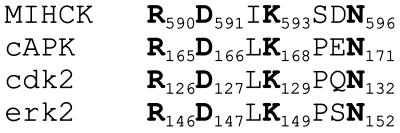Abstract
Phosphorylation of Ser-627 is both necessary and sufficient for full activity of the expressed 35-kDa catalytic domain of myosin I heavy chain kinase (MIHCK). Ser-627 lies in the variable loop between highly conserved residues DFG and APE at a position at which a phosphorylated Ser/Thr also occurs in many other Ser/Thr protein kinases. The variable loop of MIHCK contains two other hydroxyamino acids: Thr-631, which is conserved in almost all Ser/Thr kinases, and Thr-632, which is not conserved. We determined the effects on the kinase activity of the expressed catalytic domain of mutating Ser-627, Thr-631, and Thr-632 individually to Ala, Asp, and Glu. The S627A mutant was substantially less active than wild type (wt), with a lower kcat and higher Km for both peptide substrate and ATP, but was more active than unphosphorylated wt. The S627D and S627E mutants were also less active than phosphorylated wt, i.e., acidic amino acids cannot substitute for phospho-Ser-627. The activity of the T631A mutant was as low as that of the S627A mutant, whereas the T632A mutant was as active as phosphorylated wt, indicating that highly conserved Thr-631, although not phosphorylated, is essential for catalytic activity. Asp and Glu substitutions for Thr-631 and Thr-632 were inhibitory to various degrees. Molecular modeling indicated that Thr-631 can hydrogen bond with conserved residue Asp-591 in the catalytic loop and that similar interactions are possible for other kinases whose activities also are regulated by phosphorylation in the variable loop. Thus, this conserved Thr residue may be essential for the activities of other Ser/Thr protein kinases as well as for the activity of MIHCK.
Acanthamoeba myosin I heavy chain kinase (MIHCK) activates amoeba myosin I molecules by phosphorylating a single Ser or Thr residue within the subfragment 1 portion of their single heavy chains (refs. 1 and 2; for review, see ref. 3). Acanthamoeba MIHCK also phosphorylates the light chain of, and activates, smooth muscle myosin II (3, 4). MIHCK is a member of the p21-activated protein kinase (PAK) family (5). All PAKs have highly homologous catalytic domains (6–8) and most, including MIHCK (9), have been shown to be activated by the small GTP-binding proteins Rac and Cdc42. MIHCK and typical mammalian and yeast Paks have similar substrate specificities in vitro including the ability to phosphorylate and activate Acanthamoeba and Dictyostelium myosin I molecules (10, 11).
MIHCK is a 79.3-kDa protein with a 35-kDa C-terminal catalytic domain (5, 12). The activity of native MIHCK is enhanced ∼50-fold by phospholipid (and plasma membrane)-stimulated autophosphorylation incorporating 8–10 mol of phosphate per mol of protein (13–16). The expressed 35-kDa catalytic domain is also regulated by autophosphorylation (17); phosphorylation of a single Ser residue [Ser-627 in the sequence of the full-length protein (9)] is both necessary and sufficient for full activity of the expressed catalytic domain. Ser-627 is also phosphorylated in the fully active native kinase (17).
Ser-627 of MIHCK lies within the variable region between residues DFG and APE that are highly conserved in all protein kinases (for review, see ref. 18). Phosphorylation within this variable region regulates the activities of several other kinases (Fig. 1) including Pak (19, 20), cAMP-dependent protein kinase (cAPK), extracellular signal-regulated protein kinase (erk2), cyclin-dependent protein kinase (cdk2), and insulin receptor kinase (for review, see refs. 21 and 22). Ser-627 of MIHCK is at precisely the same position as the regulatory phospho-Thr of cAPK and Pak (Fig. 1), 11 residues N-terminal to the conserved Glu in the APE sequence, but the position of the phosphorylated residue is not identical in all regulated kinases (Fig. 1), and phosphorylation of more than one site within this region may be required for full activity of some of them (for review, see refs. 21–23). In addition to Ser-627, the variable loop of MIHCK contains two other hydroxyamino acids: Thr-631 and Thr-632 (5). Almost all Ser/Thr protein kinases have a Thr at the position of Thr-631 in MIHCK, 7 residues N-terminal to the Glu of the conserved APE sequence, where almost all Tyr protein kinases have a Pro (18).
Figure 1.
Sequence of the variable loop between conserved DFG and APE for MIHCK and other kinases discussed in the text. The arrows indicate the residues in MIHCK that were replaced by Ala, Asp, and Glu. The residues that are conserved in all protein kinases, DFG and APE, and the Thr that is conserved in all Ser/Thr kinases residues are in boldface type. Residues that have been shown to be phosphorylated are double underlined. See text for references.
In this article, we report the effects on the activity of the expressed catalytic domain of MIHCK of individually substituting Ala, Asp, and Glu for each of the three hydroxyamino acids in the variable loop. The results confirm the importance of Ser-627 for MIHCK activity and, further, show that highly conserved Thr-631, which is not phosphorylated, is also required for full enzymatic activity of the MIHCK catalytic domain.
MATERIALS AND METHODS
DNA Preparation.
PCR (Pfu polymerase, Perkin–Elmer) were used to prepare the DNA coding for each of the mutants of the MIHCK catalytic domain, as described for the wild-type (wt) catalytic domain (5). The internal primer for the Ser mutants consisted of bases 514–543 and the primer for both Thr mutants was bases 527–556, with appropriate changes. The wt DNA sequence for this region is 514CAGGACAAACGCGCGTCTGTGGTGGGCACGACGTACTGGATGG556 (the bases coding for Ser-627, Thr-631, and Thr-632 are underlined). The final PCR products were cloned into pBlueBacHis vector (Invitrogen) between BglII and HindIII sites, which provided an N-terminal His-tag. The DNA sequence of the mutated region was confirmed for all mutants.
Protein Expression and Purification.
Insect Sf9 cells were cotransfected with plasmid and viral Autographa californica multiple nuclear polyhedrosis virus DNA by using the Bac-N-Blue transfection kit (Invitrogen), and plaque assays and viral stock amplifications were done according to standard procedures. The wt and mutated MIHCK catalytic domains were expressed in Sf9 cells and purified as described for the wt catalytic domain (5). The purity of each of the mutant proteins was similar to that of the expressed wt protein (5), as determined by SDS/PAGE (data not shown). Myosin IC was expressed in Sf9 cells and purified as described (24). Protein concentrations were determined by the Bio-Rad protein assay.
Mass Spectrometry.
The amino acid sequences of all of the mutated regions, the total sequence of T631E, the extent of phosphorylation of the peptides of interest, and the identification of the phosphorylated residue were all determined by mass spectrometry of proteolytic digests of proteins separated by PAGE, as described (ref. 17 and S. Zhang, C.J.H., P. R. Romano, J.S., H.B., E. G. Hinnebusch, and J.Q., unpublished results).
Phosphorylation of Proteins.
To obtain fully autophosphorylated proteins, the purified wt and mutant MIHCK catalytic domains (0.1–0.25 mg/ml) were incubated at 30°C in 50 mM imidazole (pH 7.0) containing 2.5 mM ATP, 3.5 mM MgCl2, 2 mM EGTA, and BSA (0.2 mg/ml). The extent of autophosphorylation was determined by mass spectrometry as described above. The T631E mutant (which autophosphorylated extremely slowly, if at all) was phosphorylated by incubating it with either wt catalytic domain or native full-length MIHCK that had been fully activated by autophosphorylation. T631E was incubated with wt catalytic domain, 95:5 (mol/mol), under the same conditions as for autophosphorylation of the other mutants. Kinase activity was measured immediately after incubation and was corrected for the activity of wt catalytic domain, which was incubated and assayed in parallel. Alternatively, T631E (0.2 mg/ml) was incubated at 30°C with fully phosphorylated native MIHCK (0.03 mg/ml) for 45 min in 10 mM imidazole (pH 7.0) containing 2.5 mM ATP, 5 mM MgCl2, and BSA (0.2 mg/ml). After incubation, samples were mixed with Ni-NTA (nitrilotriacetic acid) resin (1 μl of packed resin per μg of protein) that was washed three times with 10 mM imidazole (pH 7.0) containing 500 mM NaCl and 10 mM 2-mercaptoethanol to remove the full-length MIHCK. T631E was eluted with 125 mM imidazole (pH 7.0) containing 100 mM NaCl, 10 mM 2-mercaptoethanol and 15% glycerol. Kinase activity was measured immediately after elution. As controls, MIHCK and T631E were separately incubated, processed, and assayed under the same conditions, and their activities subtracted from the activity of phosphorylated T631E. Protein concentrations were measured by scanning Coomassie blue-stained gels of the proteins eluted from Ni-NTA.
Kinase Assays.
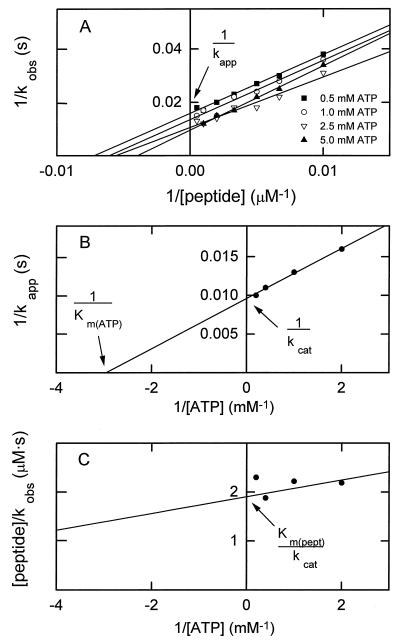
Routine assays were performed with either the 9-residue synthetic peptide PC9 [GRGRSSVYS, the sequence of the phosphorylation site of myosin IC (2)] or myosin IC as substrates. The MIHCK catalytic domain was incubated at 30°C with either 200 μM PC9 or 4 μM myosin IC in 50 mM imidazole (pH 7.0) containing 2.5 mM [γ-32P]ATP (30,000 cpm/nmol), 3.5 mM MgCl2, 2 mM EGTA, and BSA (0.2 mg/ml). The reaction was stopped with acetic acid (when the substrate was PC9) or SDS (when the substrate was myosin IC), and the extent of substrate phosphorylation was determined as described for PC9 (14) and myosin IC (13). The time of incubation was varied, depending on the enzyme activity, so that no more than 20% of the substrate was phosphorylated. For determination of the catalytic constants, kcat, Km(peptide), and Km(ATP), incubations were at 30°C in 50 mM imidazole (pH 7.0) containing 1 mM free Mg2+, 2 mM EGTA, BSA (0.2 mg/ml), and various concentrations of PC9 and [γ-32P]MgATP (30,000 cpm/nmol), as described below. The enzyme concentrations varied between 1 and 38 μg/ml and the length of incubations varied between 15 s and 2 min, depending on the mutant, so that no more than 20% of the peptide substrate was phosphorylated. Kinase activity was measured as a function of peptide concentration at four concentrations of MgATP. The intercept at infinite peptide concentration of the double-reciprocal plot of enzymatic activity (kobs) vs. peptide concentration (Fig. 2A) gave the value for kapp for each ATP concentration. The double-reciprocal plot of kapp vs. ATP concentration (Fig. 2B) gave the true value for kcat (the specific activity at infinite concentration of both peptide and ATP) and the Km(ATP) (the concentration of ATP at half-maximal specific activity at infinite peptide concentration). The intercept of the double-reciprocal plot of the slopes of the lines in Fig. 2A vs. ATP concentration (Fig. 2C) gave the value of Km(peptide)/kcat, from which Km(peptide) could be calculated. The kcat values are reported in units of s−1 (mol of peptide phosphorylated per s per mol of enzyme).‖
Figure 2.
Illustration of the method used for determining kcat, Km(peptide), and Km(ATP) for expressed wt and mutant catalytic domain of myosin I heavy chain kinase. The data shown are for the T632A mutant (Table 1). (A) A double reciprocal plot of the enzymatic activities (kobs) determined as a function of peptide concentration at four concentrations of ATP. The slope of the line for 5.0 mM ATP (but not kcat) was consistently less than the slopes at the three lower concentrations of ATP, suggesting the possibility of substrate (ATP) inhibition that is reversed by high concentrations of the second substrate (peptide). (B) A double reciprocal plot of the values for kapp estimated from A as a function of the ATP concentration. (C) A double-reciprocal plot of the slopes of the lines in A as a function of ATP concentration.
Modeling.
The look program (Molecular Applications Group, San Diego) was used to generate model structures of the MIHCK catalytic domain by using crystal structures of other kinases. The insightii program (Molecular Simulations, Palo Alto, CA) was used to view the original and modeled structures and to measure the distances between atoms.
RESULTS AND DISCUSSION
Phosphorylation State of Purified Catalytic Domains.
The extent of phosphorylation of the expressed proteins was estimated from the relative amounts of the phosphorylated and unphosphorylated tryptic peptides (17), as determined by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Ser-627 of wt, T631A, and T632A were 100% phosphorylated and T631D, T632D, and T632E were 70%, 70%, and 80% phosphorylated, respectively, as isolated. As expected, all of the Ser-627 mutants were completely unphosphorylated as was T631E. When incubated with ATP, phosphorylation of T631D and T632D increased to 95% and 80%, respectively, in 20 min and T632E was 100% phosphorylated in 10 s. As determined by mass spectrometry, there was no detectable autophosphorylation of T631E even after 20 min. Autophosphorylation did not significantly affect the kinase activity of any of the mutants when assayed at substrate concentrations of 2.5 mM ATP and 200 μM PC9. The kinetic constants reported in this article were obtained for the proteins as isolated. Except for T631E (see below), this would cause no more than a 30% error in the calculated values for kcat, which is appreciably less than the differences among the mutant enzymes.
Comparison of the Activities of the Expressed wt Catalytic Domain and Native Full-Length Kinase.
The kcat and Km(peptide) values for the expressed wt catalytic domain, 71 s−1 and 0.15 mM, respectively, were both higher than the values of 11 s−1 and 50 μM for full-length MIHCK (14) but are consistent with the activity of the catalytic domain obtained by proteolysis of phosphorylated native kinase.** Previously, we had reported that the N-terminal region of unphosphorylated native kinase strongly inhibits the activity of the C-terminal catalytic domain (25). The differences in kcat and Km(peptide) between the fully phosphorylated native MIHCK and the phosphorylated wt catalytic domain suggest that this inhibition, although weaker, also occurs in the fully phosphorylated enzyme.
Effect of Mutations on the Kinase Activity of the Expressed Catalytic Domain.
Ser-627. As expected, the S627A mutant had significantly lower catalytic activity than wt (Table 1) due about equally to a lower kcat and higher Km(peptide) and Km(ATP) values resulting in a 40-fold decrease in the ratio of kcat/Km(peptide) and 25-fold decrease in the ratio of kcat/Km(ATP). However, the decrease in the ratio kcat/Km(peptide) is an order of magnitude less than that caused by substituting Ala for the homologous Thr-197 of cAPK (26), which suggests that regulation by phosphorylation is less tight for the MIHCK catalytic domain than for cAPK (of course, regulation of full-length MIHCK might be more pronounced).
Table 1.
Kinetic constants for wt and mutants of the expressed catalytic domain of MIHCK
| Protein | kcat, s−1 | Km(peptide), mM | Km(ATP), mM | kcat/Km(peptide), s−1/mM | kcat/Km(ATP), s−1/mM |
|---|---|---|---|---|---|
| wt | 71 | 0.15 | 0.16 | 473 | 444 |
| S627A | 10 | 0.81 | 0.57 | 12 | 18 |
| S627D | 8 | 1.10 | 0.49 | 7 | 16 |
| S627E | 37 | 1.21 | 0.80 | 31 | 46 |
| T631A | 18 | 1.56 | 1.10 | 12 | 16 |
| T631D | 2 | 0.86 | 0.56 | 2 | 4 |
| T631E | 0.36 | 2.40 | 2.80 | 0.2 | 0.1 |
| T632A | 105 | 0.20 | 0.30 | 525 | 350 |
| T632D | 23 | 0.64 | 0.68 | 36 | 34 |
| T632E | 23 | 0.25 | 0.69 | 92 | 33 |
Data were obtained as explained in Materials and Methods and illustrated in Fig. 2 for the T632A mutant.
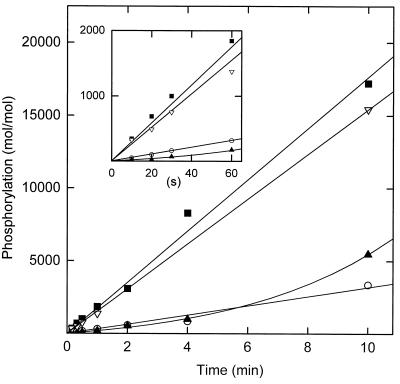
Because of its relatively high activity, it was of interest to compare the activity of S627A to the activity of unphosphorylated wt. However, autophosphorylation of the unphosphorylated wt was far too rapid, under the required assay conditions, to determine its kcat and Km values. Instead, the activities of phosphorylated, dephosphorylated, and rephosphorylated wt and S627A were compared at a single concentration of ATP [2.5 mM, which is substantially higher than Km(ATP) for both wt and S627A] and a single concentration of peptide [1 mM PC9, which is much higher than Km(peptide) for wt but about equal to the value for S627A]. The enzyme concentrations were as low as possible to minimize the rate of autophosphorylation [which is intermolecular (15)] of the dephosphorylated wt. As expected, the activity of the dephosphorylated wt increased during the incubation (Fig. 3) due to its autophosphorylation. At early times (Fig. 3 Inset), S627A was about twice as active as dephosphorylated wt. Phosphorylated wt was about 12 times more active than dephosphorylated wt [consistent with the ratio of about 15:1 published previously (17)].
Figure 3.
Comparison of the time courses of phosphorylation of peptide PC9 by the following MIHCK catalytic domains: ▪, phosphorylated wt (12 μg/ml); ▴, dephosphorylated wt (4 μg/ml); ▿, rephosphorylated wt (3.5 μg/ml); ○, the S627A mutant (42 μg/ml). Kinase was dephosphorylated and rephosphorylated as described (17). The peptide concentration was 1 mM and ATP was 2.5 mM. Activity is reported as mol of peptide phosphorylated per mol of catalytic domain. See text for details.
The catalytic constants for S627D were essentially the same as for S627A; kcat for S627E was about 4-fold higher than kcat for S627A but their Km values were similar. The same results were obtained for two independent clones of S627E. Therefore, at least in the expressed catalytic domain, neither Asp nor Glu can fully substitute for the regulatory phospho-Ser.
Thr-631.
Substitution of Ala, Asp, or Glu for the conserved Thr-631 was at least as inhibitory as substitution of these three amino acids for Ser-627 (Table 1). T631A had about the same activity as S627A with essentially identical values for both kcat/Km(peptide) and kcat/Km(ATP). T631D was less active than S627D and T631E was much less active than S627E, principally because of lower kcat values. The results for T631E were confirmed with three preparations. The entire protein sequence of T631E was confirmed by mass spectrometry to be certain that inadvertent mutations elsewhere in the molecule were not responsible for the very low activity.
As mentioned, Ser-627 was not phosphorylated in the expressed T631E and was not autophosphorylated even after prolonged incubation with ATP. To determine whether the unphosphorylated Ser-627, rather than the substitution of Glu for Thr, might have accounted for its very low activity, T631E was incubated with exogenous kinase to phosphorylate Ser-627. As determined by mass spectrometry, T631E was fully phosphorylated on Ser-627 after incubation for 10 min with wt catalytic domain and after incubation for 45 min with native kinase. The activity of fully phosphorylated T631E was still too low, however, for accurate determination of its kcat and Km values. Therefore, the activity of phosphorylated T631E was compared with the activities of unphosphorylated T631E and wt under a single set of conditions, 2.5 mM ATP and 0.4 mM peptide. Under these conditions, phosphorylated T631E was 4- to 10-fold more active than unphosphorylated T631E, thus approaching the activity of T631D, but still much less active than wt (data not shown). These results suggest that substituting Glu for Thr-631 inhibits autophosphorylation of the expressed catalytic domain even more than it inhibits substrate phosphorylation.
Similar results were obtained when the activities of T631A and wt were compared with myosin IC as substrate. Under a single set of conditions, 4 μM myosin IC and 2.5 mM ATP, T631A was about 10-fold less active than wt, 0.15 s−1 and 1.6 s−1, respectively.
Thr-632.
In contrast to Thr-631, substitution of Ala for Thr-632 had no significant effect on kinase activity (Table 1); however, as for Thr-631, substitution of Asp and Glu for Thr-632 reduced kcat (about 3-fold) and increased both Km values (about 4-fold).
Modeling the MIHCK Catalytic Domain.
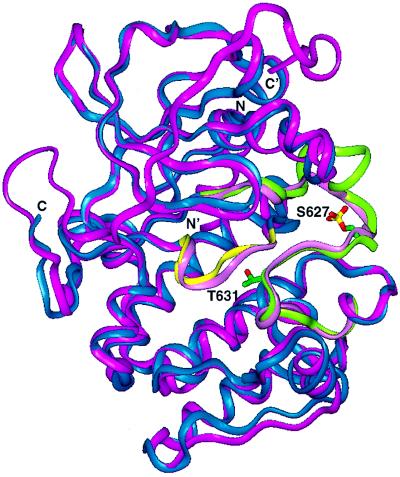
To gain some insight into which residues might interact with phospho-Ser-627 and the possible role of the conserved Thr-631, we generated the three-dimensional structure of the MIHCK catalytic domain by homology modeling to the published crystal structures of the following kinases: cAPK complexed with ATP and protein kinase inhibitor (Protein Data Base accession code 1ATP, ref. 27) or with protein kinase inhibitor only (Protein Data Base accession code 1CMK, ref. 28); cdk2, in inactive (Protein Data Base accession code 1HCL, ref. 29), partially active (Protein Data Base accession code 1FIN, ref. 30) or fully active (Protein Data Base accession code 1JST, ref. 31) conformations; erk2 (Protein Data Base accession code 1ERK, ref. 32); phosphorylase kinase (Protein Data Base accession code 1PHK, ref. 33); and casein kinase (Protein Data Base accession code 1CSN, ref. 34). The best fit‡‡ was obtained by using the 1ATP structure of cAPK as the template (Fig. 4).
Figure 4.
Structure of the catalytic domain of MIHCK (blue) modeled to the structure of the catalytic subunit of cAPK (pink). The catalytic loop and variable loop of MIHCK are yellow and green, respectively. The catalytic and variable loops of cAPK are both light pink. The positions of MIHCK phospho-Ser-627 and Thr-631 are indicated. See text for details and references.
Ser-627.
As mentioned above, Ser-627 of MIHCK is in the same position in the linear sequence as Thr-197 in cAPK (Fig. 1 and refs. 17 and 35). In the three-dimensional structure of cAPK (Fig. 4), the phosphate group of phospho-Thr-197 hydrogen bonds with Arg-165 (which immediately precedes the catalytic Asp-166 that is absolutely conserved in all kinases), with Lys-189 and with Thr-195 (in the DFG-APE variable loop; Fig. 1), and with His-87 (in the N-terminal lobe of the catalytic domain) (ref. 27; for review, see refs. 21 and 35). Similar interactions occur for the regulatory phosphoamino acid in other kinases whose structures have been determined (for review, see ref. 21).
The model of the MIHCK catalytic domain (Fig. 5) shows some of these interactions for Ser-627 and Thr-631 of MIHCK. The phosphate group on Ser-627 hydrogen bonds with Arg-590 (which immediately precedes the conserved Asp-591 in the catalytic loop) and possibly also with Arg-625 (in the DFG-APE variable loop). Phospho-Ser-627 of MIHCK is also in contact with Tyr-645 (which is C-terminal to the variable loop) but, in contrast to cAPK, the phosphate group in the model of MIHCK does not seem to interact with the N-terminal lobe of the kinase domain. The possibility that phospho-Ser-627 of MIHCK may interact with fewer residues than does phospho-Thr-197 of cAPK is consistent with the experimental observations that phospho-Ser-627 of the MIHCK catalytic domain is readily dephosphorylated by exogenous phosphatase (17) whereas phospho-Thr-197 of cAPK is not (35, 36).
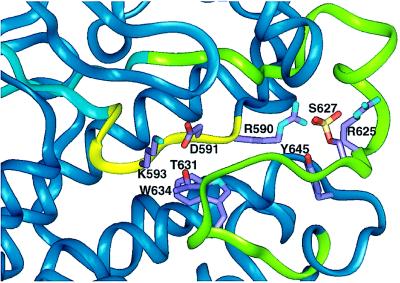
Figure 5.
Higher magnification of the region of interest in the model of MIHCK catalytic domain with residues of interest marked. The catalytic loop is in yellow and the variable loop is in green. S627 (Ser-627) is the regulatory phosphoserine, and T631 (Thr-631) is the Thr that is conserved in all Ser/Thr kinases. The distances between these and other residues of interest are discussed in the text and, for Thr-631 and the corresponding residue in other kinases whose crystal structures are available, summarized in Table 2. See text for details.
Thr-631.
The relatively low activity of all of the three point mutations of Thr-631 indicates an important, but unknown, function for this highly conserved residue (see Fig. 1). In the crystal structure of cAPK complexed with peptide substrate, Thr-201 [which corresponds to MIHCK Thr-631 (Fig. 1)] bonds to, and presumably helps ensure proper orientation of, conserved Lys-168 in the catalytic loop (ref. 37 and Fig. 6), a residue that helps anchor the peptide substrate and the γ-phosphate of ATP. In our model of the MIHCK catalytic domain (Fig. 5), Thr-631 is closer to Asp-591 (<3 Å, Table 2) than to Lys-593 (4.5 Å), which corresponds to cAPK Lys-168 (Fig. 6). The residue that corresponds to MIHCK Asp-591 in cAPK is Asp-166 (in the catalytic loop), which, like cAPK Lys-168, serves to anchor the peptide substrate and the γ-phosphate of ATP (27). In the 1ATP crystal of cAPK, Asp-166 and Thr-201 are close enough to hydrogen bond (Table 2).
Figure 6.
Sequences of the catalytic loop of MIHCK and other Ser/Thr kinases discussed in the text. The conserved residues are in boldface type.
Table 2.
Distances between conserved Thr residues in the variable loops (Fig. 1) and conserved Lys and Asp residues in the catalytic loops (Fig. 6) of the model of MIHCK (Figs. 4 and 5) and Ser/Thr kinases whose crystal structures are in the Protein Data Base
| Kinase | Description | Residues | Distance, Å |
|---|---|---|---|
| MIHCK | Model based on cAPK (1ATP) | T631–D591 | 2.9 |
| T631–K593 | 4.5 | ||
| cAPK (1ATP) | Active; complexed with PKI and ATP; phosphorylated; closed | T201–D166 | 2.8 |
| T201–K168 | 3.0 | ||
| cAPK (1CMK) | Active; complexed with PKI; phsphorylated; open | T201–D166 | 4.1 |
| T201–K168 | 2.7 | ||
| cdk2 (1JST) | Active; complexed with cyclin; phosphorylated | T165–D127 | 3.8 |
| T165–K129 | 3.0 | ||
| cdk2 (1FIN) | Partially active; complexed with cyclin and ATP; unphosphorylated | T165–D127 | 7.0 |
| T165–K129 | 5.2 | ||
| cdk2 (1HCL) | Inactive; uncomplexed; unphosphorylated | T165–D127 | 7.8 |
| T165–K129 | 7.4 | ||
| erk2 (1ERK) | Low activity; uncomplexed; unphosphorylated | T188–D147 | 3.0 |
| T188–K149 | 2.7 |
Distances shown in boldface type are equal to or less than 3 Å and can form hydrogen bonds. The Protein Data Base accession code is in parentheses. PKI, protein kinase inhibitor. One-letter amino acid code is used for residues.
Similar interactions are also possible for the two other kinases whose crystal structures have been determined and that require phosphorylation of a Ser or Thr in the DFG-APE variable loop for full activity (Fig. 1): the conserved Thr residue in the variable loops of the activated form of cdk2 (1JST) and erk2 (1ERK) is close enough to interact with either the conserved Asp or Lys in their catalytic loops (Figs. 1 and 6 and Table 2). On the other hand, the conserved Thr and the catalytic loop are too far apart for hydrogen bonding in the structures of kinases that are not regulated by phosphorylation in the variable loop, i.e., phosphorylase kinase (1PHK), casein kinase (1CSN) and twitchin kinase (1KOA, ref. 38), data not shown. It is not excluded, however, that similar interactions may also be important for these latter enzymes as the conserved Thr may be close enough to hydrogen bond with a residue in the catalytic loop at a stage of their catalytic cycles that is not represented in the crystal structures obtained to date. For example, hydrogen bonding is possible in the structure of fully active cdk2 (1JST) but not in the structures of partially active cdk2 (1FIN) or completely inactive cdk2 (1HCL).
In the model of MIHCK, conserved Thr-631 can also interact with Trp-634 (Fig. 5). Because this is a hydrophobic interaction, however, its contribution to the protein structure is likely to be less specific than the potential hydrogen bonding of Thr-631 to Asp-591. The residues in Pak and Ste20 that correspond to MIHCK Trp-634 is also Trp but it is Tyr in cAPK, cdk2, and erk2 (Fig. 1).
Possible Implications for Other Kinases.
We have found that Thr-631, which is highly conserved in the variable loop in Ser/Thr kinases, is important for the activity of the MIHCK catalytic domain and that it is not phosphorylated. It was previously suggested (37) that the corresponding Thr in cAPK hydrogen bonds to a conserved Lys in the catalytic loop. The analysis in this article of the crystal structures of cAPK, cdk2, and erk2 and the MIHCK model suggests that hydrogen bonding of the conserved Thr in the variable loop to the conserved Lys or conserved Asp in the catalytic loop may be of general importance for the activity of Ser/Thr kinases. The reported decreases in activity of several other Ser/Thr kinases caused by mutation of this conserved Thr (39–41) are consistent with this proposal.
Although some other Ser/Thr kinases have either a Ser or Thr residue at a position corresponding to Ser-627 of MIHCK, this residue is not highly conserved. It is present, however, in all members of the PAK family. In agreement with our results for Ser-627 of MIHCK, phosphorylation of the corresponding hydroxyamino acid of Pak from brain and placenta correlates with enhanced kinase activity (19, 20) but, in apparent conflict with our results, the double substitution of Ala for Thr-772 and Thr-773 in yeast Ste20 (one of which corresponds to Ser-627 in MIHCK) was found not to block kinase function in vivo (39). Quantitative analyses of kinase activities in vitro are necessary to determine if the catalytic activity of the double-Ala mutant of Ste20 might be significantly less than wt activity, as we find for the S627A mutant of MIHCK catalytic domain, although sufficient to carry out the essential functions of Ste20 in vivo.
Acknowledgments
We thank Dr. P. Boon Chock (National Heart, Lung, and Blood Institute) for advice on analyzing the kinetics of two-substrate reactions.
ABBREVIATIONS
- cAPK
cyclic nucleotide-dependent protein kinase
- cdk2
cyclin-dependent kinase
- erk2
extracellular signal-regulated protein kinase
- MIHCK
myosin I heavy chain kinase
- Pak
p-21-activated protein kinase
- wt
wild type
Footnotes
In previous papers (14, 15), we used the unit μmol per min per mg for kcat (incorrectly designated as Vmax in those papers). The values for kcat reported previously can be compared with those in this paper by multiplying the values for the MIHCK catalytic domain (Mr 38,000, including the poly-His tag) by 0.63 and the values for the full-length kinase (Mr 79,300) by 1.32.
The catalytic domain obtained by proteolysis was twice as active as the full-length kinase when assayed with a substrate concentration of 200 μM PC9 (12). The activity of the expressed wt catalytic domain with 200 μM PC9 (approximately its Km, Table 1) would be 35 s−1 (half of kcat) or about three times the activity of the native kinase.
The model score for cAPK was 102 [this is a statistically based “threading” score (Profiles 3D, Molecular Simulations)]. The model scores for other crystals varied between 71 and 95. The expected score for the native MIHCK structure is 138.
References
- 1.Pollard T D, Korn E D. J Biol Chem. 1977;248:4691–4697. [PubMed] [Google Scholar]
- 2.Brzeska H, Lynch T J, Martin B M, Korn E D. J Biol Chem. 1989;264:19340–19348. [PubMed] [Google Scholar]
- 3.Brzeska H, Korn E D. J Biol Chem. 1996;271:16983–16986. doi: 10.1074/jbc.271.29.16983. [DOI] [PubMed] [Google Scholar]
- 4.Hammer J A, III, Sellers J, R, Korn E, D. J Biol Chem. 1984;259:3224–3229. [PubMed] [Google Scholar]
- 5.Brzeska H, Szczepanowska J, Hoey J, Korn E D. J Biol Chem. 1996;271:27056–27062. doi: 10.1074/jbc.271.43.27056. [DOI] [PubMed] [Google Scholar]
- 6.Lim L, Manser E, Leung T, Hall C. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 7.Sells M A, Chernoff J. Trends Cell Biol. 1997;7:162–167. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- 8.Lee S-F, Egelhoff T T, Mahasneh A, Côté G. J Biol Chem. 1996;271:27044–27048. doi: 10.1074/jbc.271.43.27044. [DOI] [PubMed] [Google Scholar]
- 9.Brzeska H, Young R, Knaus U G, Korn E D. Mol Biol Cell. 1997;8:40a. (abstr.). [Google Scholar]
- 10.Brzeska H, Knaus U G, Wang Z-Y, Bokoch G M, Korn E D. Proc Natl Acad Sci USA. 1997;94:1092–1095. doi: 10.1073/pnas.94.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C, Lee S-F, Furmaniak-Kazmierczak E, Côté G P, Thomas D Y, Leberer E. J Biol Chem. 1996;271:31787–31790. doi: 10.1074/jbc.271.50.31787. [DOI] [PubMed] [Google Scholar]
- 12.Brzeska H, Martin B M, Korn E D. J Biol Chem. 1996;271:27049–27055. doi: 10.1074/jbc.271.43.27049. [DOI] [PubMed] [Google Scholar]
- 13.Brzeska H, Lynch T J, Korn E D. J Biol Chem. 1990;265:3591–3594. [PubMed] [Google Scholar]
- 14.Brzeska H, Lynch T J, Martin B M, Corigliano-Murphy A, Korn E D. J Biol Chem. 1990;265:16138–16144. [PubMed] [Google Scholar]
- 15.Wang Z-Y, Brzeska H, Baines I C, Korn E D. J Biol Chem. 1995;270:27969–27976. doi: 10.1074/jbc.270.46.27969. [DOI] [PubMed] [Google Scholar]
- 16.Kulesza-Lipka D, Brzeska H, Baines I C, Korn E D. J Biol Chem. 1993;268:17995–18001. [PubMed] [Google Scholar]
- 17.Szczepanowska J, Zhang X, Herring C J, Qin J, Korn E D, Brzeska H. Proc Natl Acad Sci USA. 1997;94:8503–8508. doi: 10.1073/pnas.94.16.8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanks S K, Quinn A M, Hunter T. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 19.Benner G E, Dennis P B, Masaracchia R A. J Biol Chem. 1995;270:21121–21128. doi: 10.1074/jbc.270.36.21121. [DOI] [PubMed] [Google Scholar]
- 20.Manser E, Huang H-Y, Loo T-H, Chen X-Q, Dong J-M, Leung T, Lim L. Mol Cell Biol. 1997;17:1129–1143. doi: 10.1128/mcb.17.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, L. N., Noble, M. E. M. & Owen, D. J. (1996) Cell 149–158. [DOI] [PubMed]
- 22.Hanks S K, Hunter T. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 23.Taylor S S, Radzio-Andzelm E. Structure. 1994;2:345–355. doi: 10.1016/s0969-2126(00)00036-8. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z Y, Wang F, Sobota A, Korn E D, Hammer J A., III Mol Biol Cell. 1997;8:370a. (abstr.). [Google Scholar]
- 25.Brzeska H, Martin B M, Kulesza-Lipka D, Baines I C, Korn E D. J Biol Chem. 1992;267:4949–4956. [PubMed] [Google Scholar]
- 26.Adams J A, McGlone M L, Gibson R, Taylor S S. Biochemistry. 1995;34:2447–2454. doi: 10.1021/bi00008a007. [DOI] [PubMed] [Google Scholar]
- 27.Zheng J, Knighton D R, Ten Eyck L F, Karlsson R, Xuong N-H, Taylor S S, Sowadski J M. Biochemistry. 1993;32:2154–2161. doi: 10.1021/bi00060a005. [DOI] [PubMed] [Google Scholar]
- 28.Zheng J, Knighton D R, Xuong N-H, Taylor S S, Sowadski J M, Ten Eyck L-F. Protein Sci. 1993;2:1559–1573. doi: 10.1002/pro.5560021003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulze-Gahmen U, Brandsen J, Jones H D, Morgan D O, Meijer L, Vesely J, Kim S-H. Proteins Struct Funct. 1995;22:378–391. doi: 10.1002/prot.340220408. [DOI] [PubMed] [Google Scholar]
- 30.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich N P. Nature (London) 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 31.Russo A A, Jeffrey P D, Pavletich N P. Nat Struct Biol. 1996;3:696–700. doi: 10.1038/nsb0896-696. [DOI] [PubMed] [Google Scholar]
- 32.Zhang F, Strand A, Robbins D, Cobb M H, Goldsmith E J. Nature (London) 1994;367:704–711. doi: 10.1038/367704a0. [DOI] [PubMed] [Google Scholar]
- 33.Owen D J, Noble M E M, Garman E F, Papageorigiou A C, Johnson L N. Structure. 1995;3:467–482. doi: 10.1016/s0969-2126(01)00180-0. [DOI] [PubMed] [Google Scholar]
- 34.Xu R-M, Carmel G, Sweet R M, Kuret J, Cheng X. EMBO J. 1995;14:1015–1023. doi: 10.1002/j.1460-2075.1995.tb07082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor S S, Knighton D R, Zheng J, Ten Eyck L F, Sowadski J M. Annu Rev Cell Biol. 1992;8:429–462. doi: 10.1146/annurev.cb.08.110192.002241. [DOI] [PubMed] [Google Scholar]
- 36.Steinberg R A, Cauthron R D, Symcox M M, Shuntoh H. Mol Cell Biol. 1993;13:2332–2341. doi: 10.1128/mcb.13.4.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madhusudan, Trafny E A, Xuong N-H, Adams J A, Ten Eyck L F, Taylor S S, Sowadski J M. Protein Sci. 1994;3:176–187. doi: 10.1002/pro.5560030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobe B, Heierhorst J, Feil S C, Parker M W, Benian G M, Weiss K R, Kemp B E. EMBO J. 1996;15:6810–6821. [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C, Whiteway M, Thomas D Y, Leberer E. J Biol Chem. 1995;270:15984–15992. doi: 10.1074/jbc.270.27.15984. [DOI] [PubMed] [Google Scholar]
- 40.Siow Y L, Kalmar G B, Sanghera J S, Tai G, Oh S S, Pelech S L. J Biol Chem. 1997;272:7586–7594. doi: 10.1074/jbc.272.12.7586. [DOI] [PubMed] [Google Scholar]
- 41.Palaty C H, Kalmar G, Tai G, Oh S, Amankawa L, Affolter M, Aebersold R, Pelech S L. J Biol Chem. 1997;272:10514–10521. doi: 10.1074/jbc.272.16.10514. [DOI] [PubMed] [Google Scholar]