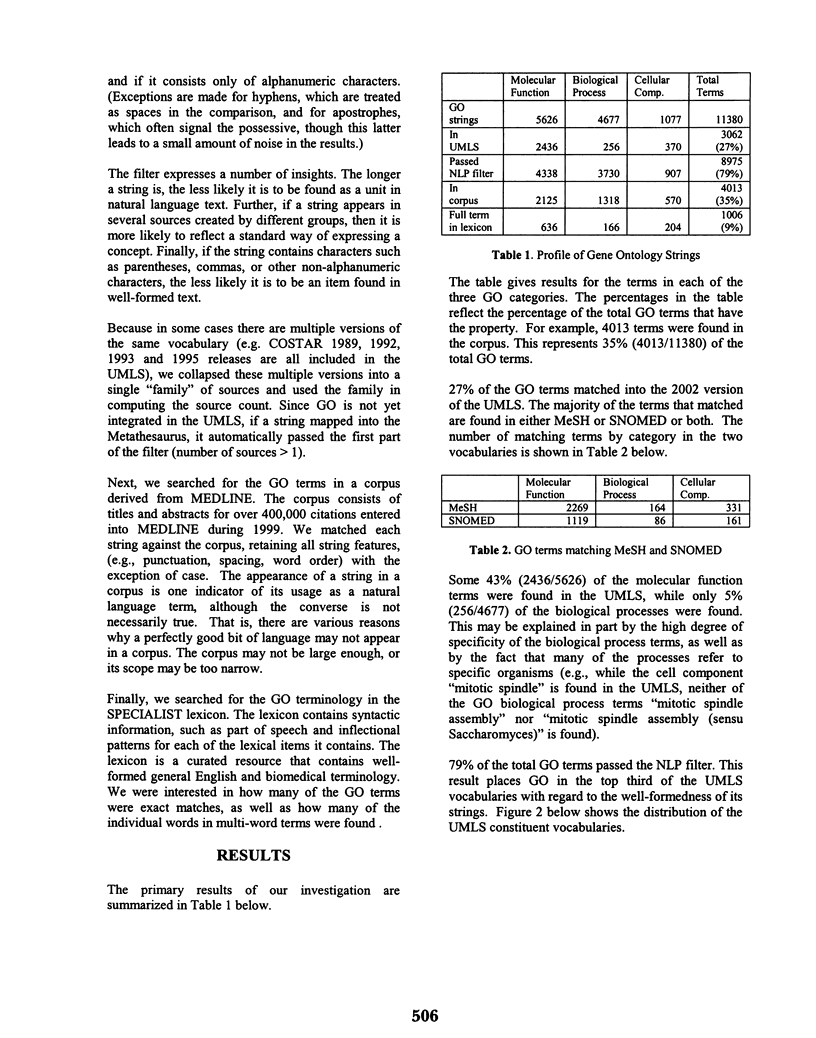
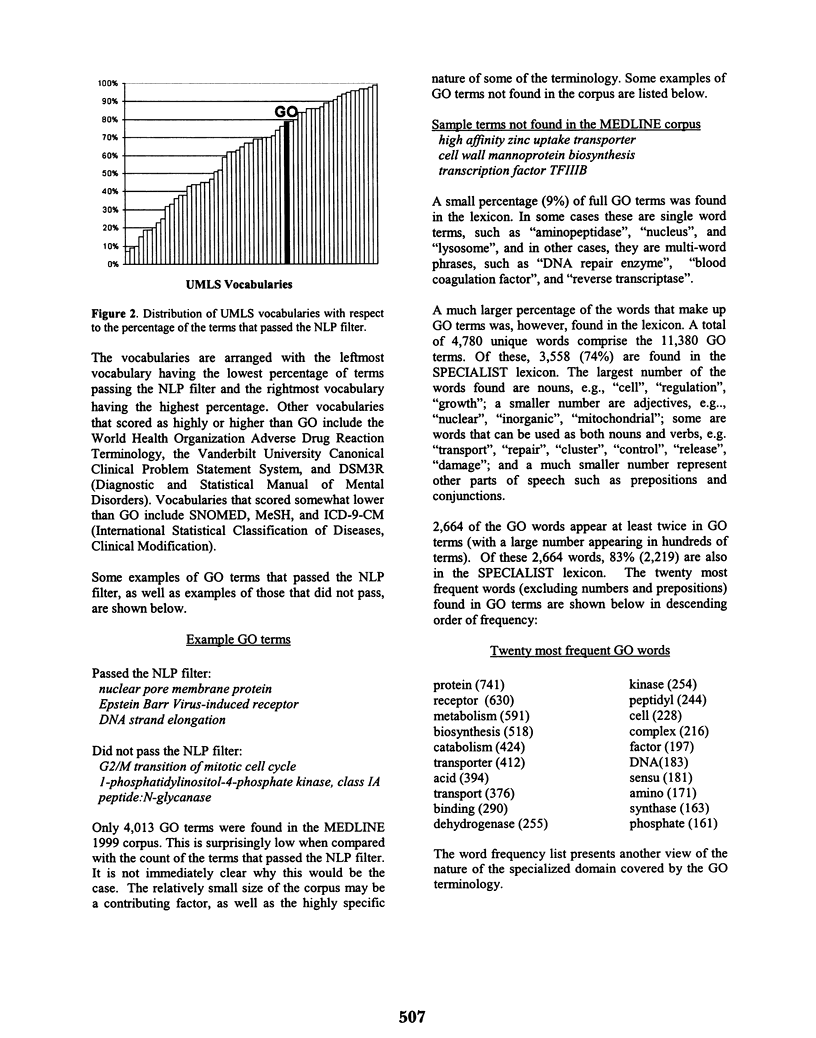
Abstract
The Gene Ontology (GO) is a construct developed for the purpose of annotating molecular information about genes and their products. The ontology is a shared resource developed by the GO Consortium, a group of scientists who work on a variety of model organisms. In this paper we investigate the nature of the strings found in the Gene Ontology and evaluate them for their usefulness in natural language processing (NLP). We extend previous work that identified a set of properties that reliably identifies natural language phrases in the Unified Medical Language System (UMLS). The results indicate that a large percentage (79%) of GO terms are potentially useful for NLP applications. Some 35% of the GO terms were found in a corpus derived from the MEDLINE bibliographic database, and 27% of the terms were found in the current edition of the UMLS.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dwight Selina S., Harris Midori A., Dolinski Kara, Ball Catherine A., Binkley Gail, Christie Karen R., Fisk Dianna G., Issel-Tarver Laurie, Schroeder Mark, Sherlock Gavin. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO). Nucleic Acids Res. 2002 Jan 1;30(1):69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium Creating the gene ontology resource: design and implementation. Genome Res. 2001 Aug;11(8):1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn U., Romacker M., Schulz S. How knowledge drives understanding--matching medical ontologies with the needs of medical language processing. Artif Intell Med. 1999 Jan;15(1):25–51. doi: 10.1016/s0933-3657(98)00044-x. [DOI] [PubMed] [Google Scholar]
- McCray A. T., Bodenreider O., Malley J. D., Browne A. C. Evaluating UMLS strings for natural language processing. Proc AMIA Symp. 2001:448–452. [PMC free article] [PubMed] [Google Scholar]
- McCray A. T. The nature of lexical knowledge. Methods Inf Med. 1998 Nov;37(4-5):353–360. [PubMed] [Google Scholar]
- Rzhetsky A., Koike T., Kalachikov S., Gomez S. M., Krauthammer M., Kaplan S. H., Kra P., Russo J. J., Friedman C. A knowledge model for analysis and simulation of regulatory networks. Bioinformatics. 2000 Dec;16(12):1120–1128. doi: 10.1093/bioinformatics/16.12.1120. [DOI] [PubMed] [Google Scholar]
- Stevens R., Goble C. A., Bechhofer S. Ontology-based knowledge representation for bioinformatics. Brief Bioinform. 2000 Nov;1(4):398–414. doi: 10.1093/bib/1.4.398. [DOI] [PubMed] [Google Scholar]


