Abstract
Ascorbate (AsA) is the most abundant antioxidant in plant cells and a cofactor for a large number of key enzymes. However, the mechanism of how AsA levels are regulated in plant cells remains unknown. The Arabidopsis (Arabidopsis thaliana) activation-tagged mutant AT23040 showed a pleiotropic phenotype, including ozone resistance, rapid growth, and leaves containing higher AsA than wild-type plants. The phenotype was caused by activation of a purple acid phosphatase (PAP) gene, AtPAP15, which contains a dinuclear metal center in the active site. AtPAP15 was universally expressed in all tested organs in wild-type plants. Overexpression of AtPAP15 with the 35S cauliflower mosaic virus promoter produced mutants with up to 2-fold increased foliar AsA, 20% to 30% decrease in foliar phytate, enhanced salt tolerance, and decreased abscisic acid sensitivity. Two independent SALK T-DNA insertion mutants in AtPAP15 had 30% less foliar AsA and 15% to 20% more phytate than wild-type plants and decreased tolerance to abiotic stresses. Enzyme activity of partially purified AtPAP15 from plant crude extract and recombinant AtPAP15 expressed in bacteria and yeast was highest when phytate was used as substrate, indicating that AtPAP15 is a phytase. Recombinant AtPAP15 also showed enzyme activity on the substrate myoinositol-1-phosphate, indicating that the AtPAP15 is a phytase that hydrolyzes myoinositol hexakisphosphate to yield myoinositol and free phosphate. Myoinositol is a known precursor for AsA biosynthesis in plants. Thus, AtPAP15 may modulate AsA levels by controlling the input of myoinositol into this branch of AsA biosynthesis in Arabidopsis.
As the most abundant antioxidant in plant tissues, ascorbate (ascorbic acid [AsA]) protects cells and organelles from oxidative damage by scavenging reactive oxygen species (ROS; Noctor and Foyer, 1998) and is important for plant growth, stress resistance, and controlling flowering time and the onset of senescence (Pavet et al., 2005; Barth et al., 2006). AsA also serves as a cofactor for several important enzymes (Conklin and Barth, 2004) and functions in maintaining redox homeostasis, hormone and cell wall biosynthesis, photoprotection, and cell division (Smirnoff and Wheeler, 2000). In addition to being a powerful antioxidant and an enzyme cofactor, AsA is a signaling molecule involved in the regulation of plants' response to the environment, such as to ozone, photooxidative conditions, and pathogen attack (Pastori et al., 2003; Conklin and Barth, 2004).
To achieve these multiple functions, the ability to synthesize AsA through multiple pathways utilizing a variety of precursors would be an advantageous strategy for higher plants. The alternative pathways reveal a more complex picture of AsA biosynthesis than had been expected (Valpuesta and Botella, 2004). Entry points for the biosynthesis of AsA that have been characterized include GDP-Man and l-Gal (Wheeler et al., 1998), myoinositol and d-GlcUA (Lorence et al., 2004), d-GalUA (Agius et al., 2003), and l-gulose (Wolucka and Van Montagu, 2003). All genes of the GDP-Man pathway have been characterized, including those encoding GDP-d-Man pyrophosphorylase (Conklin et al., 1999), GDP-Man-3′,5′-epimerase (Wolucka and Van Montagu, 2003), l-Gal dehydrogenase (Gatzek et al., 2002), l-galactono-1,4-lactone dehydrogenase (Imai et al., 1998), l-Gal-1-P phosphatase (Laing et al., 2004), and, most recently, two genes that encode enzymes that convert GDP-l-Gal to l-Gal-1-P (Dowdle et al., 2007; Laing et al., 2007; Linster et al., 2007).
Feeding of l-gulose and methyl d-GalA to Arabidopsis (Arabidopsis thaliana) cell suspension cultures increases the AsA content effectively (Davey et al., 1999). Overexpression of a mammalian l-gulonolactone oxidase resulted in 4- to 7-fold increase in the AsA content (Jain and Nessler, 2000); however, it is unclear how this enzyme works in plants. Although myoinositol is not considered as a major precursor to AsA, constitutive expression of a myoinositol oxygenase enhanced the AsA content of the Arabidopsis leaves 2- to 3-fold (Lorence et al., 2004). In contrast, the attempts to metabolically engineer the d-Man/l-Gal pathway by overexpression of pathway enzymes have not been successful (Gatzek et al., 2002).
Although the function of these pathways in different plant tissues and developmental stages is not known, the presence of such a biosynthetic network may be important for plants to survive in a constantly fluctuating environment. The AsA level in plants appears to be under strict control. Compared to the characterization of the enzymatic steps in the AsA biosynthesis network, relatively little is known about how these pathways are regulated and how AsA biosynthesis is controlled by the availability of precursor substrates. Moreover, the coordination and cross talk between different branches of the AsA biosynthesis network are virtually unknown. Knowledge of AsA biosynthesis and its regulation would advance approaches to the metabolic engineering of this important antioxidant.
Activation tagging (AT), utilizing four copies of an enhancer from the cauliflower mosaic virus 35S promoter, is a technique useful in plant functional genetics to create mutants in which the T-DNA is spliced into the genome at random sites (Hayashi et al., 1992; Weigel et al., 2000). Analysis of gain-of-function mutants created through AT can provide insight into a gene's function, and such mutants have played a pivotal role in dissecting cytokinin (Kakimoto, 1996), brassinosteroid (Neff et al., 1999), and light (Nakazawa et al., 2001) signaling. To further understand the role and regulation of AsA in oxidative stress tolerance, we utilized an ozone screen of AT Arabidopsis lines (Weigel et al., 2000) to select gain-of-function mutants. The screen selects for high AsA mutants because there is a positive correlation between total foliar AsA and ozone tolerance (Conklin et al., 1996). From approximately 3,000 AT lines, an ozone-tolerant phenotype, with elevated leaf AsA, was isolated. Here, we show that the high AsA level in the mutant was caused by activated expression of AtPAP15, a gene belonging to the major group of Arabidopsis purple acid phosphatases (PAPs). In this study, we have determined phosphatase activities against a variety of substrates both in vivo from plant crude extracts and in vitro as a glutathione S-transferase (GST) fusion protein in bacteria and His-tag protein in yeast (Saccharomyces cerevisiae). We have also examined the impact of AtPAP15 expression on plant growth and responses to environmental factors, including salt and osmotic stress and abscisic acid (ABA) sensitivity. A possible mechanism for the function of AtPAP15 in AsA biosynthesis is proposed.
RESULTS
Isolation of an Activation-Tagged Mutant with High Foliar AsA
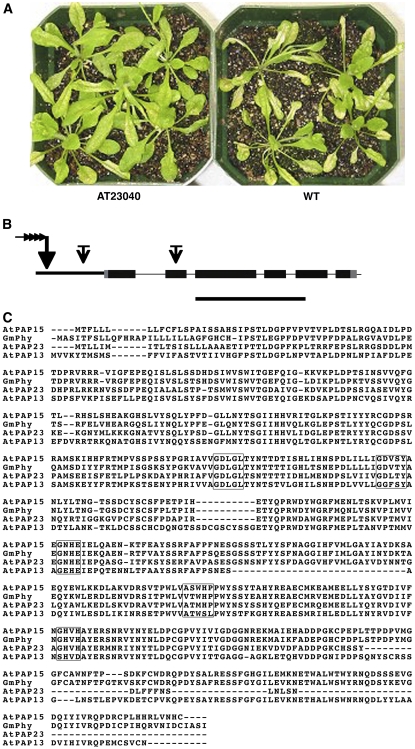
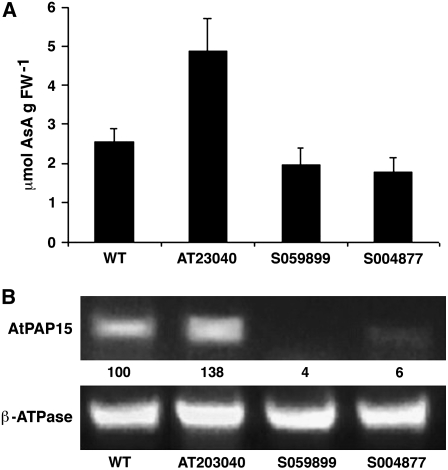
Arabidopsis mutant lines used for screening were developed with the pSKI015 AT vector (Weigel et al., 2000) and kindly provided by the Arabidopsis Biological Resource Center (ABRC; Columbus, OH). Introduction of the T-DNA into the genome can cause increased gene expression near the site of integration in an orientation-independent manner. A high concentration ozone screen of 4-week-old plants identified an oxidant-tolerant phenotype (AT23040) from approximately 3,000 independent T1 lines (Fig. 1A). Under normal growth conditions, the total foliar AsA in AT23040 was approximately 2-fold that in wild-type plants (Fig. 2A). A Southern blot, using a probe of a 1,576-bp DNA fragment complementary to pSKI015 (bases 1–1,576), indicated that AT23040 contained a single T-DNA insert (data not shown).
Figure 1.
Phenotype of the AT mutant AT23040 (Col-0) showing increased tolerance to ozone and AtPAP15 gene structure and amino acid alignment. A, Leaf injury to AT23040 after ozone treatment at 400 ppb for 4 h was considerably less than to 3-week-old wild-type plants. B, Structure of the AtPAP15 gene. Black and gray boxes indicate exons and noncoding regions, respectively. The long arrow with four small arrowheads indicates the AT pSK1015 insertion site, and short arrows with bars indicate the T-DNA insertion sites in the SALK mutants. Bar = 1,000 bp. C, Alignment of the amino acid sequences of AtPAP15, GmPhy (Hegeman and Grabau, 2001), AtPAP13, and AtPAP23 (Li et al., 2002). Boxes show the consensus motifs that coordinately bind the metal ions Fe-Mn/Zn. The alignment was made with ClustalW.
Figure 2.
Foliar AsA and expression of AtPAP15 in AT mutant and T-DNA insertion lines. A, Total foliar AsA in the wild type (WT), the AT mutant (AT23040), and two independent insertion mutants (S059899 and S004877). Bars represent the mean (n = 4) and 1 sd. B, Relative RT-PCR of AtPAP15 expression in the wild type and mutant lines. β-ATPase was used as the loading control. PCR cycles were 28 for AtPAP15 and 25 for β-ATPase. RT-PCR products were detected by staining with ethidium bromide. The AtPAP15 signal was first normalized to the β-ATPase signal and then compared with the wild type. Relative AtPAP15 intensities are analyzed with ImageJ (http://rsb.info.nih.gov/ij/) and shown under each lane as the percentage of signal relative to that in the wild type.
Identification of AtPAP15 by Plasmid Rescue and Analysis of Gene Structure
To identify the AT insertion position and the gene responsible for the mutant phenotype, the site of the T-DNA insertion was determined by plasmid rescue (Weigel et al., 2000). The T-DNA was located in chromosome 3 in the promoter region of At3g07130 (Fig. 1B), a putative PAP, AtPAP15 (Li et al., 2002). AtPAP15 contains six exons (Fig. 1B) and encodes a 533-amino acid protein with a calculated molecular mass of 60.4 kD. AtPAP15 contains eight possible glycosylation sites and a probable 27-amino acid N-terminal signal sequence targeting the protein to the cellular endomembrane system. Sequence alignment with two other AtPAPs and a soybean (Glycine max) PAP indicated high homology of five conserved domains that are involved in the coordinate binding of metal ions, FeIII-MnII/ZnII, at the reaction center of the enzyme (Fig. 1C). Sequence homology of AtPAP15 with the GmPAP, which is a phytase (Hegeman and Grabau, 2001), was 74%, suggesting that the Arabidopsis gene product may be able to hydrolyze phosphate esters of phytate, releasing phosphate and myoinositol.
AtPAP15 Insertion Mutants Have Decreased Foliar AsA, Whereas Pro35S:PAP15 Mutants Have Increased Foliar AsA
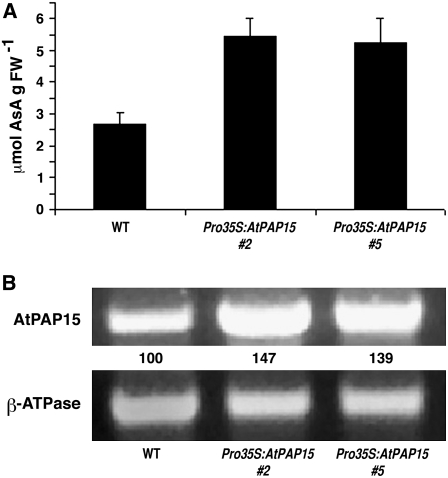
Two T-DNA insertion mutants (S004877 and S059899) were obtained from the SALK T-DNA collection (ABRC). In the former mutant, the insertion was located in the promoter region of At3g07130 and in the latter mutant, in exon 2 (Fig. 1B). Total foliar AsA in homozygous lines of the two insertion mutants was approximately 30% lower than in wild-type plants (Fig. 2A). This contrasts with the 2-fold increase in foliar AsA in the AT mutant (Fig. 2A). Relative reverse transcription (RT)-PCR indicated slight (S004877) to no (S059899) gene expression in the knockout mutants and a >30% increase in expression in the AT mutant (Fig. 2B). The trivial RT-PCR signal in S004887 may be due to the T-DNA insertion location in the promoter region that did not completely inactivate transcription. To demonstrate that AtPAP15 was responsible for increased gene expression that leads to increased foliar AsA, homozygous Pro-35S:AtPAP15 mutant lines were developed. Two lines had leaf AsA levels 2-fold higher compared to wild-type plants (Fig. 3A). Relative RT-PCR indicated that AtPAP15 gene expression was more than 30% higher in the Pro-35S:AtPAP15 mutants (Fig. 3B). These changes in AsA in the various mutants indicate that expression of AtPAP15 functions to positively influence foliar AsA.
Figure 3.
Total foliar AsA levels and expression of AtPAP15 in Pro-35S:AtPAP15 homozygous lines. A, AsA levels in leaves of the wild type and two Pro-35S:AtPAP15 lines, 35S:AtPAP15 (no. 2) and 35S:AtPAP15 (no. 5). The error bar represents the sd from three independent assays. B, Relative transcript abundance of AtPAP15 in two overexpressing lines determined by RT-PCR. β-ATPase was used as the RT-PCR loading control. PCR cycles were 28 for AtPAP15 and 25 for β-ATPase. RT-PCR products were detected by staining with ethidium bromide. The AtPAP15 signal was first normalized to the β-ATPase signal and then compared with the wild type. Relative AtPAP15 intensities are analyzed with ImageJ (http://rsb.info.nih.gov/ij/) and shown under each lane as the percentage of signal relative to that in the wild type.
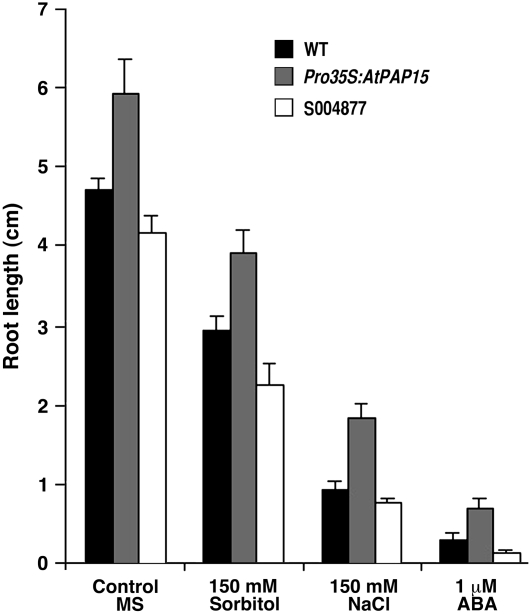
AtPAP15 Expression Stimulates Shoot Growth, Enhanced Salt and Osmotic Stress Tolerance, and ABA Insensitivity
Previous studies have demonstrated a correlative relationship between foliar AsA levels and shoot growth. The vtc1-1 mutant (low AsA) grows more slowly than wild-type plants (Conklin et al., 1996; Veljovic-Jovanovic et al., 2001); however, the phenotype is restored when gulono-1,4-lactone oxidase is constitutively expressed in the mutant (Radzio et al., 2003). When grown under unstressed conditions on Murashige and Skoog (MS) plates, Pro-35S:AtPAP15 showed a 20% to 30% increase in root length, whereas the knockout line S004877 showed a 5% to 15% decrease in root growth compared to the wild type (Fig. 4). To explore whether the changes of AsA levels result in altered abiotic stress tolerance, the growth response of Pro-35S:PAP15 and two independent T-DNA insertion lines was analyzed. The percentage of seed germination of all mutant lines on MS control plates was identical to the wild type. Root growth of Pro-35S:AtPAP15 on MS plates supplemented with 150 mm sorbitol showed 20% increased growth compared to the wild type under osmotic stress (Fig. 4). The Pro-35S:AtPAP15 was even more tolerant to salt stress than osmotic stress, as root length doubled compared to the wild type when seedlings were grown on 150 mm NaCl (Fig. 4). In contrast, in the knockout mutant S004877, root length was only 70% to 75% of the wild type under salt and osmotic stress. Furthermore, on MS plates supplemented with 1 μm ABA, Pro-35S:AtPAP15 exhibited more than 100% greater root length than the wild type, while S004877 root length was only 30% of the wild type (Fig. 4). Moreover, under 1 μm ABA, the final percent germination of wild-type seeds was <70%, S004877 was <50%, and Pro-35S:AtPAP15 was >90%.
Figure 4.
Pro-35S:AtPAP15 overexpressor and T-DNA knockout mutants have altered salt, osmotic stress, and ABA sensitivities. Seeds were germinated and grown on MS plates as controls, MS plates with 150 mm sorbitol, MS plates with 150 mm NaCl, and MS plates with 1 μm ABA. Compared to the wild type, Pro-35S:AtPAP15 had increased tolerance to salt stress and osmotic stress and decreased ABA sensitivity, whereas the T-DNA knockout mutant S004877 was less tolerant to these conditions. Bars represent mean (n = 4).
In Vitro Assays Show That AtPAP15 Is a Phytase
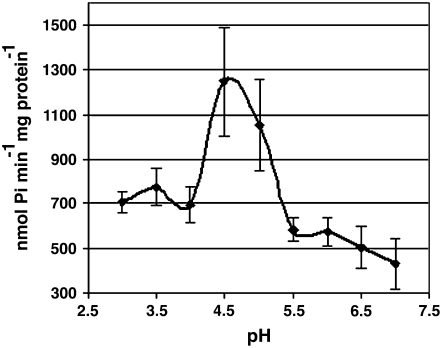
To investigate whether AtPAP15 possesses acid phosphatase activity, we expressed a His-tagged recombinant AtPAP15 protein in yeast and a GST-tagged recombinant AtPAP15 in a bacterial system for activity assays. The recombinant proteins were purified by a metal affinity column and a GST affinity column, respectively, and assayed for phosphatase activity toward several substrates. GST:AtPAP15 is produced as a nonglycosylated protein because it is expressed in bacteria. However, AtPAP15:His-6, expressed in yeast, may contain carbohydrate moiety, as predicted by potential glycosylation sites (http://www.cbs.dtu.dk/services/) in the primary amino acid sequence. GST:AtPAP15 has enzyme activities toward several substrates, which is consistent with studies of AtPAP23, which was also expressed in Escherichia coli as a GST:fusion protein and demonstrated phosphatase activity (Zhu et al., 2005). GST:AtPAP15 and AtPAP15:His-6 activities were tested against the standard phosphatase substrate p-nitrophenyl phosphate (pNPP), phytate, ATP, Glc-6-P, and d-myoinositol-1-phosphate [Ins(1)P1; Table I]. It was found that both recombinant proteins exhibited similar phosphatase activities. Phosphate hydrolysis activity was high with phytate and pNPP, intermediate with Ins(1)P1, and negligible with Glc-6-P and ATP. These results demonstrate that AtPAP15 is able to degrade phytate and Ins(1)P1 and other substrates as a phytase. Assays were conducted over a range of pH to optimize conditions for detecting enzyme activity. Under our assay conditions, the optimum hydrolysis of phytate was recorded at pH 4.5, in accordance with phytase activity data previously reported for soybean and maize (Fig. 5; Laboure et al., 1993; Hegeman and Grabau, 2001).
Table I.
Phosphatase activity of recombinant bacterial GST fusion protein GST:AtPAP15 and His-tagged protein AtPAP15:His-6 expressed in yeast toward various substrates
Values represent mean (n = 4).
| Substrate | Enzyme Activity
|
|
|---|---|---|
| GST:AtPAP15 | AtPAP15:His-6 | |
| nmol Pi min−1 mg protein−1 | ||
| pNPP | 546 ± 40 | 308 ± 26 |
| Glc-6-P | 12 ± 4 | 70 ± 18 |
| ATP | 52 ± 18 | 8 ± 3 |
| Phytate | 711 ± 112 | 751 ± 107 |
| Ins(1)P1 | 207 ± 32 | 200 ± 71 |
Figure 5.
Phytase activity of recombinant At PAP15 is pH dependant. Mean enzyme activity (n = 3) was measured between pH 3.0 and 7.0 using recombinant protein cleaved from GST. Bars represent ± 1 sd.
In Vivo Assays Indicate AtPAP15 Is a Phytase and Overexpression of AtPAP15 Decreases Phytate in Leaves
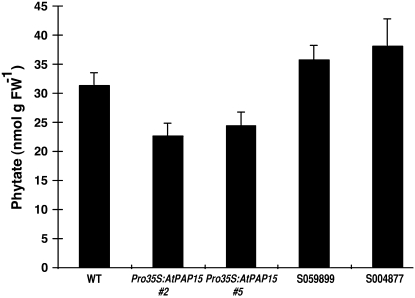
Partially purified AtPAP15 from crude extracts of wild-type and mutant foliar tissue containing the same amount of protein was tested for enzyme activity using three different phosphate esters (Table II). Pro-35S:AtPAP15 showed the highest phosphatase activity toward all substrates tested, approximately 70%, 80%, and 90% higher than the wild type with substrates pNPP, sodium pyrophosphate (NaPP), and phytate, respectively. Phytase activity in the AtPAP15 knockout line was <20% of the wild type, with similar phosphatase activity toward pNPP and NaPP. To investigate the impact of phytase activity in vivo, 3-week-old plants of the wild type, two lines of AtPAP15 overexpressors, and two lines of T-DNA knockouts mutants were analyzed for foliar phytate levels by HPLC. In each of the overexpression lines, phytate levels were lower than in the wild type, with reductions of about 20% to 30%, while two T-DNA knockouts lines, S04788 and S059899, showed increased phytate of between 15% and 20% (Fig. 6) compared to the wild type.
Table II.
A Pro-35S:AtPAP15 mutant has higher phytase activity than wild-type plants, whereas a knockout mutant (Atpap15) has lower phytase activity
Plant protein extracts were partial purified on a concanavalin A column. Phytase activity was measured as released Pi (A355). Values represent mean (n = 3).
| Substrate | Enzyme Activity
|
||
|---|---|---|---|
| Wild Type | Pro-35S:AtPAP15 | Atpap15 | |
| nmol Pi min−1 mg protein−1 | |||
| pNPP | 925 ± 56 | 1,577 ± 100 | 976 ± 101 |
| Phytate | 68 ± 3 | 129 ± 21 | 12 ± 8 |
| NaPP | 1,760 ± 691 | 3,219 ± 206 | 2,058 ± 363 |
Figure 6.
Foliar phytate content in wild-type control, two Pro-35S:AtPAP15 lines (AtPAP15 [no. 2] and AtPAP15 [no. 5]), and T-DNA knockout mutants. Phytate was measured by HPLC in leaves of 3-week-old plants. Bars represent mean (n = 3).
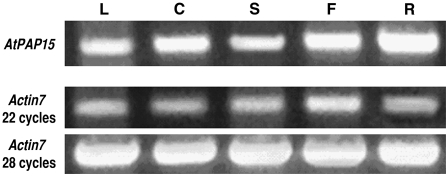
AtPAP15 Is Ubiquitously Expressed
RT-PCR experiments revealed that the AtPAP15 transcript is produced ubiquitously in Arabidopsis. The AtPAP15 transcript was detected in all organs tested including leaves, cotyledons, stems, flowers, and roots (Fig. 7). Little differences in AtPAP15 transcripts were seen across all tissues except roots, which showed the highest expression levels.
Figure 7.
Expression of AtPAP15 is highest in roots but present in all tissues tested. Relative transcript abundance of AtPAP15 in leaf (L), cotyledon (C), stem (S), flower (F), and root (R) of wild-type Arabidopsis plants determined by RT-PCR. Numbers of PCR cycles were 28 for AtPAP15. RT-PCR products were detected by staining with ethidium bromide.
DISCUSSION
Here, we describe an activation-tagged Arabidopsis mutant with elevated foliar AsA and characterized AtPAP15, a PAP that exhibits phytase activity. AtPAP15 represents one of the 29 Arabidopsis PAPs (AtPAP1–AtPAP29) that form the metallophosphatase family, enzymes that contain a dinuclear center in their active site (Que and True, 1990; Vincent et al., 1991; Li et al., 2002). PAPs have been shown in vitro to nonspecifically catalyze the release of phosphate from numerous phosphate esters and anhydrides at a pH range of 4 to 7 (Klabunde et al., 1996). PAPs are suggested to act as multifunctional enzymes and may play an important role in plant growth and development under both normal and stressed conditions (Zhu et al., 2005). Several PAPs are regulated by phosphorous starvation (Haran et al., 2000), implying that they are important in phosphate acquisition. The expression of two PAPs, isolated from cultured tobacco cells, increased during cell wall regeneration in protoplasts and may have some role in the production of cellulose microfibrils (Kaida et al., 2003). AtPAP17 displays peroxidase activity and has been suggested to be involved in ROS metabolism during senescence (del Pozo et al., 1999). A PAP from kidney bean (Phaseolus vulgaris) may function in an antioxidant role to prevent formation of ROS in seeds (Klabunde et al., 1995). AtPAP23, which shares the highest amino acid sequence similarity with AtPAP15 (62%) in Arabidopsis, is found predominantly expressed in flowers, but its function is unknown (Zhu et al., 2005).
The function of AtPAP15 has not been studied previously, but based on its amino acid sequence similarity with a soybean phytase, GmPHY (75%), AtPAP15 was predicted to act as a phytase. Both in vitro and in vivo enzyme assays demonstrated phytase activity of AtPAP15 (Tables I and II). HPLC analysis of leaf phytate content in Pro-35S:AtPAP15 plants showed an approximately 20% decrease compared to the wild type. Phytase is a special type of phosphohydrolase with the capability of initiating dephosphorylation of phytate. Because phytate is the predominant inositol phosphate present in seeds and roots, phytase might be responsible for the hydrolysis of phytate in germinating seeds and in endodermis cells of primary roots (Hubel and Beck, 1996). Although phytases in seeds, leaves, and roots share remarkable similarity, the physiological function of phytase in nonreproductive tissue is unclear (Laboure et al., 1993; Hubel and Beck, 1996). Phytate may function as a universal transitory of phosphorus storage and inositol product in both reproductive and nonreproductive plant tissue. Phytase, irrespective of its location, may play a role in releasing inositol derivatives and free phosphate.
Phytate (myoinositol-1,2,3,4,5,6-hexakisphosphate), the product of inositol phosphorylation, represents a storage form for both inositol and phosphorus. Phytate is found in relatively large quantities in Arabidopsis seeds (24.5 ± 3.5 μmol g−1 in dry seeds) and much less in other plant tissues and organs, including leaves, pollen, roots, and tubers (8–45 μmol g FW−1; Bentsink et al., 2003; Raboy, 2003). Phytate is synthesized from Glc-6-P through an initial conversion to Ins(3)P1 by Ins(3)P1 synthase (Shukla et al., 2004) or from myoinositol catalyzed by a myoinositol kinase (Shi et al., 2005). Phytate and inositol derivatives are important to plant signaling functions. Phytate is also known to act in regulating stomatal closure in response to ABA (Lemtiri-Chlieh et al., 2000). High transcript levels of Ins(3)P1 synthase in photosynthetic tissue (Keller et al., 1998) indicate that phytate might be actively synthesized in leaves. The relatively high inorganic phosphate (Pi) levels, high expression of phytate biosynthesis genes, and low accumulation of phytate in leaves suggest that AtPAP15 could be involved in the constant hydrolysis of phytate, releasing myoinositol and free phosphate.
Myoinositol and derivatives play important roles in several metabolic pathways. We have identified myoinositol as an initial substrate for AsA biosynthesis (Lorence et al., 2004), while others have shown it to be a precursor for the synthesis of the nucleotide sugar UDP-GlcUA, which participates in the production of residues of plant cell walls (Loewus and Murthy, 2000; Kanter et al., 2005). Moreover, as a component of phospholipids, myoinositol is involved in several signaling pathways (Lehle, 1990). Despite these essential functions of myoinositol in plant biology, little is known about the partitioning of this molecule between different cells, tissues, or subcellular compartments. The first plant myoinositol transporters, MITR1 and MITR2 (Mesembryanthemum INOSITOL TRANSPORTER), were described as tonoplast proteins catalyzing the inositol-dependent efflux of Na+ ions from vacuoles (Nelson et al., 1999; Chauhan et al., 2000), where phytate is largely accumulated. Free myoinositol is regarded as a ubiquitous constituent of plant tissues and is a more widespread compound in a variety of life forms than generally realized (Loewus and Murthy, 2000). Dephosphorylation of Ins(3)P1 by a specific myoinositol monophosphatase is regarded as the major route to free myoinositol in plants. However, the concurrent availability of Ins(3)P1, Ins(3)P1 synthase, and myoinositol monophosphatase can be limiting; thus, other sources involving recovery from myoinositol-containing compounds, such as phytate, might be important as well. The utilization of myoinositol and phosphorylated derivatives in several different functional roles probably requires a complex recycling system in which numerous enzymes, cell compartments, and biochemical reactions are involved and from which other cellar processes, such as AsA biosynthesis, are supported.
In this study, we have shown that plants with high expression of AtPAP15 have increased foliar AsA, decreased foliar phytate, more tolerance to abiotic stresses including ozone and excess salt, and decreased sensitivity to ABA. The expression pattern of AtPAP15 established that transcripts were present in all tissue types, with the highest levels in the roots, which is in agreement with a previous study characterizing the expression of 28 AtPAPs in various organs (Zhu et al., 2005) as well as with the quantitative estimate of AtPAP15 expression using massively parallel signature sequencing expression profile (http://mpss.udel.edu/at/; Brenner et al. 2000; Meyers et al., 2004).
The high expression in roots suggests that AtPAP15 may function in recycling of phosphate from the phosphate ester pool. High AsA and AtPAP15 transcript levels in the activation-tagged mutant point toward a possible role of the phytate in AsA synthesis. Low AsA in AtPAP15 knockouts and high AsA in Pro-35S:PAP15 mutants suggest that AtPAP15 might be important for maintaining the AsA pool in addition to the major AsA biosynthesis pathway through Man. Because phytase sequentially hydrolyzes phytate, releasing free myoinositol and Pi, AtPAP15 could function to increase AsA through the myoinositol pathway by increasing the supply of this substrate. As the most abundant myoinositol phosphate in plant cells, phytate accumulates predominately in vacuoles (Mitsuhashi et al., 2005). AtPAP15 might be one of a large number of hydrolytic enzymes found in the vacuole or tonoplast. Localization of AtPAP15 to vacuole is suggested by its pH optimum of 4.5 and is supported by the cellular localization PSORT prediction (Nakai and Horton, 1999).
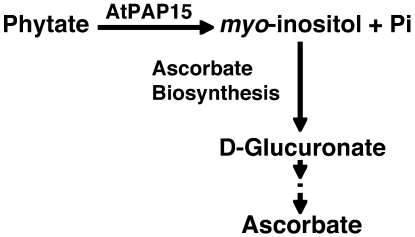
The relationship between AtPAP15 expression and elevated AsA levels may involve a shared biosynthesis pathway of AsA and glucuronate-derived cell wall polysaccharides. In the myoinositol oxygenase pathway to uronosyl and pentose subunits of pectin and hemicelluloses, myoinositol is converted to glucuronate and then phosphorylated by an as-yet-uncharacterized kinase (Loewus and Murthy, 2000). In the AsA synthesis pathway, glucuronate is reduced to gulonate by an as-yet-uncharacterized glucuronate reductase (Lorence et al., 2004). Overexpression of myoinositol oxygenase (Lorence et al., 2004) or glucono-1,4-lactone oxidase (Radzio et al., 2003) increased foliar AsA in Arabidopsis. Overexpression of myoinositol oxygenase also increased the incorporation of labeled myoinositol into cell matrix polysaccharides (Kanter et al., 2005). AtPAP15 could therefore function to increase both AsA and cell wall synthesis by supplying additional myoinositol from phytate. In this research, we have reported a novel phytase, AtPAP15, which may increase the availability of free myoinositol for AsA (Fig. 8) and cell wall biosynthesis and provide support for a possible additional AsA synthesis pathway (phytate to AsA via myoinositol).
Figure 8.
A possible mechanism of AtPAP15 function in AsA biosynthesis. AtPAP15 catalyzes the degradation of phytate and to produce myoinositol and free Pi. AtPAP15 may promote the biosynthesis of AsA through d-glucuronate by increasing the supply of myoinositol.
It is reasonable to assume that an intricate regulatory system exists to control AsA levels in the various cellular compartments in response to developmental cues and to environmental conditions. Thus, in this complex AsA network, different pathways are likely to be subjected to different regulatory mechanisms in order to survive in a constantly fluctuating environment. To our knowledge, this is the first report indicating that cross-talk occurs between phytate and AsA metabolism.
MATERIALS AND METHODS
Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col-0) was used for comparison with mutant plants. Growth conditions were 16-h days at 22°C and 8-h nights at 16°C, under 100 to 150 μmol m−2 s−1 photosynthetically active radiation. The activation-tagged plants containing pSKI015 (Basta resistance) were screened with 0.1% Basta before ozone treatment. The homozygous S004788 and S059899 mutants were in Col-0 and identified as segregating lines in T3 seeds provided by the SALK Institute for Genomic Analysis Laboratory. Homozygous lines were developed by kanamycin selection and confirmed by PCR (the specific gene cannot be amplified in a homozygous line due to the T-DNA insertion). Binary T-DNA vectors were introduced into Agrobacterium tumefaciens strain GV3101 (Koncz and Schell, 1986). Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998). The Pro-35S:AtPAP15 transgene was introduced into the wild-type Col-0 plants. Seedlings were selected on MS (Murashige and Skoog, 1962) medium containing 500 mg L−1 carbenecillin and 25 mg L−1 hygromycin. Both primary transformants and their progeny were used for RT-PCR and AsA assays.
Ozone Fumigation Screen of Activation-Tagged Mutants
Activation-tagged plants were exposed to O3 at concentrations of 400 to 500 nL L−1 for 4 h in a continuously stirred tank reactor. Ozone was generated from O2 by U.V. discharge (Osmonics) and delivered to the chambers by flow meters. O3 concentrations in the chambers were monitored with a TECO UV photometric O3 analyzer (Thermo Electron) and regulated through the flow meters. Tolerant plants were selected for analysis to identify lines with elevated AsA levels.
Leaf Tissue AsA Measurement
AsA content of leaves was measured by the AsA oxidase assay (Rao and Ormrod, 1995). Plant extracts were made from tissue frozen in liquid nitrogen, ground in 6% (w/v) meta-phosphoric acid, and centrifuged at 15,000g for 15 min. Reduced AsA was determined by measuring the decrease in A265 (extinction coefficient of 14.3 cm−1 mm−1) after the addition of 1 unit of AsA oxidase (Sigma) to 1 mL of the reaction mixture containing the plant extract in 100 mm potassium phosphate, pH 6.9. Oxidized AsA was measured by the increase in A265 after addition of 1 μL of 0.2 mm dithiothreitol and incubating at room temperature for 15 min. Total AsA was the sum of AsA and oxidized AsA.
DNA Preparation and Plasmid Rescue
Genomic DNA from the activation-tagged mutant was prepared with a DNA extraction kit (Qiagen). For plasmid rescue, 3 to 5 μg genomic DNA was digested overnight with EcoRI (Promega). After phenol-chloroform extraction, digested DNA was self-ligated overnight at 4°C with T4 DNA ligase in a total volume of 150 μL. Ligated DNA was precipitated with ethanol and was transformed into electroporation-competent Escherichia coli DH10B cells (Invitrogen) by electroporation. Plasmid DNA extracted from kanamycin-resistant clones was sequenced with the primer SKI015: GCAAGAACGGAATGCGCG.
Constructs
For the overexpression of AtPAP15, an NcoI-XbaI fragment containing the Pro-35S and an XbaI-HindIII fragment containing the AtPAP15 transcription terminator were excised from and cloned in the binary vector pCAMBIA1300. The AtPAP15 coding region was amplified from a cDNA library (ABRC) with primers AtPAP15-F (5′-CCCATGGATGACGTTTCTACTACTT-3′) and AtPAP15-R (5′-CCTCTAGATTAGCAATGGTTAACAAGG-3′) under the following conditions: denaturation at 94°C for 3 min, followed by 30 cycles of 94°C for 50 s, 50°C for 50 s, and 72°C for 1 min using Taq Polymerase (Promega). The amplified fragment was cloned into the pGEM-T Easy vector (Promega) sequenced to verify its integrity and subcloned into pRTL2 (Restrepo et al., 1990) to create the Pro-35S∷AtPAP15 construct.
Expression and Purification of GST:AtPAP15 Fusion Protein in E. coli
The coding region cDNA of AtPAP15 was amplified with primers PAP15pGEX-F (5′-CGAATTCCATGACGTTTCTACTACTT-3′) and PAP15PGEX-R (5′-GCGGCCGCTTAGCAATGGTTAACA-3′) and cloned into the bacteria expression vector pGEX-4T-3 through EcoRI and NotI. The construct was transformed into E. coli cells (strain BL21) by heat shock. Expression of GST:AtPAP15 was induced with 1 mm isopropylthio-β-galactoside. Cells were pelleted by centrifugation at 4°C and resuspended in phosphate-buffered saline (PBS; 140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.3), followed by mild sonication. Triton X-100 was added to a final concentration of 1% then mixed gently at room temperature for 30 min. The supernatant was collected by centrifuging the crude extract at 10,000g for 5 min at 4°C. Lysate from induced expression of GST:AtPAP15 fusion protein in E. coli was applied to glutathione-agarose prepacked columns (Sigma). Protein bound to the column was eluted with 10 mm reduced glutathione in 50 mm PBS, pH 7.5, and exchanged with 50 mm Tris-HCl, pH 7.0, using a 10-kD molecular mass cutoff centrifugal filter device (Millipore).
Expression and Purification of AtPAP15:His-6 Fusion Protein in Yeast
AtPAP15 was subcloned into the BamHI and XbaI sites of the yeast (Saccharomyces cerevisiae) expression vector pYES2/CT (Invitrogen) under the control of a Gal-inducible promoter. A His-6 tag was introduced at the C-terminal end of AtPAP15. Yeast strain INVSc1 MATα hishis3Δ1 leu2 trp1-289 ura3-52 (Invitrogen) was transformed with the AtPAP15-pYES2/CT or the positive control pYES2/CT/lacZ vector by the lithium polyethylene glycol method (Invitrogen), and uracil-based selection was used to screen for transformants. A single colony was selected, and transformed yeast cells were grown overnight at 30°C in SC (synthetic complete) selective medium without uracil (6.7 g L−1, yeast nitrogen base, 1.92 g L−1 yeast synthetic dropout media without uracil [Sigma]) containing 2% raffinose. To induce AtPAP15 expression, cells were centrifuged at 1,500g for 5 min, and cell pellet was resuspended to OD600 = 0.4 with induction medium (6.7 g L−1, yeast nitrogen base, 1.92 g L−1 yeast synthetic dropout media without uracil [Sigma]) containing 2% Gal for 18 h. Cells were collected by centrifugation and resuspended in Y-PER yeast protein extraction reagent (Pierce) then mixed gently at room temperature for 20 min. The lysate was collected by centrifugation and was applied to a column packed with His-Catch metal-chelating cellulose (Bioline). Protein bound to the column was eluted with 500 mm imidazole in 50 mm PBS, pH 7.5, and exchanged with 50 mm Tris-HCl, pH 7.0, using a 10-kD molecular mass cutoff centrifugal filter device (Millipore).
The expression of recombinant proteins was confirmed by immunoblot analysis. Purified recombinant proteins were separated on an SDS gel and transferred onto a nitrocellulose membrane. The recombinant proteins were detected by incubation with a monoclonal antibody against the His-6 epitope. Untransformed yeast cells were used as negative control, and yeast cells transformed with pYES2/CT/lacZ were used as positive control.
Plant Tissue Extraction and Concanavalin A Sepharose Column Chromatograph
Approximately 75 mg of 3-week-old leaf tissue was ground in liquid N2 and homogenized in 1 mL of extract buffer (50 mm Tris-HCl, pH 7.4, containing 1 mm reduced glutathione, 5 mm AsA, and 5 mm DTT). The extracts were centrifuged (20,000g, 10 min at 4°C) and the pellets were discarded. The supernatant was applied to a concanavalin A column (GE Healthcare) equilibrated with binding buffer (50 mm Tris-HCl, pH 7.4, containing 500 mm NaCl), and the column was incubated at 4°C for 2.5 h. After washing off unbound materials, bound glycoprotein was eluted with 200 mm methyl α-d-mannopyranoside in binding buffer. Sample purity was assessed by SDS-PAGE. Crude extract and eluant were used in enzyme assay as described.
Phosphatase Assays
Pi released by acid phosphatase activity was measured by a modification of the ammonium molybdate assay (Heinonen and Lahti, 1981). Partially purified, recombinant GST:AtPAP15 was incubated with 0.4 mm substrate of either sodium phytate (Sigma), pNPP, ATP, Glc-6-P, or Ins(1)P1 (Cayman Chemical) in a 100-μL reaction volume of 100 mm NaOAc, pH 4.5, at 37°C for 30 min. Freshly prepared AAM solution (1 mL of 2:1:1 acetone:10 mm ammonium molybdate:5 m H2SO4) was then added to the reaction, vortexed, and examined at 355 nm. A standard curve was generated for quantification of Pi using a 50 ppm stock of NaH2PO4 in 50 mm Tris-HCl, pH 7.5, to generate standard concentrations from 1 to 10 nmol of Pi. Identification of the optimal pH for enzyme activity used phytate as the substrate and purified GST:AtPAP15 in 100 mm sodium acetate at pH 3.0 to 7.0 in 0.5 pH increments.
HPLC Analysis of Phytate
Phytate content of leaves was determined by HPLC (Chiera et al., 2004). Leaf tissue (100 mg) was ground in liquid nitrogen and was added into a 2-mL Eppendorf tube containing 1 mL of 0.5 n HCl. The samples were incubated overnight at room temperature on a rotary shaker and were centrifuged at 15,000g for 30 min. The supernatant was exchanged twice with equal volume of methanol:chloroform (1:1) and centrifuged for 5 min. The supernatant was dried in a speed vac and dissolved in 100 μL of water, and 30 μL was analyzed using an IonPac AS7 anion-exchange column with an Ion-Pac AG7 guard column (Dionex) that was previously equilibrated with 10 mm 1-methylpiperazine, pH 4.0. Inositol phosphates were eluted from the column with a linear 45-min gradient from 100% 10 mm 1-methylpiperazine to 100% 0.5 m NaCl in 10 mm 1-methylpiperazine, pH 4.0, at a flow rate of 1.0 mL min−1. The column eluent was mixed with a colorimetric reagent containing 50 mm 1-methylpiperazine, 0.015% FeCl3, 0.15% sulfosalicylic acid, pH 4.0, at a rate of 1.0 mL min−1 using a mixing tee before detecting the presence of phytate at 465 nm. The amount of phytate in the sample was calculated from a standard curve of sodium phytate and was reported as nanomoles of phytate per gram fresh weight.
Gene Expression by RT-Mediated PCR
For expression analysis, approximately 100 mg of tissues from leaves, stem, roots, flower, and cotyledon was harvested and frozen immediately in liquid nitrogen. Total RNA was extracted with the RNeasy plant mini kit (Qiagen). Crude RNA preparations were treated with 10 units of RNase-free DNase I (Promega) and further purified according to the RNeasy plant mini kit protocol. For RT-mediated PCR studies, cDNA was synthesized from 1.5 μg of DNA-free RNA template using an oligo(dT) primer and Superscript Reverse Transcriptase (Ambion). One-tenth volume of each cDNA was used as a template for PCR amplification. To determine whether comparable amounts of RNA had been used, β-ATPase or actin7 primers were used as control (Kinoshita et al., 1992). PCR reactions were conducted using the following thermal profile: denaturation at 94°C for 4 min, followed by 25 to approximately 30 cycles of 94°C for 45 s, 50°C for 45 s, and 72°C for 1 min, with a 10-min terminal extension step at 72°C. Number of PCR cycles for the transcripts investigated was determined by testing between 20 and 30 to the linear range. PCR products were detected on 1.0% agarose gels infiltrated with ethidium bromide.
Abiotic Stress Assays
For salt, osmotic stress response, and ABA sensitivity assays, 20 to 30 surface-sterilized wild-type and mutant seeds were sown on plates containing MS media with or without 150 mm NaCl (salt stress), 150 mm sorbitol (osmotic stress), or 1 μm ABA. Three replicate plates were used per treatment. Seedlings were grown on vertical plates, and root length was measured 15 d after germination.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AF448726 (AtPAP15), AF272346 (GmPhy), AY390530 (AtPAP23), and AF492665 (AtPAP13).
Acknowledgments
We thank Dr. Elizebeth Grabau for kindly providing us the construct containing AtPAP15. We gratefully acknowledge Dr. Argelia Lorence and Dr. Javad Torabinejad for stimulating discussion. Amy Vance and Laura Nessler are also thanked for their valuable technical assistances.
This work was supported by the Interagency Metabolic Engineering Program (National Science Foundation IPB/MCB grant no. 0118612 and U.S. Department of Agriculture NRICG grant no. 2002–35321–11600), and by the Hatch Project (grant no. VA–135644).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Craig L. Nessler (cnessler@vt.edu).
Open Access articles can be viewed online without a subscription.
References
- Agius F, González-Lamonthe R, Caballero JL, Muñoz-Blanco J, Botella MA, Valpuesta V (2003) Engineering increased vitamin C levels in plants by over-expression of a D-galacturonic acid reductase. Nat Biotechnol 21 177–181 [DOI] [PubMed] [Google Scholar]
- Barth C, Tullio MD, Conklin PL (2006) The role of ascorbic acid in the control of flowering time and the onset of senescence. J Exp Bot 57 1657–1665 [DOI] [PubMed] [Google Scholar]
- Bentsink L, Yuan K, Koornneef M, Vreugdenhil D (2003) The genetics of phytate and phosphate accumulation in seeds and leaves of Arabidopsis thaliana, using natural variation. Theor Appl Genet 106 1234–1243 [DOI] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al (2000) Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat Biotechnol 18 630–634 [DOI] [PubMed] [Google Scholar]
- Chauhan S, Forsthoefel N, Ran Y, Quigley F, Nelson DE, Bohnert HJ (2000) Na+/myoinositol symporters and Na+/H+-antiport in Mesembryanthemum crystallinum. Plant J 24 511–522 [DOI] [PubMed] [Google Scholar]
- Chiera JM, Finer JJ, Grabau EA (2004) Ectopic expression of a soybean phytase in developing seeds of Glycine max to improve phosphorus. Plant Mol Biol 56 895–904 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Conklin PL, Barth C (2004) Ascorbic acid, a familiar small molecule intertwined in the response of plants to ozone, pathogens, and the onset of senescence. Plant Cell Environ 27 959–970 [Google Scholar]
- Conklin PL, Norris SR, Wheeler GL, Williams EH, Smirnoff N, Last RL (1999) Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin C) biosynthesis. Proc Natl Acad Sci USA 96 4198–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin PL, Williams EH, Last RL (1996) Environmental stress sensitivity of an ascorbic acid-deficient Arabidopsis mutant. Proc Natl Acad Sci USA 93 9970–9974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MW, Gilot C, Persiau G, Østergaard J, Huan Y, Bauw GC, Van Montagu MC (1999) Ascorbate biosynthesis in Arabidopsis cell suspension culture. Plant Physiol 121 535–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Pena A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19 579–589 [DOI] [PubMed] [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N (2007) Two genes in Arabidopsis thaliana encoding GDP-l-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J 52 673–689 [DOI] [PubMed] [Google Scholar]
- Gatzek S, Wheeler GL, Smirnoff N (2002) Antisense suppression of l-galactose dehydrogenase in Arabidopsis thaliana provides evidence for its role in ascorbate synthesis and reveals light modulated l-galactose synthesis. Plant J 30 541–553 [DOI] [PubMed] [Google Scholar]
- Haran S, Logendra S, Seskar M, Bratanova M, Raskin I (2000) Characterization of Arabidopsis acid phosphatase promoter and regulation of acid phosphatase expression. Plant Physiol 124 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H, Czaja I, Lubenow H, Schell J, Walden R (1992) Activation of a plant gene by T-DNA tagging: auxin-independent growth in vitro. Science 258 1350–1353 [DOI] [PubMed] [Google Scholar]
- Hegeman CE, Grabau EA (2001) A novel phytase with sequence similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiol 126 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen JK, Lahti RJ (1981) A new and convenient colorimetric determination of inorganic orthophosphate and its application to the assay of inorganic pyrophosphate. Anal Biochem 113 313–317 [DOI] [PubMed] [Google Scholar]
- Hubel F, Beck E (1996) Maize root phytase: purification, characterization, and localization of enzyme activity and its putative substrate. Plant Physiol 112 1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Karita S, Shiratori G, Hattori M, Nunome T, Oba K, Hirai M (1998) L-Galactono-gamma-lactone dehydrogenase from sweet potato: purification and cDNA sequence analysis. Plant Cell Physiol 39 1350–1358 [DOI] [PubMed] [Google Scholar]
- Jain AK, Nessler CL (2000) Metabolic engineering of an alternative pathway for ascorbic acid biosynthesis in plants. Mol Breed 6 73–78 [Google Scholar]
- Kaida R, Sage-Ono K, Kamada H, Okuyama H, Syono K, Kaneko TS (2003) Isolation and characterization of four cell wall purple acid phosphatase genes from tobacco cells. Biochim Biophys Acta 1625 134–140 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (1996) CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science 274 982–985 [DOI] [PubMed] [Google Scholar]
- Kanter U, Usadel B, Guerineau F, Li Y, Pauly M, Tenhaken R (2005) The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta 221 243–254 [DOI] [PubMed] [Google Scholar]
- Keller R, Brearley CA, Trethewey RN, Müller-Röber B (1998) Reduced inositol content and altered morphology in transgenic potato plants inhibited for 1D-myo-inositol 3-phosphate synthase. Plant J 16 403–410 [Google Scholar]
- Kinoshita T, Imamura J, Nagai H, Shimotohno K (1992) Quantification of gene expression over a wide range by the polymerase chain reaction. Anal Biochem 206 231–235 [DOI] [PubMed] [Google Scholar]
- Klabunde T, Sträter N, Fröhlich R, Witzel H, Krebs B (1996) Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures. J Mol Biol 259 737–748 [DOI] [PubMed] [Google Scholar]
- Klabunde T, Sträter N, Krebs B, Witzel H (1995) Structural relationship between the mammalian Fe(III)-Fe(II) and the Fe(III)-Zn(II) plant purple acid phosphatases. FEBS Lett 367 56–60 [DOI] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 383–396 [Google Scholar]
- Laboure AM, Gagnon J, Lescure AM (1993) Purification and characterization of a phytase (myo-inositol-hexakisphosphate phosphohydrolase) accumulated in maize (Zea mays) seedlings during germination. Biochem J 295 413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing WA, Bulley S, Wright M, Cooney J, Jensen D, Barraclough D, MacRae E (2004) A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. Proc Natl Acad Sci USA 101 16976–16981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing WA, Wright MA, Cooney J, Bulley SM (2007) The missing step of the L-galactose pathway of ascorbate biosynthesis in plants, an L-galactose guanyltransferase, increases leaf ascorbate content. Proc Natl Acad Sci USA 104 9534–9539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehle L (1990) Phosphatidyl inositol metabolism and its role in signal transduction in growing plants. Plant Mol Biol 15 647–658 [DOI] [PubMed] [Google Scholar]
- Lemtiri-Chlieh F, MacRobbie EA, Brearley CA (2000) Inositol hexakisphosphate is a physiological signal regulating the K+-inward rectifying conductance in guard cells. Proc Natl Acad Sci USA 97 8687–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhu H, Liu K, Liu X, Leggewie G, Udvardi M, Wang D (2002) Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277 27772–27781 [DOI] [PubMed] [Google Scholar]
- Linster CL, Gomez TA, Christensen KC, Adler LN, Young BD, Brenner C, Clarke SG (2007) Arabidopsis VTC2 encodes a GDP-L-galactose phosphorylase, the last unknown enzyme in the Smirnoff-Wheeler pathway to ascorbic acid in plants. J Biol Chem 282 18879–18885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewus FA, Murthy PPN (2000) myo-Inositol metabolism in plants. Plant Sci 150 1–19 [Google Scholar]
- Lorence A, Chevone BI, Mendes P, Nessler CL (2004) myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134 1200–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Lee DK, Vu TH, Tej SS, Edberg SB, Matvienko M, Tindell LD (2004) Arabidopsis MPSS. An online resource for quantitative expression analysis. Plant Physiol 135 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi N, Ohnishi M, Sekiguchi Y, Kwon Y, Chang Y, Chung S, Inoue Y, Reid RJ, Yagisawa H, Mimura T (2005) Phytic acid synthesis and vacuolar accumulation in suspension-cultured cells of Catharanthus roseus induced by high concentration of inorganic phosphate and cations. Plant Physiol 138 1607–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15 473–497 [Google Scholar]
- Nakai K, Horton P (1999) PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24 34–36 [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25 213–221 [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, et al (1999) BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Koukoumanos M, Bohnert HJ (1999) Myo-inositol-dependent sodium uptake in ice plant. Plant Physiol 119 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49 249–279 [DOI] [PubMed] [Google Scholar]
- Pastori G, Kiddle G, Antoniw J, Bernard S, Veljovic-Jovanovic S, Verrier PJ, Noctor G, Foyer CH (2003) Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell 15 939–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavet V, Olmos E, Kiddle G, Mowla S, Kumar S, Antoniw J, Alvarez ME, Foyer CH (2005) Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol 139 1291–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que LJ, True AE (1990) Dinuclear iron- and manganeso-oxo sites in biology. Prog Inorg Chem 38 97–200 [Google Scholar]
- Raboy V (2003) Myo-inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64 1033–1043 [DOI] [PubMed] [Google Scholar]
- Radzio JA, Lorence A, Chevone BI, Nessler CL (2003) L-Gulono-1,4-lactone oxidase expression rescues vitamin C deficient Arabidopsis (vtc) mutants. Plant Mol Biol 53 837–844 [DOI] [PubMed] [Google Scholar]
- Rao M, Ormrod DP (1995) Ozone pressure decreases UVB sensitivity in a UVB-sensitive flavonoid mutant of Arabidopsis. Photochem Photobiol 61 71–78 [DOI] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang H, Hazebroek J, Ertl DS, Harp T (2005) The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J 42 708–719 [DOI] [PubMed] [Google Scholar]
- Shukla S, Vantoai TT, Pratt RC (2004) Expression and nucleotide sequence of an INS (3) P1 synthase gene associated with low-phytate kernels in maize (Zea mays L.). J Agric Food Chem 52 4565–4570 [DOI] [PubMed] [Google Scholar]
- Smirnoff N, Wheeler GL (2000) Ascorbic acid in plants: biosynthesis and function. Crit Rev Biochem Mol Biol 35 291–314 [DOI] [PubMed] [Google Scholar]
- Valpuesta V, Botella MA (2004) Biosynthesis of L-ascorbic acid in plants: new pathways for an old antioxidant. Trends Plant Sci 9 573–577 [DOI] [PubMed] [Google Scholar]
- Veljovic-Jovanovic SD, Pignocchi C, Noctor G, Foyer CH (2001) Low ascorbic acid in the vtc-1 mutant of Arabidopsis is associated with decreased growth and intracellular redistribution of the antioxidant system. Plant Physiol 127 426–435 [PMC free article] [PubMed] [Google Scholar]
- Vincent JB, Olivier-Lilley GL, Averill BA (1991) Proteins containing oxo-bridged dinuclear iron centers: a bioinorganic perspective. Chem Rev 90 1447–1467 [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al (2000) Activation tagging in Arabidopsis. Plant Physiol 122 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler GL, Jones MA, Smirnoff N (1998) The biosynthetic pathway of vitamin C in higher plants. Nature 393 365–369 [DOI] [PubMed] [Google Scholar]
- Wolucka BA, Van Montagu M (2003) GDP-mannose 3′,5′-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J Biol Chem 278 47483–47490 [DOI] [PubMed] [Google Scholar]
- Zhu H, Qian W, Lu X, Li D, Liu X, Liu K, Wang D (2005) Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominately transcribe in flowers. Plant Mol Biol 59 581–594 [DOI] [PubMed] [Google Scholar]