Abstract
The γ134.5 protein of herpes simplex virus type 1 (HSV-1) functions to block the shutoff of protein synthesis involving double-stranded RNA-dependent protein kinase (PKR). In this process, the γ134.5 protein recruits cellular protein phosphatase 1 (PP1) to form a high-molecular-weight complex that dephosphorylates eIF-2α. Here we show that the γ134.5 protein is capable of mediating eIF-2α dephosphorylation without any other viral proteins. While deletion of amino acids 1 to 52 from the γ134.5 protein has no effect on eIF-2α dephosphorylation, further truncations up to amino acid 146 dramatically reduce the activity of the γ134.5 protein. An additional truncation up to amino acid 188 is deleterious, indicating that the carboxyl-terminal domain alone is not functional. Like wild-type HSV-1, the γ134.5 mutant with a truncation of amino acids 1 to 52 is resistant to interferon, and resistance to interferon is coupled to eIF-2α dephosphorylation. Intriguingly, this mutant exhibits a similar growth defect seen for the γ134.5 null mutant in infected cells. Restoration of the wild-type γ134.5 gene in the recombinant completely reverses the phenotype. These results indicate that eIF-2α dephosphorylation mediated by the γ134.5 protein is required for HSV response to interferon but is not sufficient for viral replication. Additional functions or activities of the γ134.5 protein contribute to efficient viral infection.
The γ134.5 gene of herpes simplex virus type 1 (HSV-1) strain F encodes a protein of 263 amino acids consisting of a large amino-terminal domain, a linker region of triplet repeats (AlaThrPro), and a carboxyl-terminal domain (13). The triplet repeats are a constant feature of the γ134.5 protein, but the number of repeats varies among different strains (13). Studies suggest that the number of triplet repeats in the γ134.5 protein may affect the ability of HSV to invade the central nervous system from the peripheral tissue (2, 29). The carboxyl-terminal domain is essential to prevent the shutoff of protein synthesis in virus infection (12, 19, 20), but the role of the amino-terminal domain is unknown. It is well established that the γ134.5 protein is essential for viral virulence. HSV mutants that fail to express the γ134.5 protein are incapable of multiplying and causing encephalitis in experimental animal models (10, 27, 35, 37).
Considerable evidence indicates that the γ134.5 protein functions, at least in part, to inhibit host interferon response mediated by the double-stranded RNA-dependent protein kinase (PKR) (7-9, 11, 12, 20, 21). Moreover, it has been demonstrated that the γ134.5 null mutant is virulent in PKR knockout mice but not in wild-type mice (10, 25). In contrast to the above observations, the γ134.5 null mutant, with an additional deletion in the US11 promoter region, inhibits PKR activity but nevertheless is avirulent in experimental mice (30). Similarly, the γ134.5 null mutant, with a secondary mutation outside the US11 promoter region, only partially restored virulence. Recent experiments showed that the γ134.5 protein blocks the surface expression of major histocompatibility complex class II molecules in HSV-1-infected cells, which is believed to impair the functions of CD4+ T cells (34). Interestingly, when expressed in mammalian cells, the γ134.5 protein is distributed both in the nucleus and cytoplasm (6, 28). In fact, the γ134.5 protein bears nuclear import and export signals that direct its shuttling between the cytoplasm, nucleus, and nucleolus (6). This dynamic process is likely to be required for the different activities associated with the γ134.5 protein during viral infection (6).
The most extensively characterized function of the γ134.5 protein is its ability to inhibit the antiviral action of PKR. In cells infected with HSV-1, viral DNA replication leads to the activation of PKR that phosphorylates the α subunit of translation initiation factor 2 (eIF-2α) and thereby inhibits translation initiation (9, 11). As a countermeasure, the γ134.5 protein is expressed by HSV to prevent the shutoff of protein synthesis (11). In doing so, the γ134.5 protein interacts with cellular protein phosphatase 1 (PP1) by its carboxyl-terminal domain, forming a high-molecular-weight complex that dephosphorylates eIF-2α (20, 21). Currently, it remains unknown whether additional viral or cellular proteins are present in this complex.
Previous studies suggest that the carboxyl terminus of the γ134.5 protein consists of a PP1 binding domain and an effector domain, which is functionally interchangeable with the corresponding domain of cellular protein GADD34/MyD116 (8, 19, 20). GADD34/MyD116 belongs to a family of proteins induced under conditions of genotoxic stress, growth arrest, differentiation, and apoptosis (18, 23, 26, 31, 32, 39). Like the γ134.5 protein, GADD34/MyD116 associates with proliferating cell nuclear antigen (PCNA), a cellular protein required for DNA replication and cell cycle control (5). The PP1 binding domain of the γ134.5 protein contains 12 amino acids with a signature sequence motif of (Arg/Lys)(Val/Ile)XaaPhe found in many PP1 binding proteins (14, 15, 20). These PP1 binding proteins perform diverse functions, such as cell division, mRNA splicing, glycogen metabolism, and neurotransmission (1, 14, 15, 22, 33, 36, 38). The effector domain of the γ134.5 protein consists of 59 amino acids, but its precise role is not yet understood. It appears the concerted action of these two subdomains is critical for eIF-2α dephosphorylation (8, 20). However, the minimal unit required for eIF-2α dephosphorylation has not yet been defined. The objective of this study was to further analyze the activities of the γ134.5 protein.
Defining the minimal functional module of the γ134.5 protein that dephosphorylates eIF-2α.
In HSV-1-infected cells, the γ134.5 protein recruits PP1 to form a high-molecular-weight complex that dephosphorylates eIF-2α (20). However, it is not known whether the activity of this complex requires additional viral proteins. To address this issue, monolayers of HeLa cells (Tet-off) were transfected with a plasmid encoding the wild-type γ134.5 protein driven by a tetracycline-inducible promoter. As a control, a plasmid encoding the mutant γ134.5 protein with Val193Glu and Phe195Leu substitutions was also included in the experiment. Previous studies have shown that this mutant is unable to mediate eIF-2α dephosphorylation in virus-infected cells (8, 20).
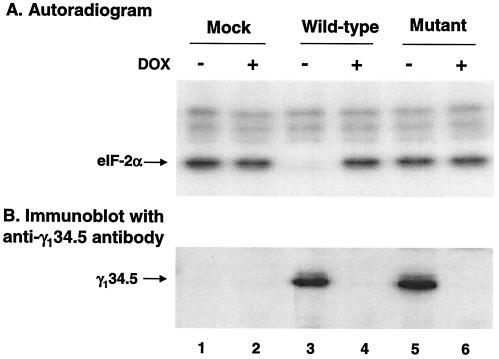
Cells were grown with or without doxycycline (1 μg/ml) for 36 h. Lysates prepared from the transfected cells were reacted with 32P-labeled eIF-2 and subjected to electrophoresis for autoradiography. As shown in Fig. 1A, in cells transfected with the wild-type γ134.5 gene, eIF-2α remained phosphorylated in the presence of doxycycline, but it became dephosphorylated in the absence of doxycycline. Dephosphorylation of eIF-2α correlated with the expression of the wild-type γ134.5 protein (Fig. 1B, lanes 3 and 4). In contrast, in cells transfected with the mutant γ134.5 gene, eIF-2α remained phosphorylated regardless of the expression of the γ134.5 protein (Fig. 1A, lanes 5 and 6). Similarly, eIF-2α remained phosphorylated in mock-transfected cells (Fig. 1A, lanes 1 and 2). Western blot analysis with anti-γ134.5 serum showed that both the wild-type and mutant γ134.5 proteins were expressed at similar levels in the absence of doxycycline (Fig. 1B). We conclude from these experiments that when expressed alone in HeLa cells, wild-type but not the mutant γ134.5 protein mediates dephosphorylation of eIF-2α. This finding extends previous observations that eIF-2α phosphatase is activated in cells infected with wild-type virus but not with the γ134.5 null mutants (20, 21). These data suggest that the γ134.5 protein is a viral component required for the γ134.5-PP1 complex, which functions in the absence of other HSV proteins.
FIG. 1.
(A) eIF-2α phosphatase activity in cells transfected with the γ134.5 gene. HeLa cells (Tet-off) grown in Dulbecco modified Eagle medium with (+) or without (−) doxycycline (DOX) (1 μg/ml) were transfected with either pGF9912 expressing the wild-type γ134.5 protein or pGF9913 in which Val193Glu and Phe195Leu substitutions were made in the γ134.5 protein. Thirty-six hours after transfection, cells were harvested, and lysates were prepared. Aliquots of lysates were incubated with 32P-labeled eIF-2, subjected to electrophoresis, and processed for autoradiography (8). (B) Expression of the γ134.5 protein. Cell extracts prepared from transfected cells were subjected to electrophoresis on a sodium dodecyl sulfate-12% polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with anti-γ134.5 antibody (6).
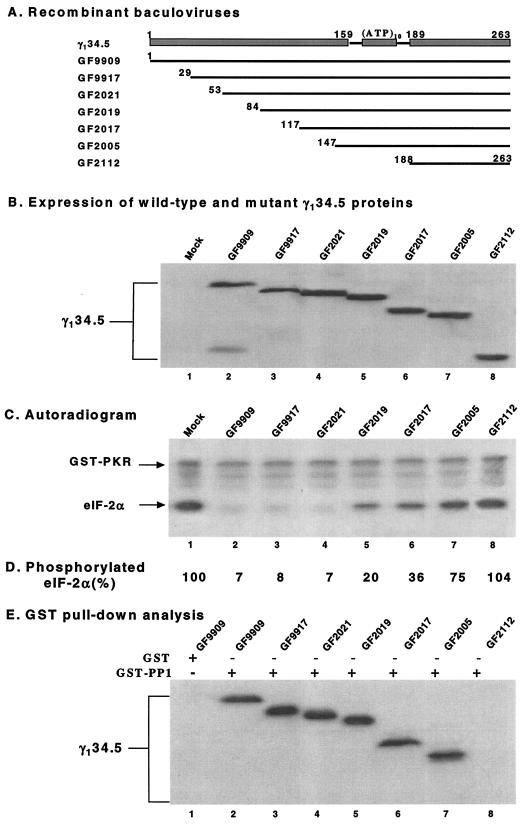
Prior studies have demonstrated that the carboxyl terminus of the γ134.5 protein is required for eIF-2α dephosphorylation (8). To address whether the amino-terminal or central domain is involved in this process, a series of truncations were introduced into the γ134.5 protein. We took advantage of the baculovirus system in which the γ134.5 protein mediates efficient eIF-2α dephosphorylation (10). As shown in Fig. 2A, recombinant baculoviruses were generated to express mutant forms of the γ134.5 protein with a series of nested deletions from amino acids 1 to 188. Western blot analysis demonstrated that these mutants expressed the truncated forms of the γ134.5 protein at levels comparable to that of the wild-type γ134.5 protein (Fig. 2B).
FIG. 2.
(A) Schematic diagram of recombinant baculoviruses expressing wild-type and mutant forms of the γ134.5 protein. The virus designations are indicated to the left of the γ134.5 protein constructs. The shaded bars at the top represent the domain structure of the γ134.5 protein. (ATP)10 represents the triplet repeats of AlaThrPro, which connects the amino-terminal domain and the carboxyl-terminal domain. Thin lines indicate the coding regions retained in the wild-type γ134.5 protein or deletion mutants. The numbers denote the first and last amino acids in theconstructs. (B) Expression of wild-type γ134.5 and mutant forms of the γ134.5 protein. Lysates of Sf9 cells which were mock infected or infected with the indicated viruses were subjected to electrophoresis on a denaturing 12% polyacrylamide gel, transferred to a nitrocellulose membrane, and reacted with anti-His tag antibody (Qiagen Inc.) (8). (C) Activity of the γ134.5 mutants in Sf9 cells. Sf9 cells were mock infected or infected with the indicated recombinant baculoviruses at 5 PFU per cell. At 48 h after infection, cells were harvested, and S10 fractions were prepared as described previously (8). Aliquots of each lysate were then processed for eIF-2α dephosphorylation assay. (D) Quantitation of the phosphorylated eIF-2α in panel C. The numbers indicate the percentages of phosphorylated eIF-2α remaining after incubation with the cell lysates relative to that in mock-infected cell lysate. (E) Interaction of PP1 with the γ134.5 mutants. Sf9 cells infected with baculoviruses expressing wild-type γ134.5 or mutant forms of the γ134.5 protein were harvested at 48 h after infection. The lysates were prepared and then incubated with GST-PP1 (+) bound to beads at 4°C. After the beads were washed, the protein complexes were electrophoretically separated and processed for immunoblotting with anti-γ134.5 antibody (8).
We next examined the ability of these mutants to modulate the activity of eIF-2α phosphatase. Aliquots of lysate from cells mock infected or infected with each virus were reacted with 32P-labeled eIF-2α and processed for autoradiography (8). As shown in Fig. 2C, lysates of cells mock infected exhibited no eIF-2α phosphatase activity (lane 1), whereas lysates of cells infected with GF9909 (wild type), GF9917 (with amino acids 1 to 28 deleted [Δ1-28]), or GF2021 (Δ1-52) displayed a high level of eIF-2α phosphatase activity (lanes 2, 3, and 4, respectively). There was no detectable difference between these mutants and the wild-type γ134.5 protein. However, lysates of cells infected with GF2019 (Δ1-83), GF2017 (Δ1-116), or GF2005 (Δ1-146) exhibited a significant decrease in eIF-2α phosphatase activities (lanes 5, 6, and 7, respectively). Lysates of cells infected with GF2112 (Δ1-187) completely lost eIF-2α phosphatase activity (lane 8). Same results were observed when serially diluted cell lysates were used for eIF-2α phosphatase assay (data not shown).
Phosphorimage analysis showed that detectable 32P-labeled eIF-2α was less than 10% after reaction with lysates of cells infected with GF9909, GF9917 and GF2021. In contrast, the level of 32P-labeled eIF-2α remained between 20 and 75% for GF2019, GF2017, and GF2005, respectively (Fig. 2D). These results indicated that deletions up to amino acids 52 in the amino terminus of the γ134.5 protein did not affect eIF-2α dephosphorylation. However, additional truncations to amino acid 146 in the γ134.5 protein reduced eIF-2α dephosphorylation by 75%. A further deletion to amino acid 187 was deleterious.
To examine whether deletions in the γ134.5 protein affected its binding to PP1, we performed a glutathione S-transferase (GST) pull-down experiment (8). GST-PP1 expressed from Escherichia coli was incubated with lysates of HeLa cells infected with GF9909, GF9917, GF2021, GF2019, GF2017, GF2005, or GF2112. The protein complexes were electrophoretically separated and processed for immunoblot analysis with anti-γ134.5 antibody. The results in Fig. 2E show that GST-PP1, but not GST, bound to the wild-type γ134.5 protein. Moreover, GST-PP1 bound to all mutants except GF2112.
Our data indicated that GF2005, GF2019, and GF2017 were able to mediate eIF-2α dephosphorylation, but they retained only a small fraction of the activity of the wild-type γ134.5 protein. Although these mutants were capable of interacting with PP1, it is likely that truncations may cause improper assembly or arrangement of the γ134.5-PP1 complex with reduced activity. Thus, while it is not essential, the amino-terminal domain of the γ134.5 protein may facilitate eIF-2α dephosphorylation. Early studies established that the carboxyl-terminal domain of the γ134.5 protein is essential for eIF-2α dephosphorylation, which is required to prevent the shutoff of protein synthesis (8, 12, 19, 20). It has been shown that removal of amino acids up to 257 from the carboxyl extreme had no effect on eIF-2α dephosphorylation, whereas additional deletions of the carboxyl terminus were deleterious (8). Given that truncation up to amino acid 52 from the amino terminus of the γ134.5 protein did not have any effect on eIF-2α dephosphorylation and PP1 binding, it is reasonable to conclude that the domain containing amino acids 53 to 258 of the γ134.5 protein constitutes a functional entity, which is capable of mediating efficient eIF-2α dephosphorylation. It is also notable that truncation up to amino acid 188 from the animo terminus completely abolished eIF-2α dephosphorylation. This indicates that deletions of both the amino terminus and the triplet repeats abolished the activity of the γ134.5 protein. This phenotype is attributed to the disruption of the γ134.5-PP1 interaction. These data indicate that the carboxyl terminus of the γ134.5 protein is required but is not sufficient to mediate eIF-2α dephosphorylation.
Analysis of the activity of γ134.5 mutants within the context of the HSV genome.
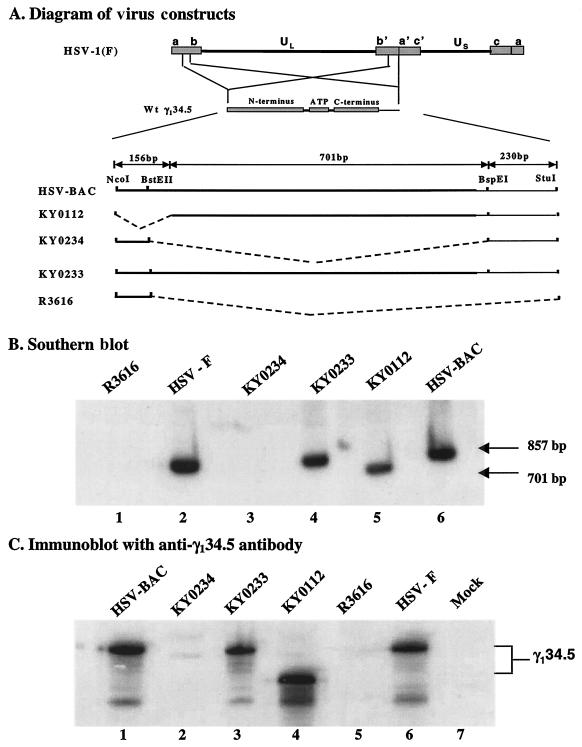
Data in Fig. 2C indicate that deletion of amino acids 1 to 52 in the γ134.5 protein had no effect on dephosphorylation of eIF-2α. In order to analyze the effect of this deletion in the context of the HSV genome, a recombinant virus KY0112 was constructed by using the bacterial artificial chromosome (BAC) system (24). As controls, this mutant was further engineered to either restore or delete the full-length γ134.5 gene, yielding KY0233 and KY0234, respectively (Fig. 3A). To verify the virus constructs, Southern blot analysis was performed after NcoI and BspEI digestion of viral DNA (12). As seen in Fig. 3B, KY0112, which contains a deletion from amino acids 1 to 52 of the γ134.5 protein, gave rise to a 701-bp fragment. Parental HSV-BAC and HSV-1(F) yielded an 857-bp fragment (16, 24). Similarly, KY0233, which contains the restored wild-type γ134.5 gene, yielded an 857-bp fragment. No bands were detected for KY0234 and R3616 due to deletion of the γ134.5 gene.
FIG. 3.
(A) Schematic representation of the genome structure of HSV-1 and its derivatives. HSV-1(F) is the prototype strain used in this laboratory (16). The location of the wild-type (Wt) γ134.5 gene is shown in the expanded portions of the inverted repeat sequences b and b′. The thick lines under the enlarged region of the γ134.5 gene denote wild-type HSV-BAC virus and the mutant viruses. Restriction sites and fragment sizes are indicated. The broken lines represent sequences deleted from the γ134.5 gene. Recombinant virus R3616 lacks 1,000 bp from the coding region of the HSV-1(F) γ134.5 gene (10). HSV-BAC is the parental virus used to construct mutant KY0112, in which the region coding amino acids 1 to 52 of the γ134.5 gene was deleted (24). KY0112 was used to construct KY0233 and KY0234, respectively. KY0233 has the restored wild-type γ134.5 gene, whereas KY0234 has the deleted γ134.5 gene. (B) Autoradiographic image of recombinant and parental viral DNAs. Confluent Vero cells were infected with the indicated viruses at 10 PFU per cell. At 18 h postinfection, infected cells were harvested. Viral DNA was prepared and digested by NcoI and BspEI, electrophoretically separated on 0.8% agarose gels, and transferred to nitrocellulose sheets. The γ134.5 gene was detected by hybridization to electrophoretically separated digests of the viral DNA with a 32P-labeled BstEII-BspEI fragment of the γ134.5 gene, followed by exposure to Kodak X-ray film (19). (C) Expression of the γ134.5 protein and its derivatives. Vero cells were either mock infected or infected with the indicated viruses at 10 PFU per cell. At 18 h after infection, the cells were harvested, solubilized, subjected to polyacrylamide gel electrophoresis, transferred to nitrocellulose sheets, and reacted with anti-γ134.5 antibody (6).
To examine expression of the γ134.5 protein, Western blot analysis was performed with an anti-γ134.5 antibody (11). Figure 3C shows that in Vero cells, HSV-BAC, HSV-1(F), and KY0233 expressed the full-length γ134.5 protein, and the smaller bands likely result from proteolytic cleavage of the full-length γ134.5 protein (lanes 1, 3, and 6). KY0112 expressed a truncated γ134.5 protein with the expected size (lane 4). The γ134.5 protein was not detected in R3616 and KY0234. Taken together, these experiments indicate that the recombinant viruses constructed contain the expected γ134.5 gene derivatives.
Next we measured growth properties of the recombinant viruses. In this series of experiments, monolayers of cells were infected with HSV-1(F), R3616, HSV-BAC (wild-type γ134.5), KY0112 (Δ1-52), KY0234 (Δγ134.5), or KY0233 (restoration of wild-type γ134.5) at 5 PFU per cell. At 24 h postinfection, the cells were harvested, and virus yields were measured. As shown in Table 1, in mouse 10T1/2 cells, HSV-1(F) and HSV-BAC replicated efficiently, as they expressed the wild-type γ134.5 protein. Under this experimental condition, viral yield reached up to 9.5 × 106 PFU/ml. In contrast, both R3616 and KY0234 replicated less efficiently, with titers reaching 1.4 × 105 and 4.2 × 105 PFU/ml, respectively. This decrease in viral replication is attributed to the deletion of the entire γ134.5 gene. Interestingly, KY0112, which has a deletion of amino acids 1 to 52 in the γ134.5 protein, had a titer of only 1.8 × 105 PFU/ml. This mutant exhibited a growth defect similar to the defect observed for R3616 or KY0234. Notably, recombinant KY0233 displayed efficient replication similar to HSV-1(F) or HSV-BAC, as this virus has the restored wild-type γ134.5 gene. As indicated in Table 1, the growth trend for these recombinants is similar in human neuroblastoma SK-N-SH cells. However, it seems that KY0112 replicated better than KY0234 (sixfold) in SK-N-SH cells.
TABLE 1.
Replication of the wild-type HSV-1(F) and γ134.5 mutantsa
| Virus | Virus replication (titer) in cell line:
|
|||
|---|---|---|---|---|
| 10T1/2 | SK-N-SH | HeLa | Vero | |
| HSV-1(F) | 9.5 × 106 | 3.3 × 107 | 3.1 × 107 | 1.8 × 108 |
| R3616 | 1.4 × 105 | 4.2 × 104 | 2.6 × 106 | 1.9 × 107 |
| HSV-BAC | 8.0 × 106 | 3.5 × 107 | 2.4 × 107 | 2.0 × 108 |
| KY0234 | 4.2 × 105 | 6.9 × 104 | 1.6 × 106 | 1.5 × 107 |
| KY0112 | 1.8 × 105 | 4.4 × 105 | 2.8 × 106 | 1.1 × 107 |
| KY0233 | 1.0 × 107 | 1.5 × 107 | 2.5 × 107 | 9.3 × 107 |
Monolayers of cells were infected with each virus at 5 PFU per cell at 37°C. Twenty-four hours postinfection, cells were harvested, freeze-thawed three times, and titrated on Vero cells at 37°C. The data are averages from duplicate samples.
We also examined viral growth properties in Vero cells and HeLa cells. Although the general growth patterns were similar to those seen in mouse 10T1/2 or human SK-N-SH cells, there were smaller differences in the viral titers of wild-type and mutant viruses in these cell lines. As shown in Table 1, in Vero cells, all viruses expressing the wild-type γ134.5 grew to more than 1.8 × 108 PFU/ml, whereas the γ134.5 deletion mutants (R3616 and KY0234) grew to titers of 1.5 ×107 to 1.9 ×107 PFU/ml. Nevertheless, in this cell line, growth of KY0112 was similar to that of R3616 or KY0234. There was generally a 10-fold decrease in viral yield. Similar results were also obtained in HeLa cells. Taken together, these experiments indicate that deletion of amino acids 1 to 52 decreased viral replication, albeit to different extents, in infected cells.
It is interesting that the growth defect associated with the γ134.5 mutant KY0112 (Δ1-52) varies in different cell lines. We speculate that these cells may express different levels of cellular inhibitors of viral infection. Conversely, they may express different levels of host factors required for viral infection. Consistent with these ideas, it has been reported that mutations in oncogenes that constitutively activate the Ras signaling pathway promote cell permissiveness to HSV infection (17). In addition, the growth phase of the cell may affect viral replication. In mouse fibroblast 3T6 cells, replication of HSV that fails to express the γ134.5 protein is restricted in resting cells but less so in actively dividing cells (3). In vitro studies showed that the γ134.5 protein interacts with PCNA, a cellular protein required for DNA replication and cell cycle control (5). It is believed that the interaction of the γ134.5 protein with PCNA may release cells from growth arrest and facilitate viral replication in HSV-infected cells (5). At this point, the nature of the defect associated with the γ134.5 mutant KY0112 is not clear, but it is conceivable that deletion of amino acids 1 to 52 in the γ134.5 protein may alter the interactions between the γ134.5 mutant and host cells that are critical for viral replication.
Since deletion of amino acids 1 to 52 in the γ134.5 protein altered viral replication, next we assessed whether the deletion affected viral response to interferon. In this experiment, monolayers of Vero cells were left untreated or pretreated with alpha interferon for 20 h and subsequently infected with a serial dilution of HSV-BAC, KY0112, KY0234, or KY0233. At 96 h after infection, the numbers of plaques were determined, and the results are presented as the ratio of the number of plaques without and with interferon treatment. The data summarized in Table 2 indicate that plaque formation for wild-type HSV-BAC (wild-type γ134.5) was reduced by alpha interferon only slightly (threefold), exhibiting an interferon-resistant phenotype. However, plaque formation for the γ134.5 deletion mutant KY0234 was reduced dramatically. This reduction is approximately 1,000-fold compared to the wild-type virus. Importantly, plaque formation for KY0112 (Δ1-52) was reduced only fourfold, which is the same as that of HSV-BAC or the repaired virus KY0233. These data demonstrate that deletion of amino acids 1 to 52 in the γ134.5 protein has no effect on viral response to interferon.
TABLE 2.
Effect of alpha interferon on plaque formation of viruses in Vero cellsa
| Virus | γ134.5 gene | Virus titer ratiob |
|---|---|---|
| HSV-BAC | Wild-type | 3.2 ± 0.6 |
| KY0234 | Deletion of entire coding region | 2,000 ± 250 |
| KY0112 | Deletion of amino acids 1 to 52 | 4.5 ± 0.5 |
| KY0233 | Restoration of wild-type γ134.5 gene | 3.0 ± 1.0 |
Vero cells were untreated or pretreated with 1,000 U of human alpha interferon (Sigma) per ml for 20 h. Cells were then infected with a serial dilution of the indicated viruses. The numbers of plaques were counted, and virus titers were determined.
Ratio of viral titer in the absence of alpha interferon to the viral titer in the presence of alpha interferon. The data are the means ± standard deviations for three independent experiments.
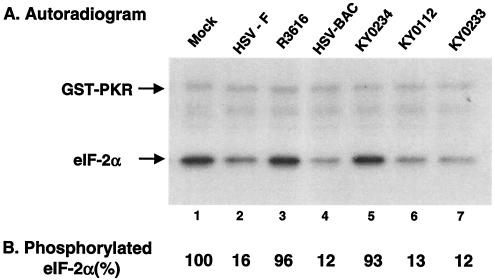
On the basis of the above analysis, we also examined eIF-2α phosphatase activity in vitro. Monolayers of HeLa cells were either mock infected or infected with wild-type or mutant viruses at 20 PFU per cell. At 15 h after infection, cell lysates were prepared to analyze eIF-2α phosphatase activity. As shown in Fig. 4, lysates of cells infected with KY0112 displayed eIF-2α phosphatase activity similar to that seen for wild-type HSV-1(F), HSV-BAC, or KY0233 in which the wild-type γ134.5 gene was restored (Fig. 4, lanes 2, 4, 6, and 7). In contrast, lysates of cells mock infected or infected with the γ134.5 deletion virus R3616 or KY0234 displayed no eIF-2α phosphatase activity (Fig. 4, lanes 1, 3, and 5). Western blot analysis with anti-γ134.5 antibody showed that the γ134.5 protein and its derivative are present at comparable levels in lysates of cells infected with HSV-1(F), HSV-BAC, KY0112, or KY0233 (data not shown). These experiments demonstrated that amino acids 53 to 263 of the γ134.5 protein are sufficient to mediate eIF-2α dephosphorylation, which is consistent with the observation that KY0112 is resistant to interferon.
FIG. 4.
(A) eIF-2α phosphatase activity in HSV-infected cells. Confluent HeLa cells were mock infected or infected with the indicated viruses at 10 PFU per cell. At 16 h postinfection, cells were harvested, and S10 fractions were prepared (21). Aliquots of each lysate were then reacted with 32P-labeled eIF-2 at 34°C. The reaction was stopped after 2 min of incubation. Samples were then separated electrophoretically on a denaturing 12% polyacrylamide gel and subjected to autoradiography (8). (B) Quantitation of the phosphorylated eIF-2α. Phosphorylated eIF-2α in each lane in panel A was quantitated after eIF-2α phosphatase assays with the phosphorimage system (ImageQuant software). The numbers are the percentages of phosphorylated eIF-2α remaining after incubation with the cell lysates relative to that of unreacted eIF-2α. The data are averages from two independent experiments.
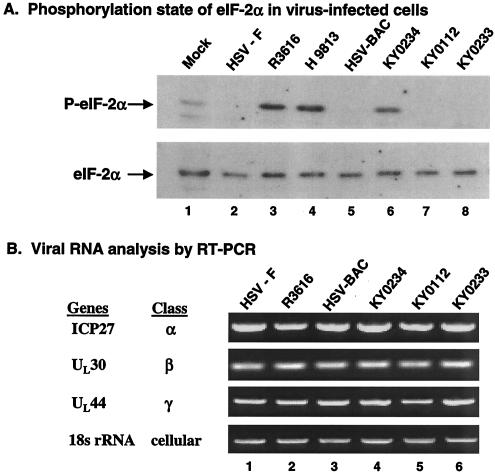
To explore whether in vitro eIF-2α phosphatase activity correlates with the phosphorylation status of eIF-2α in virus-infected cells, we performed Western blot analysis. Specifically, monolayers of mouse 10T1/2 cells were mock infected or infected with HSV-1(F), R3616, HSV-BAC, KY0112, KY0234, KY0233, or H9813, which has Val193Glu and Phe195Leu substitutions in the PP1 binding motif, at 5 PFU per cell (8). At 20 h postinfection, cell lysates were prepared and processed for immunoblotting with antibodies against phosphorylated and total eIF-2α. As shown in Fig. 5A, eIF-2α was present at comparable levels in mock-infected and virus-infected cells. A small amount of phosphorylated eIF-2α was present in mock-infected cells. Interestingly, eIF-2α remained unphosphorylated in cells infected with HSV-1(F), HSV-BAC, KY0112, or KY0233 (Fig. 5A, lanes 2, 5, 7, and 8, respectively). Essentially, viruses expressing the wild-type γ134.5 protein prevented eIF-2α phosphorylation, yet the KY0112 mutant with a deletion of amino acids 1 to 52 of the γ134.5 protein did not induce eIF-2α phosphorylation. In contrast, eIF-2α became phosphorylated in cells infected with R3616, KY0234, or H9813 (Fig. 5A, lanes 3, 4, and 6), which resulted from the failure to express the γ134.5 protein or from point mutations in the PP1 binding motif in the γ134.5 protein. These results correlate well with eIF-2α phosphatase activity analyzed in vitro.
FIG. 5.
(A) Phosphorylation state of eIF-2α in intact cells. Mouse 10T1/2 cells were mock infected or infected with the indicated viruses at 5 PFU per cell. At 20 h postinfection, cells were harvested. The lysates were prepared and processed for Western blotting with antibodies against phosphorylated eIF-2α (P-eIF-2α) and total eIF-2α in the same membrane (Cell Signaling Tech, Inc.). (B) Viral gene transcription pattern in 10T1/2 cells. Cells were infected with the indicated viruses as described above, and total RNA was extracted using an RNeasy kit by Qiagen. Equal amounts of RNA from the samples were then subjected to RT-PCR amplification of specific viral cDNAs (ICP27, UL30, and UL44). As a control, the cellular 18s rRNA was included in the assay. The kinetic classes of selected genes are indicated to the left of the blots. The data are representative of three independent experiments.
An important finding emerging from these experiments is that in HSV-infected cells, eIF-2α dephosphorylation mediated by the γ134.5 protein is tightly coupled to viral resistance to interferon, but not necessarily to efficient viral replication. Consistent with the data from the baculovirus system, the γ134.5 mutant KY0112 with a deletion of amino acids 1 to 52 showed full activity in mediating eIF-2α dephosphorylation. In correlation with these results, KY0112 exhibited an interferon-resistant phenotype like wild-type virus (Table 2). These data strongly support the notion that functional interaction of the γ134.5 protein and PKR is involved in HSV resistance to interferon. However, it is surprising to find that replication of the γ134.5 mutant KY0112 was defective in infected cells. In this respect, it resembled the γ134.5 null mutant. As restoration of the wild-type γ134.5 gene completely reversed the phenotype, the decrease in viral replication for KY0112 is attributable only to a defect in the γ134.5 gene. Although the molecular basis remains unknown, it appears that the region containing amino acids 1 to 52 of the γ134.5 protein is required for efficient viral replication. A simple explanation is that this region represents a distinct functional module. However, an alternative interpretation is that the region containing amino acids 1 to 52 has an indirect role. Deletion of this region may distort conformation of the γ134.5 protein, which indirectly disrupts one or more activities associated with the γ134.5 protein.
To test whether deletions in the γ134.5 gene altered gene expression, we measured the levels of RNA transcript for ICP27 (α gene), UL30 (β-gene), and UL44 (γ gene) in cells infected with viruses. Mouse 10T1/2 cells were infected with HSV-1(F), R3616, HSV-BAC, KY0112, KY0234, or KY0233 at 5 PFU per cell. At 20 h postinfection, total RNA was extracted and subjected to reverse transcription-PCR (RT-PCR) amplification. As shown in Fig. 5B, ICP27, UL30, and UL44 mRNAs were expressed at comparable levels in all virus-infected cells. These results indicated that deletions of the γ134.5 gene have no effect on the expression of mRNA in HSV-infected mouse 10T1/2 cells. Since RT-PCR analysis failed to detect major differences in mRNA expression between wild-type and mutant viruses in 10T1/2 cells, it is likely that the defect is at a step(s) after mRNA expression. Given that dephosphorylation of eIF-2α is essential for viral replication (8), these results suggest that efficient viral replication requires additional activities of the γ134.5 protein.
The γ134.5 protein of HSV is crucial for viral neurovirulence in vivo (10, 27, 35, 37). Accumulating evidence suggests that the γ134.5 protein is involved in different processes during HSV infection (2-4, 6, 11, 21, 28, 34). A remarkable property of this viral factor is to recruit PP1, forming a high-molecular-weight-complex that dephosphorylates eIF-2α and thereby evades the host antiviral response. The fact that eIF-2α phosphatase activity is not necessarily sufficient for efficient viral replication is consistent with the proposal that an additional function(s) associated with the γ134.5 protein contributes to viral infection.
Acknowledgments
We thank Bernard Roizman for providing HSV-1(F) and R3616 and Brian Horsburgh and Frank Tafaro for providing HSV-BAC plasmid. We are grateful to Melissa Cerveny for critical reading of the manuscript.
This work was supported in part by grant AI 46665 (B.H.) from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Aitken, A., and P. Cohen. 1982. Isolation and characterisation of active fragments of protein phosphatase inhibitor-1 from rabbit skeletal muscle. FEBS Lett. 147:54-58. [DOI] [PubMed] [Google Scholar]
- 2.Bower, J. R., H. Mao, C. Durishin, E. Rozenbom, M. Detwiler, D. Rempinski, T. L. Karban, and K. S. Rosenthal. 1999. Intrastrain variants of herpes simplex virus type 1 isolated from a neonate with fatal disseminated infection differ in the ICP34.5 gene, glycoprotein processing, and neuroinvasiveness. J. Virol. 73:3843-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, S. M., J. Harland, A. R. MacLean, J. Podlech, and J. B. Clements. 1994. Cell type and cell state determine differential in vitro growth of non-neurovirulent ICP34.5-negative herpes simplex virus types 1 and 2. J. Gen. Virol. 75:2367-2377. [DOI] [PubMed] [Google Scholar]
- 4.Brown, S. M., A. R. MacLean, J. D. Aitken, and J. Harland. 1994. ICP34.5 influences herpes simplex virus type 1 maturation and egress from infected cells in vitro. J. Gen. Virol. 75:3679-3686. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S. M., A. R. MacLean, E. A. McKie, and J. Harland. 1997. The herpes simplex virus virulence factor ICP34.5 and the cellular protein MyD116 complex with proliferating cell nuclear antigen through the 63-amino-acid domain conserved in ICP34.5, MyD116, and GADD34. J. Virol. 71:9442-9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, G., M. E. Brett, and B. He. 2002. Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the γ134.5 protein of herpes simplex virus type 1. J. Virol. 76:9434-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, G., M. E. Brett, and B. He. 2001. Val193 and Phe195 of the γ134.5 protein of herpes simplex virus 1 are required for viral resistance to interferon α/β. Virology 290:115-120. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, G., M. Gross, M. E. Brett, and B. He. 2001. AlaArg motif in the carboxyl terminus of the γ134.5 protein of herpes simplex virus type 1 is required for the formation of a high-molecular-weight complex that dephosphorylates eIF-2α. J. Virol. 75:3666-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou, J., J. J. Chen, M. Gross, and B. Roizman. 1995. Association of a Mr 90, 000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2α and premature shutoff of protein synthesis after infection with γ134.5- mutants of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA 92:10516-10520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou, J., E. R. Kern, R. J. Whitley, and B. Roizman. 1990. Mapping of herpes simplex virus-1 neurovirulence to γ134.5, a gene nonessential for growth in culture. Science 250:1262-1266. [DOI] [PubMed] [Google Scholar]
- 11.Chou, J., and B. Roizman. 1992. The γ134.5 gene of herpes simplex virus 1 precludes neuroblastoma cells from triggering total shutoff of protein synthesis characteristic of programmed cell death in neuronal cells. Proc. Natl. Acad. Sci. USA 89:3266-3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou, J., and B. Roizman. 1994. Herpes simplex virus 1 γ134.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Proc. Natl. Acad. Sci. USA 91:5247-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou, J., and B. Roizman. 1990. The herpes simplex virus 1 gene for ICP34.5, which maps in inverted repeats, is conserved in several limited-passage isolates but not in strain 17syn+. J. Virol. 64:1014-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen, P. T. 2002. Protein phosphatase 1—targeted in many directions. J. Cell Sci. 115:241-256. [DOI] [PubMed] [Google Scholar]
- 15.Egloff, M. P., D. F. Johnson, G. Moorhead, P. T. Cohen, P. Cohen, and D. Barford. 1997. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 16:1876-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 17.Farassati, F., A. D. Yang, and P. W. Lee. 2001. Oncogenes in Ras signalling pathway dictate host-cell permissiveness to herpes simplex virus 1. Nat. Cell Biol. 3:745-750. [DOI] [PubMed] [Google Scholar]
- 18.Grishin, A. V., O. Azhipa, I. Semenov, and S. J. Corey. 2001. Interaction between growth arrest-DNA damage protein 34 and Src kinase Lyn negatively regulates genotoxic apoptosis. Proc. Natl. Acad. Sci. USA 98:10172-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, B., J. Chou, D. A. Liebermann, B. Hoffman, and B. Roizman. 1996. The carboxyl terminus of the murine MyD116 gene substitutes for the corresponding domain of the γ134.5 gene of herpes simplex virus to preclude the premature shutoff of total protein synthesis in infected human cells. J. Virol. 70:84-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, B., M. Gross, and B. Roizman. 1998. The γ134.5 protein of herpes simplex virus 1 has the structural and functional attributes of a protein phosphatase 1 regulatory subunit and is present in a high molecular weight complex with the enzyme in infected cells. J. Biol. Chem. 273:20737-20743. [DOI] [PubMed] [Google Scholar]
- 21.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirano, K., F. Erdodi, J. G. Patton, and D. J. Hartshorne. 1996. Interaction of protein phosphatase type 1 with a splicing factor. FEBS Lett. 389:191-194. [DOI] [PubMed] [Google Scholar]
- 23.Hollander, M. C., Q. Zhan, I. Bae, and A. J. Fornace, Jr. 1997. Mammalian GADD34, an apoptosis- and DNA damage-inducible gene. J. Biol. Chem. 272:13731-13737. [DOI] [PubMed] [Google Scholar]
- 24.Horsburgh, B. C., M. M. Hubinette, D. Qiang, M. L. MacDonald, and F. Tufaro. 1999. Allele replacement: an application that permits rapid manipulation of herpes simplex virus type 1 genomes. Gene Ther. 6:922-930. [DOI] [PubMed] [Google Scholar]
- 25.Leib, D. A., M. A. Machalek, B. R. Williams, R. H. Silverman, and H. W. Virgin. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. USA 97:6097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lord, K. A., B. Hoffman-Liebermann, and D. A. Liebermann. 1990. Sequence of MyD116 cDNA: a novel myeloid differentiation primary response gene induced by IL6. Nucleic Acids Res. 18:2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacLean, A., L. Robertson, E. McKay, and S. M. Brown. 1991. The RL neurovirulence locus in herpes simplex virus type 2 strain HG52 plays no role in latency. J. Gen. Virol. 72:2305-2310. [DOI] [PubMed] [Google Scholar]
- 28.Mao, H., and K. S. Rosenthal. 2002. An N-terminal arginine rich cluster and a proline-alanine-threonine repeat region determines the cellular localization of the herpes simplex virus type-1 ICP34.5 protein and its ligand, protein phosphatase 1. J. Biol. Chem. 277:11423-11431. [DOI] [PubMed] [Google Scholar]
- 29.Mao, H., and K. S. Rosenthal. 2003. Strain-dependent structural variants of herpes simplex virus type 1 ICP34.5 determine viral plaque size, efficiency of glycoprotein processing, and viral release and neuroinvasive disease potential. J. Virol. 77:3409-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohr, I., D. Sternberg, S. Ward, D. Leib, M. Mulvey, and Y. Gluzman. 2001. A herpes simplex virus type 1 γ34.5 second-site suppressor mutant that exhibits enhanced growth in cultured glioblastoma cells is severely attenuated in animals. J. Virol. 75:5189-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novoa, I., H. Zeng, H. P. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2α. J. Cell Biol. 153:1011-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novoa, I., Y. Zhang, H. Zeng, R. Jungreis, H. P. Harding, and D. Ron. 2003. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22:1180-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang, P. M., J. A. Bondor, K. M. Swiderek, and A. A. DePaoli-Roach. 1991. Molecular cloning and expression of the regulatory (RG1) subunit of the glycogen-associated protein phosphatase. J. Biol. Chem. 266:15782-15789. [PubMed] [Google Scholar]
- 34.Trgovcich, J., D. Johnson, and B. Roizman. 2002. Cell surface major histocompatibility complex class II proteins are regulated by the products of the γ134.5 and UL41 genes of herpes simplex virus 1. J. Virol. 76:6974-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valyi-Nagy, T., M. U. Fareed, J. S. O'Keefe, R. M. Gesser, A. R. MacLean, S. M. Brown, J. G. Spivack, and N. W. Fraser. 1994. The herpes simplex virus type 1 strain 17+ γ34.5 deletion mutant 1716 is avirulent in SCID mice. J. Gen. Virol. 75:2059-2063. [DOI] [PubMed] [Google Scholar]
- 36.Van Eynde, A., S. Wera, M. Beullens, S. Torrekens, F. Van Leuven, W. Stalmans, and M. Bollen. 1995. Molecular cloning of NIPP-1, a nuclear inhibitor of protein phosphatase-1, reveals homology with polypeptides involved in RNA processing. J. Biol. Chem. 270:28068-28074. [DOI] [PubMed] [Google Scholar]
- 37.Whitley, R. J., E. R. Kern, S. Chatterjee, J. Chou, and B. Roizman. 1993. Replication, establishment of latency, and induced reactivation of herpes simplex virus γ134.5 deletion mutants in rodent models. J. Clin. Investig. 91:2837-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams, K. R., H. C. Hemmings, Jr., M. B. LoPresti, W. H. Konigsberg, and P. Greengard. 1986. DARPP-32, a dopamine- and cyclic AMP-regulated neuronal phosphoprotein. Primary structure and homology with protein phosphatase inhibitor-1. J. Biol. Chem. 261:1890-1903. [PubMed] [Google Scholar]
- 39.Zhan, Q., K. A. Lord, I. Alamo, Jr., M. C. Hollander, F. Carrier, D. Ron, K. W. Kohn, B. Hoffman, D. A. Liebermann, and A. J. Fornace, Jr. 1994. The gadd and MyD genes define a novel set of mammalian genes encoding acidic proteins that synergistically suppress cell growth. Mol. Cell. Biol. 14:2361-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]