Abstract
Two types of porcine circovirus (PCV), which differ in their pathogenicity, are known. PCV type 2 (PCV2) is the etiological agent of postweaning multisystemic wasting syndrome in swine, while PCV1 has not yet been linked to a disease. Corroborating earlier observations in PCV1, transcript mapping revealed that the rep gene of PCV2 encodes two products, the full-length protein Rep and the spliced version Rep′ and that the simultaneous expression of Rep and Rep′ proteins is essential for initiation of replication of PCV2. The interchangeability of the replication factors of PCV1 and PCV2 was examined. The rep gene products of PCV2 were not only able to bind the PCV2 origin but also the origin of PCV1 and vice versa. To investigate the competence of the Rep/Rep′ proteins to initiate replication at the heterologous origin, a new replication assay was developed. It measures the expression of a luc reporter gene present on a plasmid carrying the origin of the investigated replicon. Replication is initiated by expression of the appendant replicase from a second plasmid and results in replication of the origin plasmid coupled with an increase in the Luc activity. Using this method to compare replication of PCV1 and PCV2 in cell culture, it was shown that the Rep/Rep′ protein of PCV2 initiated replication at the origin of PCV1, as did the reciprocal combination. Our results indicate that the cis- and trans-acting replication factors of the two viruses are functionally exchangeable.
Two strains of porcine circoviruses (PCV) are known. PCV type 1 (PCV1) and PCV2 are characterized by a small, circular closed and single-stranded DNA genome (1759 and 1768 nucleotides [nt]). Since no disease has yet been linked to PCV1, it has been classified as an apathogenic virus. In contrast, PCV2 is the etiological agent of postweaning multisystemic wasting syndrome, a newly emerging disease of swine, whose characteristic symptoms include loss of weight, fever, dyspnea, and lymphadenopathy (14).
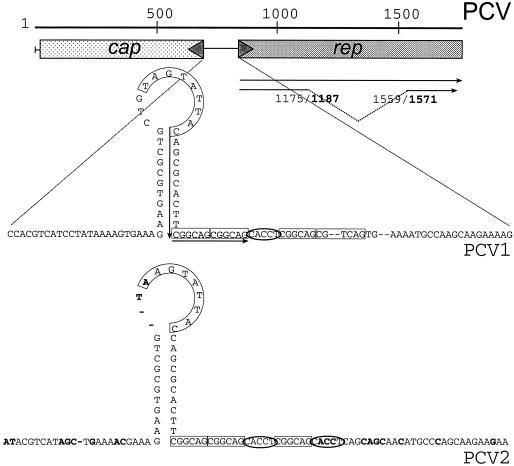
The genomic organization of PCV is simple and compact, only two major open reading frames, rep and cap, are seen (Fig. 1). The cap gene is located on the counterclockwise strand of PCV and encodes the major structural protein of the virus (12), while the rep gene directs the synthesis of the Rep proteins. Analysis of the rep gene of PCV1 has shown that differential splicing results in production of two rep isoforms, the full-length Rep protein (312 amino acids [aa]) and the spliced and thereby frame-shifted variant Rep′ (168 aa). A rapid amplification of cDNA ends (RACE) approach has demonstrated that rep and rep′ transcripts are also produced in PCV2-infected cells, plus several smaller transcripts (2). Rep and Rep′ contain three amino acid motifs, which are conserved in enzymes involved in initiation of DNA replication in the rolling-circle mechanism (4). A deoxynucleoside triphosphate-binding domain has been identified in the Rep protein, but not in Rep′ (9). The rep gene products of PCV1 have been reported to bind in vitro to double-stranded DNA (ds DNA) fragments carrying the origin of replication (18). Expression of both proteins is essential for initiation of PCV1 replication (9). Rep represses transcription of the rep gene promoter, while Rep′ does not (8).
FIG. 1.
Map of PCV1 and PCV2. A linear map of the circular genomes of PCV1 and PCV2 is shown. The origin of replication is located between the divergently transcribed cap and rep gene (gray shaded boxes). The origin is enlarged for both viruses and shows the characteristic stem-loop element and the adjacent 6-bp and 5-bp repeats (open boxes and ovals). Sequence deviations between the origins of PCV1 and PCV2 are indicated by bold letters, the minimal binding sites of the Rep and Rep′ protein in PCV1 is marked by black arrows. A stippled line indicates the rep and rep′ transcript and the position of the splice junction in PCV1 and PCV2 (bold).
The origin of replication of PCV is located between the two divergently transcribed major open reading frame (ORF). The origin of PCV1 has been mapped to an 111-bp fragment, position 728 to 838 (11). A corresponding fragment for PCV2 has been identified (7), but its activity has not yet been experimentally demonstrated. These fragments contain a potential stem-loop structure with a nonamer [5′-(T/A)AGTATTAC], a sequence not only present in all other circoviruses from avian species (19), but also found in the related virus families Geminiviridae (16) and Nanoviridae (13). These plant-affecting viruses are characterized by a single-stranded DNA genome and a rolling-circle replication mechanism, thus showing similarity to the circoviruses. Next to the stem-loop of PCV1 and PCV2, hexamer (5′-CGGCAG) and pentamer (5′-CACCT) repeats are found (Fig. 1). These elements constitute the minimal binding site (MBS) for Rep/Rep′ of PCV1, which is composed from the right part of the stem-loop and the two inner hexamers H1/H2 (18). The origin of replication and the rep genes show a high degree of homology (>85%), while the cap genes are less conserved (62%).
Mapping of the rep transcripts of PCV2 corroborated previously published results that PCV2 also encodes two rep gene products (2). The importance of the rep gene products for the replication of PCV2 was assessed. Several assays can be used to determine viral replication rates. The classical approach detects the alteration of the methylation status of a GATC-methylated input DNA after replication by restriction with Dam-dependant enzymes, e.g., DpnI (17). Although the so-called DpnI assay was for many years state-of-the-art, the result is seen as a band on an autoradiograph and can therefore be evaluated only semiquantitatively. Nowadays, replication assays often employ quantitative PCR approaches (3, 5). A new quantitative replication assay based on the expression of two reporter genes was developed to investigate the replication of PCV2. The Rep and Rep′ proteins of PCV1 and PCV2 were investigated with respect to their ability to bind in vitro to the origin of replication and to initiate replication of the heterologous virus in cell culture. The results presented in this study indicate that the rep gene products of PCV1 and PCV2 can be functionally exchanged.
MATERIALS AND METHODS
Plasmid construction.
All plasmids were constructed by cloning of PCR-generated fragments. For pRL16, a 322-bp fragment (nt 647 to 969) comprising the origin of replication of PCV1 (nt 728 to 838; GenBank accession no. Y09921) was amplified with the primers F436n and B437 (5′-GCT CTA GAT GCC AAA TAT GGT CTT CTC CG, 5′-GAG GTA CCT TTC ACT GAC GCT GCC GAG G; restriction sites underlined). The PCR product was digested with XbaI and KpnI and cloned into the vector pGL3 promoter (Promega, Mannheim, Germany) (GenBank U47298). For pRL16.2, a 324-bp fragment (nt 1635 to 195), containing the putative origin of PCV2 (GenBank AF201307) was amplified with the primers F452 and B453 (5′-GAA GAT CTT GGC GGG GGT GGA CGA, 5′-GGG GTA CCC CTC GCC AAC AAT AAA ATA ATC). The fragment was BglII and KpnI restricted and cloned into plasmid pGL3 promoter. In both plasmids, the fragment was inserted counterclockwise, to avoid transcription from Prep of PCV1 and PCV2 directed toward the luc gene. The resultant plasmids pRL16 and pRL16.2 cannot replicate unless the rep gene of PCV1 or 2 is expressed in trans by plasmid pORF4 (9) and pSVL-rep(PCV2). The latter contains the rep gene (nt 822 to 1766) of PCV2, encoding the Rep and Rep′ protein. A 968-bp DNA fragment was amplified from plasmid pREP-PCV2 (rep gene of PCV2 cloned into vector pcDNA3.1, kindly provided by D. Mahé, AFSSA, Ploufragan, France) with the primers F500 and B466 (5′-GCT CTA GAA GCA GCA ACA TGC CCA G; 5′-CGG GAT CCA AGT GAT AAA AAA GAC TCA GTA ATT). The PCR product was digested with XbaI and BamHI and cloned into expression vector pSVL (GenBank U13868). pSVL-rep′(PCV2) encodes the cDNA product of the spliced rep′ transcript. It was amplified using the same primers with cDNA synthesized from PCV2-infected PK-15 cells and subsequent cloning of the product into vector pSVL. PCR conditions were 94°C for 2 min; 30 cycles of 94°C for 15 s, 57°C for 30 s, and 72°C for 45 s; and 72°C for 7 min. For disabling synthesis of the Rep′ protein, the plasmid pSVL-rep*(PCV2) was constructed. For this purpose, plasmid pSVL-rep(PCV2) was restricted with SacI and SacII. The excised 124-bp fragment was rebuilt from 10 oligonucleotides and recloned into pSVL-rep(PCV2). The oligonucleotides were designed to replace all GT dinucleotides, which may serve as putative splice donor sites. The following modifications were introduced: T1172C, G1178A, G1188A, T1193C, G1199A, T1214C, T1218C, G1224A, and G1247A. This manipulation led to three conservative alterations of the amino acid sequence of the Rep* protein with respect to wt-Rep: V123I, V133I and V135I. RT-PCR of PK-15 cells transfected with plasmid pSVL-rep*(PCV2) revealed that in contrast to pSVL-rep(PCV2)-transfected cells, the spliced rep′ transcript was no longer detectable (data not shown).
For in vitro expression of Rep and Rep′ protein of PCV2, PCR fragments were generated from pSVL-rep(PCV2) and pSVL-rep′(PCV2) with primers F603 (5′-CGG AAT TCA TGG ACT ACA AGG ACG ACG ACG ACA AGC CCA GCA AAA AGA ATG GAA [FLAG tag shown in italics]) and B466 (sequence given above). This step fused the FLAG epitope to position 822 of the rep gene, i.e., to the amino acid adjacent to the Rep start codon. PCR fragments were restricted with EcoRI and BamHI and cloned into vector pGEM3Zf(+) (Promega) (GenBank X65306). All recombinant plasmids were sequenced to rule out PCR-induced errors.
RNA extraction, cDNA synthesis and RT-PCR.
Cells were washed with phosphate-buffered saline (PBS) and RNA was harvested 68 h postinfection using the RNeasy Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions for total RNA minipreps from animal tissues. RNA isolation included DNase I treatment with the Qiagen RNase-free DNase set protocol for 30 min at room temperature. To finally remove a small residual amount of contaminating viral single-stranded DNA, a second DNase I treatment with 50 U of DNase I (Pharmacia, Freiburg, Germany) for 45 min at room temperature and subsequent phenol extraction were performed. cDNA synthesis was done according to the SuperscriptII protocol (Life Technologies, Paisley, United Kingdom) using 1 μg total RNA and 500 ng of oligo(dT)12-18 primer. PCR was performed in a volume of 20 μl with 1/10 of a cDNA preparation. The following conditions were used (final concentrations are given): 200 nM (each) primer, 1× PCR buffer without MgCl2, 2 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate, 5% dimethyl sulfoxide, 0.75 U of TaqGold (Perkin-Elmer, Zaventem, Belgium). cDNA was amplified subsequently with primers F410 and B411 (5′-CTCAGGGACAACGGAGTGAC; 5′-CTGGGACAGCAGCCGAGGAG) designed to detect both rep transcripts, or F412 and B413 (5′-GGGAGTCTGGTGACCGTTGC; 5′-TCCGTGGACTGTTCTGTAGC), amplifying only the unspliced variant. Cycling conditions were 95°C for 12 min; 38 cycles of 95°C for 20 s, 56°C (for primers 410 and 411) or 53°C (for primers 412 and 143) for 20 s, and 72°C for 45 s; and 72°C for 10 min.
Cells and immunofluorescence assay.
Antisera detecting Rep and Rep′ of PCV2 are not yet available. Therefore, expression of Rep and Rep′ of PCV2 was investigated with antibodies specifically recognizing Rep and Rep′ of PCV1. These antibodies were raised by immunizing rabbits with the fusion proteins GST:Rep′(120-168)PCV1 and GST:Rep′(120-168)PCV1. The resultant antibodies are directed against the C-terminal moieties of Rep (aa 121 to 312) and Rep′ (aa 121 to 168), which are distinct in sequence; therefore, they react specifically (T. Finsterbusch, unpublished data).
Porcine kidney cell line PK-15 (ATCC CCL33) was grown at 37°C, 5% CO2 in Dulbecco's modified Eagle medium with 5% fetal calf serum and passaged weekly. PK-15 cells were infected in 24-well plates with 15 μl of PCV-containing supernatant with a TCID50 of 108. For the indirect immunofluorescence assay, PCV-infected cells on coverslips were fixed and permeabilized by immersing the coverslips in −20°C methanol-acetone (1:1) for 10 min and subsequent air drying. Cells were blocked for 1 h with 1% bovine serum albumin and 0.05% Tween 20 in PBS and stained for 2 h at room temperature with the primary α-GST:Rep(120-312)PCV1 serum, 1:300 or with α-GST:Rep′(120-168)PCV1 serum, 1:300 diluted in blocking buffer. The cells were washed three times with PBS and incubated with fluorescein isothiocyanate-conjugated goat α-rabbit immunoglobulin G (Dianova, Hamburg, Germany). Subsequently, the coverslips were washed, mounted, and analyzed by confocal laser scanning microscopy.
EMSA studies.
Rep and Rep′ were expressed in vitro as FLAG-fusion proteins from plasmids pGEM-rep(PCV1), pGEM-rep′(PCV1), pGEM-rep(PCV2) and pGEM-rep′(PCV2) using the TNT wheat germ extract system (Promega) as published previously (18). Electrophoretic mobility shift assays (EMSA) were performed according to previously published experimental conditions (18) using the hybridized oligonucleotides F229 and B265 (5′-AAGTGCGCTGCTGTAGTATTACCAGCGCACTTCGGCAGCGGCAGCACCTCGGCAGCGTCAG; 5-Cy5CTGACGCTGCCGAGGTGCTGCCGCTGCCGAAGTGCGCTGGTAATACTACAGCAGCGCACTT) comprising the origin of PCV1. Oligonucleotides F462 and B463 (5′-GAAGTGCGCTGTAAGTATTACCAGCGCACTTCGGCAGCGGCAGCACCTCGGCAGCAGCACCT-3′; Cy5-AGGTGCTGCCGAGGTGCTGCCGCTGCCGAAGTGCGCTGGTAATACTTACAGCGCACTTC) were used to test binding to the origin of PCV2.
Replication assay.
A new assay was developed in which the replication activity of an origin of replication and its cognate replicase can be quantified. For this purpose, the origin of interest was cloned into plasmid pGL3 promoter (Promega), in which the luc gene is constitutively expressed by the simian virus 40 (SV40) late promoter. Replication of the luc plasmid will not occur without the compatible replicase. Therefore, a basal luc expression is detected. Endogenous luc activity of the PK-15 cells and the transcriptionally silent plasmid was tested with plasmid pGL3-basic, in which the SV40 promoter is deleted. After cotransfection of the second plasmid expressing the appropriate replication initiator enzyme, the copy number of the luc plasmid should rise and correspondingly, Luc activity will increase. For standardization of transfection efficiency, plasmid pRSV-βGal expressing the lacZ gene was cotransfected. Extracts were measured for Luc and Gal activity using the Dual light kit following the manufacturer's instructions (Applied Biosystems, Weiterstadt, Germany). Luciferase activity indicated replication activity of the investigated replicon, while β-galactosidase activity was used to normalize for differences in transfection efficiency. Standardized Luc units were calculated by dividing the Luc units by the Gal units. The ratio of Luc/Gal indicated the activity of the combined origin/replicase in correlation to the Luc/Gal ratio of the nonreplicated origin. Transfections were performed in duplicate, each assay was performed three times and the mean of the data points and the standard error of the means were calculated.
This method was used to investigate the replication competence of the Rep and Rep′ proteins of PCV1 and PCV2 and their respective origins of replication. PK-15 cells were transfected with Effectene (Qiagen) using 50 ng of pRSV-βGal (6) and 100 ng of pRL16 or pRL16.2 plasmids carrying the origin of replication of PCV1 or PCV2, plus 100 ng of plasmid pORF4 or pSVL-rep(PCV2) and their derivatives, expressing the rep gene products of PCV1 and PCV2. Medium was changed after 24 h, cell extracts were prepared after 2 days and Luc/Gal activity was determined. Reporter gene expression was measured for 10 s in a Microlumat Plus LB96V (EG&G Berthold, Bad Wildbad, Germany) after addition of 1 μl of Galacton-plus substrate (diluted 1:100 in buffer B).
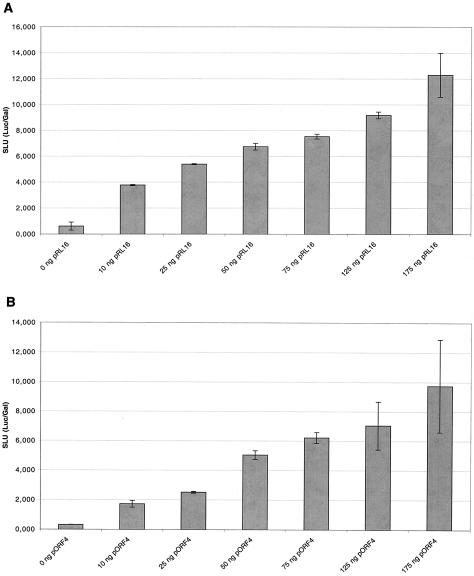
To verify that this test can be used as a quantitative assay, the correlation between the copy number of the ori/Luc or of the Rep plasmid and the Luc activity was investigated. First, a variation of the copy number of plasmid pRL16 was performed, while amounts of the Rep- and the Gal-expressing plasmids were constant. pRL16 was used in increasing amounts (0, 10, 25, 50, 75, 125, and 175 ng), plasmid pGL3-p was added to a the total amount of 175 ng. This mixture was cotransfected with 50 ng of pORF4 supplying the rep gene products of PCV1 and 25 ng of plasmid pRSV-βGal for internal standardization. Investigation of the correlation between copy number of plasmid pRL16 and Luc activity revealed a linear increase (Fig. 2A). In a reciprocal experiment the copy number of replicase plasmid pORF4 was varied versus a fixed amount of the origin plasmid pRL16. Increasing amounts of pORF4 (0, 10, 25, 50, 75, 125, and 175 ng) were supplemented with DNA of the vector pSVL to a total amount of 175 ng and cotransfected with 50 ng of plasmid pRL16 and 25 ng of plasmid pRSV-βGal. Again, a linear curve progression indicated a quantitative correlation of the copy number of the Rep plasmid and the Luc activity (Fig. 2B). Taken together, these results indicate that the copy number of the ori/Luc and the Rep plasmid is linked by a linear correlation with the Luc/Gal activity. Therefore, this test can be used to quantify replication activity in cell culture.
FIG. 2.
Correlation between copy number of the origin and replicase plasmids with the Luc/Gal activity. To verify whether the Luc/Gal assay can be used to quantify replication rate of a particular plasmid, a variation of the copy number of the origin plasmid pRL16 (A) and the replicase-expressing plasmid pORF4 (B) was performed. (A) Increasing amounts of DNA of the plasmid pRL16 were supplemented to 175 ng with vector DNA pGL3 promoter and cotransfected with 50 ng of pORF4A and 25 ng of pRSV-βGal into PK-15 cells. A Luc/Gal assay was performed. (B) A Luc/Gal assay was performed using increasing amounts of DNA of the plasmid pORF4A were supplemented to 175 ng with vector DNA pSVL, cotransfected with 50 ng of pRL16 plus 25 ng of pRSV-βGal into PK-15 cells. SLU, standardized Luc units; error bars, standard error of the mean.
RESULTS
Mapping of two rep transcripts in PCV2.
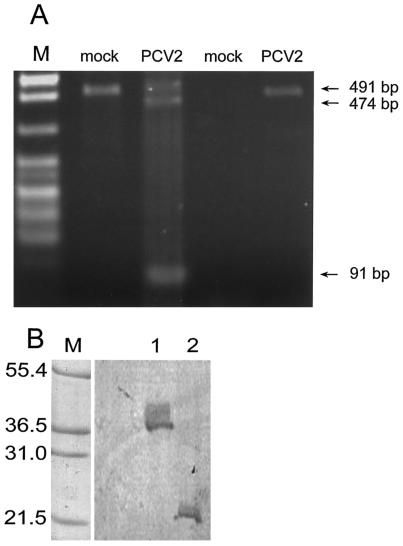
The rep genes of PCV1 and PCV2 including the potential splice and donor sites are highly conserved. We therefore assumed that rep of PCV2 might direct not only the synthesis of a full-length product but also of a differentially spliced version, corresponding to the rep′ transcript of PCV1. After isolation of RNA from PCV2 infected PK-15 cells and subsequent synthesis of cDNA, two PCRs were done: the first was performed with primers F410/B411 located up- and downstream of the putative splice acceptor and donor sites, the second used primers F412/B413 to detect the unspliced transcript. In the first case, two specific products were seen, a smaller fragment of approximately 100 bp and a longer product, approximately 480 bp in size (Fig. 3A). Using F412/B413, only one specific fragment, approximately 490 bp, was detected. PCR products were cloned and sequenced. The longer fragments showed a 474-bp sequence at position 1156 to 1629 and a 491-bp fragment at position 1209 to 1699 of the rep reading frame, while the shorter PCR fragment was the result of a splicing event in which position 1187 was joined to position 1571. In agreement with earlier results obtained for PCV1 (9) and with a RACE analysis of another isolate of PCV2 (2), this finding indicates that more than one rep transcript is produced by differential splicing of the rep mRNA of PCV2. The complete products rep and rep′ were amplified with primers F500 and B466 from the same cDNA pool and cloned into pCR2.1. Sequencing verified that two distinct products corresponding either to the unspliced or to the spliced version were obtained.
FIG. 3.
Analysis of rep gene products in PCV2-infected cells. (A) The result of an RT-PCR is shown. RNA was isolated from mock- or PCV2-infected cells and copied into cDNA. The cDNA was amplified with the primer pair F410/B411 hybridizing up- and downstream of the putative splice site of the rep transcript (lanes 2 and 3). The primer pair F412/B413 binds downstream of the splice donor and only the full-length rep transcript can be amplified (lanes 4 and 5). A sketch of the two differentially spliced rep transcripts is given in Fig. 1. (B) Expression of Rep and Rep′ in vitro. The ORF for Rep (lane 1) and Rep′ (lane 2) were cloned into plasmid pGEM3Zf(+) and expressed with the TNT wheat germ extract system using [35S]methionine. Mark 12 wide-range protein standard (showing molecular mass in kilodaltons [lane M]) was stained with amido black.
Expression of Rep and Rep′ protein in PCV-infected cells.
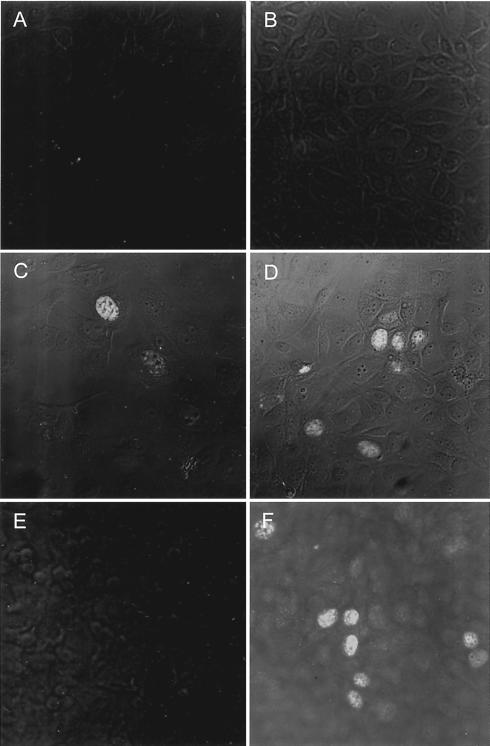
The cDNAs for Rep and Rep′ were subcloned into vector pGEM3Zf(+), thereby a FLAG tag was added to the 5′ end of the two ORF. In vitro expression of [35S]methionine-labeled Rep and Rep′ proteins of PCV2 and subsequent PAGE and autoradiography revealed an apparent molecular weight of the two proteins of approximately 36 and 20 kDa, respectively (Fig. 3B). The expression of the two proteins was investigated in an indirect immune fluorescence assay (Fig. 4). Since antisera against the PCV2 Rep proteins are not yet available, the expression of the two rep gene products of PCV2 was investigated using antisera raised against the two Rep isoforms of PCV1. These antisera have been generated by immunization with the C termini of Rep and Rep′ of PCV1, which differ in their amino acid sequence (T. Finsterbusch, manuscript in preparation). Expression of Rep was detected with the α-Rep(120-312)PCV1 antiserum in PCV1-infected PK-15 cells (Fig. 4D), as well as in PCV2-infected cells (Fig. 4F). In contrast, the α-Rep′(120-168)PCV1 antiserum detected expression of Rep′ protein in PCV1-infected PK-15 cells, but it did not react with the Rep′ protein of PCV2 (Fig. 4C and E).
FIG. 4.
Expression of Rep and Rep′ proteins in PCV-infected cells. PCV-infected PK-15 cells were probed with antisera raised against the C-terminal regions of Rep and Rep′ of PCV1. (A) Mock-infected PK-15 cells/α-Rep′(120-168); (B) mock/α-Rep(120-312)PCV1; (C) PCV1-infected PK-15 cells/α-Rep′(120-168); (D) PCV1/α-Rep(120-312)PCV1; (E) PCV2-infected PK-15 cells/α-Rep′(120-168); (F) PCV1/α-Rep(120-312)PCV1.
Replication of PCV2 is promoted by Rep plus Rep′.
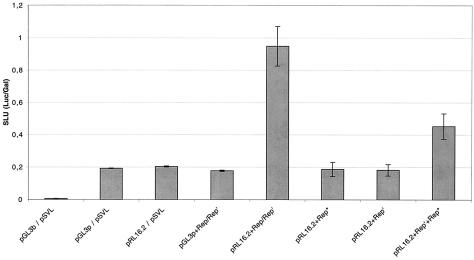
PCV1 and PCV2 express two distinct rep transcripts, which products in the case of PCV1 are essential for replication of viral DNA. To investigate whether expression of Rep and Rep′ is also a prerequisite for replication of PCV2, a replication study was performed using the Luc/Gal replication assay. Plasmids pGL3-basic and pSVL were transfected into PK-15 cells serving as a negative control, demonstrating that no endogenous luc activity was present in transfected PK-15 cells (Fig. 5, column 1). The combination of plasmid pGL3 promoter with vector pSVL served as another control indicating the level of basal luc expression from the SV40 promoter. To exclude influence of the cloned regions upon the Luc expression, plasmids pRL16.2 was cotransfected with plasmid pSVL and plasmid pGL3-p with pSVL-rep(PCV2) (columns 2 to 4). Luc expression was comparable in all experiments. Plasmid pRL16.2 carrying the putative origin of replication of PCV2 was cotransfected with plasmid pSVL-rep(PCV2), expressing Rep and Rep′ protein (column 5). The Luc/Gal ratio increased fivefold, demonstrating that rep gene products induced replication of plasmid pRL16.2. When either plasmid pSVL-rep′(PCV2) expressing Rep′ or plasmid pSVL-rep*(PCV2) expressing only the full-length product Rep were used instead of pSVL-rep(PCV2), no increase in Luc/Gal activity was seen (columns 6 and 7). This result indicates that replication of plasmid pRL16 did not occur, if only one of the two Rep isoforms was supplied. When the two plasmids pSVL-rep′(PCV2) and pSVL-rep*(PCV2) were cotransfected together with pRL16.2, a twofold increase in Luc/Gal activity was observed (column 8). This result suggests that expression of Rep plus Rep′ is an essential prerequisite for initiation of PCV2 replication.
FIG. 5.
Replication of PCV2. A Luc/Gal replication assay was performed in which the origin of PCV2 (plasmid pRL16.2) was combined with plasmids expressing the Rep and or the Rep′ protein. Plasmid pGL3b in column 1 contains a promoter-less luc gene and represents endogenous Luc activity. Plasmid pGL3p was cotransfected with pSVL to serve as an indicator for basal Luc expression in a nonreplicated system (column 2). Influence of the origin fragment exerted upon Luc activity was tested by combination of pRL16.2 with pSVL (column 3). To test reciprocally the influence of Rep and Rep′ on expression of Luc, plasmids pGL3p was cotransfected with pSVL-rep(PCV2) (column 4). The origin of replication of PCV2 is combined with the rep gene of PCV2 expressing Rep and Rep′ in column 5, pRL16.2+pSVL-rep(PCV2). The rep* gene (column 6) is encoded in plasmid pSVL-rep*(PCV2) and contains an engineered version of rep expressing only the Rep protein. pSVL-rep′(PCV2) leads to the production of Rep′ (column 7). For reconstitution of the replication activity (column 8), the proteins are supplied separately from plasmids pSVL-rep*(PCV2) and pSVL-rep′(PCV2). SLU, standardized Luc units; error bars, standard error of the mean.
Binding of Rep/Rep′ protein of PCV1 and PCV2 to the heterologous origin of replication in vitro.
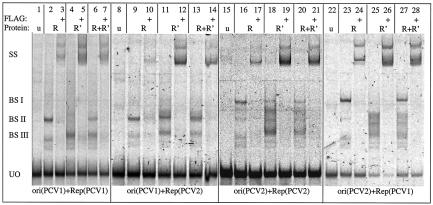
The Rep and Rep′ proteins of PCV1 recognize an MBS, whose nucleotide sequence also appears in PCV2 (18). To investigate, whether the rep gene products of PCV2 are able to bind in vitro to the PCV2 origin of replication and whether the rep gene products of one virus can bind to the origin of the other virus, gel shift assays were performed. Rep and Rep′ proteins of one PCV type were expressed in vitro with a FLAG tag and incubated either with fragments of the compatible origin of replication or with the heterologous fragment (Fig. 6). As demonstrated previously, the Rep and Rep′ proteins of PCV1 bound to a fragment, containing the PCV1 origin (lanes 2, 4, and 6). The interaction of the two rep gene products of PCV1 with the origin of PCV2 was investigated next (lanes 9, 11, and 13). After binding of Rep, Rep′ and a mixture of both PCV2 proteins to the origin of PCV1, a retarded complex is visible, indicating that the origin of PCV1 is recognized by the rep gene products of PCV1 and PCV2. An interaction was also observed when the origin of PCV2 was incubated with PCV2 rep gene products Rep and Rep′, indicating that Rep and Rep′ of PCV2 can recognize the origin of PCV2 in vitro (lanes 16, 18, and 20). Finally, the interaction of the same fragment with the PCV1 Rep and Rep′ protein was tested. As in case with the PCV2 proteins, a complex was observed after binding of Rep and the Rep′ or a mixture of Rep and Rep′ (lanes 23, 25, and 27). In summary, these experiments show that the rep gene products of PCV1 and PCV2 are capable of binding to the homologous and heterologous origin. Compared to the complexes obtained with the PCV1 origin (left part of the panel), the PCV2 origin is shifted to a higher extent, i.e., the ori PCV2/Rep PCV2 and ori2/Rep PCV1 complexes seem to be larger (right half). This is in accordance with earlier results (18), in which we showed that a longer fragment of the PCV1 origin containing all four hexamer repeats migrated faster than a shorter one in which H4 was deleted. This phenomenon was ascribed to a special interaction between the 5′ and 3′ ends of the same template investigated here again. The nature of this interaction is not yet known. This result indicates that the size of the complex seems to be influenced by the nature of the supplied oligonucleotide and not by the protein. In most lanes, more than one retarded band was observed, indicating that different protein amounts may be loaded upon the fragments. The observation that Rep and Rep′ of PCV1 can oligomerize strengthens this hypothesis (T. Finsterbusch, unpublished data). The observed band shift effect of Rep plus Rep′ in PCV1 and in PCV2 is additive, but not cooperative. The specificity of each binding reaction has been verified by induction of a supershift after addition of the α-FLAG antibody to the FLAG-tagged protein.
FIG. 6.
Replication proteins of PCV1 and PCV2 bind to the heterologous origin in vitro. FLAG-tagged Rep and Rep′ proteins of PCV1 (lanes 1 to 7 and 22 to 28) and PCV2 (lanes 8 to 21) were expressed in vitro and incubated with the ds EMSA substrates F229/B265 comprising the origin of PCV1 (lanes 1 to 14) or with F462/B463 carrying the origin of PCV2 (lanes 15 to 28). The fragments carry the putative stem-loop element plus the adjacently located hexamer and pentamer repeats. In lanes 1, 8, 15, and 22, an unprogrammed extract has been used as a negative control (u). In lanes marked with a +, the specificity of the binding reaction has been examined by induction of a supershift with the α-FLAG antibody. The letter R points out the use of the Rep protein, R′ indicates application of the Rep′ protein, while the mixture of Rep and Rep′ is given by R+R′. UO designates the unbound oligonucleotides; the band shifts are marked by BSI, BSII, and BSIII; and the supershift is indicated by SS.
Interchangeability of PCV1 and PCV2 replication components: initiation of replication occurs at the heterologous origin.
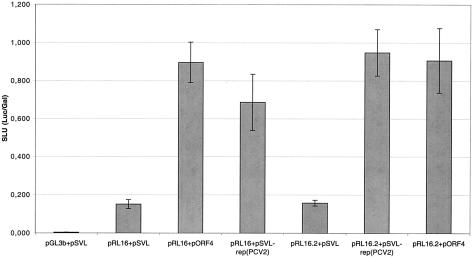
To assess whether binding of the proteins to the heterologous origin of replication is followed by initiation of DNA replication, a replication assay was performed by combining the rep gene of one type of PCV with the origin of the other virus type. Plasmids pGL3-b and pSVL served as a negative control (Fig. 7, column 1). Plasmids pRL16 and pRL16.2 were cotransfected with the plasmid pSVL to indicate the basal Luc activity expressed by the nonreplicated reporter plasmids (columns 2 and 5). Combination of these two plasmids with the cognate replicase, i.e., pRL16/pORF4 and pRL16.2/pSVL-rep(PCV2) served as a positive control (columns 3 and 6), in which the Luc/Gal activity increased fivefold. When the rep gene of PCV1 (expressed by plasmid pORF4) was combined with the plasmid pRL16.2, carrying the origin of PCV2, a comparable increase in Luc/Gal activity was observed (column 7), as well as in the inverse experiment, in which the origin of PCV1 in plasmid pRL16.1 was cotransfected with pSVL-rep(PCV2) expressing the PCV2 rep gene (column 4). The Luc/Gal assay indicates that replication at the origin of PCV2 could be initiated by the rep gene of PCV1 and vice versa. Thus, the cis- and trans-acting factors of PCV1 and PCV2 can be exchanged in vitro. Furthermore, replication activity in any combination was comparable in the cell culture system, since only a slight decrease in Luc/Gal activity was seen with the heterologous combinations.
FIG. 7.
Replication factors of PCV1 and PCV2 can be exchanged. A Luc/Gal replication assay was performed in which the origin of PCV1 was replicated by its cognate replicase proteins or by the heterologous rep gene products of PCV2 and vice versa. Plasmid pGL3b in column 1 contains a promoterless luc gene and serves as a negative control. pRL16 carries the origin of PCV1 and is combined with the vector pSVL (column 2). This experiment indicates the Luc activity of plasmid pRL16 without replication. The replication activity of ori(PCV1) is measured with cotransfected plasmids pORF4 expressing the rep gene of PCV1 (column 3) or pSVL-rep(PCV2) (column 4). The reciprocal approach with pRL16.2, carrying the origin of PCV2, is shown in the next three experiments. Column 5 indicates the Luc activity of pRL16.2 without supplementation of a rep gene. Combination of the origin of PCV2 with its rep gene is shown in column 6, while the cotransfection of ori(PCV2) with rep(PCV1) is given in column 7. SLU, standardized Luc units; error bars, standard error of the mean.
DISCUSSION
Transcription mapping of PCV2 led to the identification of two rep gene transcripts homologous to those in PCV1. The Rep protein of PCV2 has a size of 314 aa (37.5 kDa) and corresponds to the complete rep ORF. The putative Rep′ protein is produced by differential splicing in which position 1187 is joined to position 1571, resulting in a size reduction to 178 aa (20.2 kDa). The two gene products have been expressed in vitro and the apparent size of the proteins matched the predicted molecular weights. Splicing results in the induction of a reading frame shift from phase 3 to 2 resulting in sequence deviation in the C-terminal 57 aa of Rep and Rep′. These results are in agreement with a detailed analysis of PCV2 transcription published recently (2) and furthermore it indicates a clear parallel to rep gene expression in PCV1 (18). Expression of the Rep protein has been detected in PCV2-infected cells using antibodies raised specifically against the C terminus of PCV1 Rep, which differs from that of Rep′. A nuclear localization was seen for Rep, corroborating the results observed with PCV1-infected cells. However, the PCV2 isoform Rep′ did not react with α-Rep′(PCV1), indicating either that Rep′ is not expressed in PCV2-infected cells or that it is not recognized by the antiserum. We suppose that the Rep′ protein is expressed but not recognized by the antiserum due to the lower degree of sequence homology. The N-terminal region, which is identical in Rep and Rep′ proteins of one virus, shows a homology of 86% between PCV1(1-120) and PCV2(1-123). A similar value (84%) is obtained, when the C termini of Rep(PCV1)(121-312) and Rep(PCV2)(123-314) are compared. In contrast, only 64% homology is seen for the amino acid sequence of the C-terminal regions downstream of the splice junction of Rep′(PCV1)(121-158) and Rep′(PCV2)(123-168). Taking into account that the C terminus of Rep′ is very short, the failure of recognition of Rep′(PCV2) by α-Rep′(PCV1) may be attributed to this phenomenon. Therefore, final demonstration of Rep′ expression in PCV2-infected cells will require the production of a Rep′-specific antiserum which is at present not available.
Recently three additional, smaller rep transcripts have been mapped in PCV1 and PCV2 (2). Interestingly, these mRNAs were not detected in our cDNA PCR approach, probably due to the lower transcript number. The putative proteins have not yet been detected and their biological function remains unclear. Since they comprise neither the DNA-binding domain nor the P-loop, activity in replication is rather not to be expected. Nevertheless, this function of these proteins has to be evaluated in future studies.
A new replication assay was developed in which replication of a plasmid carrying a constitutively expressed luc gene plus the origin of replication of interest is driven by expression of the compatible replicase from a cotransfected plasmid. Since the correlation between copy number and Luc activity is linear, this fast, simple and reliable test for viral replication can be used to quantify replication of a replicon of interest in cell culture. A further advantage of the Luc/Gal assay is the fact that replication of a given virus can be tested after transfection of normally nonpermissive cells that cannot be naturally infected. Hopefully, this assay will be useful for investigation and quantification of other viral origins of replication and of their cognate replicases.
Using the Luc/Gal replication assay, we could confirm that the origin of PCV2 is located on a 324-bp fragment between nt 1635 to 195. This fragment is collinear to a smaller one comprising the origin of replication of PCV1. If the high degree of homology between these origin fragments of PCV1 and PCV2 is considered, this result was to be expected, and further experiments including the fine-mapping and mutagenesis of the origin of PCV1 and PCV2 are currently being performed. The replication assay indicated furthermore that neither Rep nor Rep′ of PCV2 alone could promote replication of the PCV2 origin. Corroborating earlier results investigating the replication of PCV1 using the DpnI assay (11), coexpression of Rep and Rep′ protein of PCV2 is necessary to initiate replication at the origin of PCV2. Compared to pSVL-rep(PCV2) induced replication, the activity observed with pSVL-rep′(PCV2) plus pSVL-rep*(PCV2) was lower. A similar observation was made when the replication of PCV1 was analyzed with the semiquantitative DpnI assay (9, 10). The decrease in replication activity with separate supply of Rep and Rep′ may have various reasons: Three aa have been exchanged in the engineered rep version Rep*, in which synthesis of Rep′ is prevented. Although this alteration is conservative and the secondary prediction of the Rep* protein is not changed with respect to Rep(PCV2), a decreased efficiency of Rep* in initiation of replication cannot be ruled out. Second, this effect could occur because the two proteins may have to be synthesized in a highly coordinated fashion, i.e., that synthesis of Rep and Rep′ must be coupled in status nascendii, a condition that cannot be met by synthesis from two separate plasmids. Therefore, a final description of the role of Rep and Rep′ protein cannot be given at the present moment, this question should be studied with an in vitro replication system using purified proteins.
Since the origin of replication and the rep genes of PCV1 and PCV2 are highly conserved and the MBS recognized by Rep and Rep′ is identical in sequence, the question arose, whether replication factors of PCV1 and PCV2 can be exchanged. Experiments presented in this study provided evidence that the Rep and Rep′ protein of one PCV variant could bind in vitro to the origin of the other, and moreover, that replication was initiated at the heterologous origin. Similar analyses investigating replication of a mild and a severe strain of the geminivirus Tomato leaf curl virus revealed a mutation in the amino acid sequence of the Rep proteins of the mild strain which is coupled with base pair substitution in the origin nucleotide sequence (1). This mutation results in impaired replication and lower accumulation of viral genomes of the mild strain. In contrast, our observations show that the MBS of the two viruses are identical and that the Rep proteins of both viruses, PCV1 and PCV2, can bind to the appropriate origin and to the heterologous counterpart. Together with the observation that at least in vitro the efficiency of replication of both viruses was comparable, this may be taken as a first indication that the replication strategy of PCV1 and PCV2 may not be the main factor determining the distinct pathogenicity of PCV1 and PCV2. Therefore, the virus replication rate in animals infected with PCV1 or with PCV2 should be thoroughly studied, using for example a TaqMan approach (15).
Until now a model describing the initiation of PCV replication is lacking, but at least some pieces of information can be put together. In both viruses, two Rep isoforms recognize and bind to the origin. In PCV1, the minimal binding site is H1/H2 plus the right part of the stem-loop (18). Investigation of the DNA-binding capabilities of PCV2 Rep and Rep′ and fine-mapping of the MBS revealed similar results and will be published separately (B. Mueller, unpublished data). Details of the molecular action of the two proteins and the role of the conserved nonamer are still to be worked out. When the amino acid sequence of the Rep proteins and the smaller variant Rep′ is analyzed, it becomes apparent that the N terminus of both proteins contains a DNA-binding domain which carries the protein signatures of rolling-circle mediating replicases. The C termini of Rep contains a P-loop indicating that Rep may serve as a helicase, while Rep′, in which this motif is missing, may function as a nickase.
Acknowledgments
We thank Petra Kurzendoerfer for technical assistance.
This work was supported by the European Union (Project number QLK2-CT-1999-00307) and the Deutsche Forschungsgemeinschaft (MA 2126/2-1). C.S. was supported by a grant from the Sonnenfeldstiftung, Berlin.
REFERENCES
- 1.Chatterji, A., M. Padidam, R. N. Beachy, and C. M. Fauquet. 1999. Identification of replication specificity determinants in two strains of tomato leaf curl virus from New Delhi. J. Virol. 73:5481-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung, A. K. 2003. Transcriptional analysis of porcine circovirus type 2. Virology 305:168-180. [DOI] [PubMed] [Google Scholar]
- 3.Gudima, S., J. Chang, G. Moraleda, A. Azvolinsky, and J. Taylor. 2002. Parameters of human hepatitis delta virus genome replication: the quantity, quality, and intracellular distribution of viral proteins and RNA. J. Virol. 76:3709-3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koonin, E. V., and T. V. Ilyina. 1993. Computer-assisted dissection of rolling circle DNA replication. Biosystems 30:241-268. [DOI] [PubMed] [Google Scholar]
- 5.Leung, A. Y., M. Chan, S. C. Tang, R. Liang, and Y. L. Kwong. 2002. Real-time quantitative analysis of polyoma BK viremia and viruria in renal allograft recipients. J. Virol. Methods 103:51-56. [DOI] [PubMed] [Google Scholar]
- 6.MacGregor, G. R., A. E. Mogg, J. F. Burke, and C. T. Caskey. 1987. Histochemical staining of clonal mammalian cell lines expressing E. coli beta galactosidase indicates heterogeneous expression of the bacterial gene. Somat. Cell Mol. Genet. 13:253-265. [DOI] [PubMed] [Google Scholar]
- 7.Mankertz, A., M. Domingo, J. M. Folch, P. LeCann, A. Jestin, J. Segales, B. Chmielewicz, J. Plana-Duran, and D. Soike. 2000. Characterisation of PCV-2 isolates from Spain, Germany and France. Virus Res. 66:65-77. [DOI] [PubMed] [Google Scholar]
- 8.Mankertz, A., and B. Hillenbrand. 2002. Analyses of transcription of Porcine circovirus type 1. J. Gen. Virol. 83:2743-2751. [DOI] [PubMed] [Google Scholar]
- 9.Mankertz, A., and B. Hillenbrand. 2001. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology 279:429-438. [DOI] [PubMed] [Google Scholar]
- 10.Mankertz, A., B. Hillenbrand, T. Steinfeldt, K. Hattermann, C. Schmitt, R. Çaliskan, and T. Finsterbusch. Molecular biology of porcine circovirus type 1 and type 2: analyses of gene expression and viral replication. Vet. Microbiol., in press. [DOI] [PubMed]
- 11.Mankertz, A., F. Persson, J. Mankertz, G. Blaess, and H. J. Buhk. 1997. Mapping and characterization of the origin of DNA replication of porcine circovirus. J. Virol. 71:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nawagitgul, P., I. Morozov, S. R. Bolin, P. A. Harms, S. D. Sorden, and P. S. Paul. 2000. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81:2281-2287. [DOI] [PubMed] [Google Scholar]
- 13.Randles, J. W., P. W. G. Chu, J. L. Dale, R. Harding, J. Hu, L. Katul, M. Kojima, K. M. Makkouk, Y. Sano, J. E. Thomas, and H. J. Vetten. 2000. Nanoviridae, p. 299-303. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 14.Rosell, C., J. Segales, J. Plana-Duran, M. Balasch, G. M. Rodriguez-Arrioja, S. Kennedy, G. M. Allan, F. McNeilly, K. S. Latimer, and M. Domingo. 1999. Pathological, immunohistochemical, and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J. Comp. Pathol. 120:59-78. [DOI] [PubMed] [Google Scholar]
- 15.Rovira, A., M. Balasch, J. Segales, L. Garcia, J. Plana-Duran, C. Rosell, H. Ellerbrok, A. Mankertz, and M. Domingo. 2002. Experimental inoculation of conventional pigs with porcine reproductive and respiratory syndrome virus and porcine circovirus 2. J. Virol. 76:3232-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rybicki, E. P., R. W. Briddon, J. K. Brown, C. M. Fauquet, D. P. Maxwell, B. D. Harrison, P. G. Markham, D. M. Bisaro, D. Robinson, and J. Stanley. 2000. Geminiviridae, p. 299-303. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 17.Sears, A. E., and B. Roizman. 1990. Amplification by host cell factors of a sequence contained within the herpes simplex virus 1 genome. Proc. Natl. Acad. Sci. USA 87:9441-9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steinfeldt, T., T. Finsterbusch, and A. Mankertz. 2001. Rep and Rep′ protein of porcine circovirus type 1 bind to the origin of replication in vitro. Virology 291:152-160. [DOI] [PubMed] [Google Scholar]
- 19.Todd, D., M. McNulty, A. Mankertz, P. Lukert, J. Randles, and J. Dale. 2000. Circoviridae, p. 299-303. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.