Abstract
Brome mosaic virus (BMV) is a positive-sense RNA plant virus, the tripartite genomic RNAs of which are separately packaged into virions. RNA3 is copackaged with subgenomic RNA4. In barley protoplasts coinoculated with RNA1 and RNA2, an RNA3 mutant with a 69-nucleotide (nt) deletion in the 3′-proximal region of the 3a open reading frame (ORF) was very poorly packaged compared with other RNA3 mutants and wild-type RNA3, despite their comparable accumulation in the absence of coat protein. Computer analysis of RNA secondary structure predicted two stem-loop (SL) structures (i.e., SL-I and SL-II) in the 69-nt region. Disruption of SL-II, but not of SL-I, significantly reduced RNA3 packaging. A chimeric BMV RNA3 (B3Cmp), with the BMV 3a ORF replacing that of cucumber mosaic virus (CMV), was packaged negligibly, whereas RNA4 was packaged efficiently. Replacement of the 3′-proximal region of the CMV 3a ORF in B3Cmp with the 3′-proximal region of the BMV 3a ORF significantly improved packaging efficiency, and the disruption of SL-II in the substituted BMV 3a ORF region greatly reduced packaging efficiency. These results suggest that the 3′-proximal region of the BMV 3a ORF, especially SL-II predicted between nt 904 and 933, plays an important role in the packaging of BMV RNA3 in vivo. Furthermore, the efficient packaging of RNA4 without RNA3 in B3Cmp-infected cells implies the presence of an element in the 3a ORF of BMV RNA3 that regulates the copackaging of RNA3 and RNA4.
Viral RNAs are specifically selected for packaging during viral infection. Specific packaging occurs through an interaction between viral RNAs and structural coat proteins (CPs). The specific recognition of viral RNAs by CPs plays a crucial role in diverse facets of the viral life cycle and in the packaging event. The binding of CP to specific RNA elements is required for viral protein translation during infection initiation in alfalfa mosaic virus (28) and for the regulation of translation and the initiation of RNA packaging in the RNA phages (38). In retroviruses, nucleocapsid protein is thought to stimulate genomic RNA dimerization, which is required for efficient RNA packaging, reverse transcription, and recombination (14, 31). Many plant viruses require RNA packaging for systemic spread and for cell-to-cell movement (3, 10). Therefore, the CP-RNA interaction is a critical event in the viral life cycle. Compared with the characterization of trans-acting factors such as CP in RNA packaging, however, the RNA elements involved in specific packaging have not been well characterized for many viruses. One of the best-characterized RNA elements is “origin of assembly” of tobacco mosaic virus (36, 40). cis-acting sequences for RNA packaging have also been demonstrated in several icosahedral viruses, including flock house virus (39), Sindbis virus (37), hepatitis B virus (17), the human immunodeficiency viruses (2, 14, 22), and turnip crinkle virus (32).
Brome mosaic virus (BMV) is an icosahedral plant RNA virus and is the type member of the genus Bromovirus in the family Bromoviridae in the alphavirus-like superfamily (18). The genome of BMV consists of three species of messenger sense single-stranded RNA (1). RNA1 (3.2 kb) and RNA2 (2.9 kb), which encode the 1a and 2a replicase proteins, respectively (1, 13, 19), are packaged separately into individual particles (21). RNA3 (2.1 kb), which encodes the 3a cell-to-cell movement protein (35), is packaged into a single particle together with subgenomic RNA4 (0.9 kb) (21). RNA4 is synthesized from the minus strand of RNA3 (25) and encodes CP. CP is required for packaging, cell-to-cell movement, and the systemic spread of the virus (29, 33, 34).
A highly conserved N-terminal arginine-rich motif in BMV CP plays an important role in BMV RNA packaging through RNA-CP interactions (4, 5, 33, 34). The crystallographic structure of BMV virions has been determined (23). RNA regions or elements involved in the packaging of BMV RNAs have been assigned to the coding region of BMV RNA1 by UV cross-linking and band-shift assays (11) as well as to the 3′-proximal region of the 3a open reading frame (ORF) in RNA3 (9). The tRNA-like structures (TLS) in the 3′-untranslated regions of BMV RNAs also play a crucial role in BMV RNA packaging in vitro (6). In the present study, we delimit a nucleotide sequence required for the efficient packaging of BMV RNA3 and show that 69 nucleotides (nt) in the 3′-proximal region of the BMV 3a ORF, especially a predicted stem-loop structure (30 nt), is essential for the efficient packaging of BMV RNA3. We also propose the presence of elements in the BMV 3a ORF that are involved in the regulation of the copackaging of RNA3 and RNA4.
MATERIALS AND METHODS
Plasmid clones.
The plasmids pBTF1, pBTF2, and pBTF3WSS5R25 used in this study contain the full-length cDNAs of BMV RNA1, RNA2, and RNA3, respectively (8, 9, 26).
Construction of BMV RNA3 mutant clones and in vitro transcription.
cDNA clones for BMV RNA3 mutants with deletions (BR3Ds) or with altered nucleotide sequences (stem-loop mutants [SLMs] and control mutant [CM]) (see Fig. 1 and 4) were derived from the plasmid pBTF3WSS5R25 (8). PCR-based in vitro mutagenesis (15) with appropriate combinations of oligodeoxynucleotide primers was used to construct the desired cDNAs with appropriate deletions or base substitutions. The amplified cDNA products of the BR3Ds were digested with PstI and EcoRI and were cloned into pUC119 (Takara, Otsu, Japan) at the corresponding enzyme sites. To facilitate subcloning by avoiding the possible undesired mutations derived from PCR, the cDNA clones were digested with the appropriate restriction enzymes (ClaI and EcoRI for BR3D-200 and BR3D-374; Aor51HI and EcoRI for BR3D-584; and ClaI and Aor51HI for BR3D-797, BR3D-866, and BR3D-935), and the resulting cDNA fragments were used in combination with pBTF3WSS5R25 to create BR3D clones.
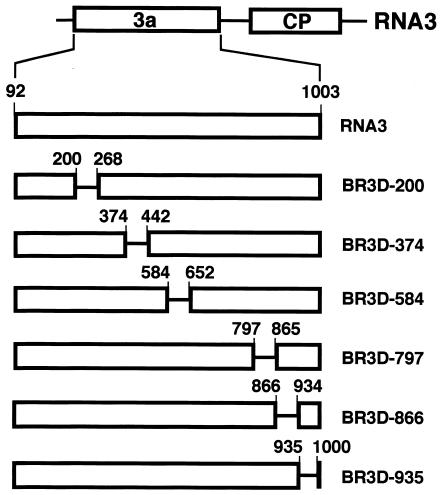
FIG. 1.
Schematic representation of the deleted regions in BMV RNA3. Nucleotide deletions (69 and 66 nt) from 3a ORF (open box) are indicated by a horizontal line with nucleotide numbers at their 5′ and 3′ ends. Wild-type RNA3 is also shown at the top of this figure. All BR3D-RNAs (BR3D-200, BR3D-374, BR3D-584, BR3D-797, BR3D-866, and BR3D-935) were constructed by in vitro mutagenesis to create in-frame deletions.
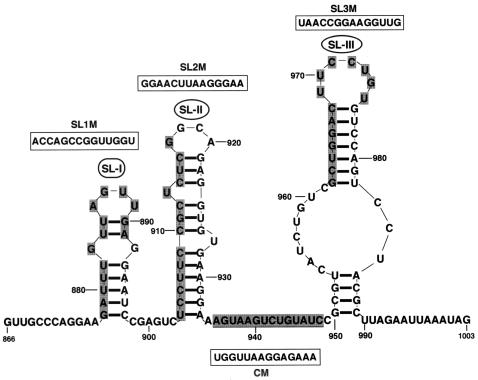
FIG. 4.
Computer-predicted stem-loop structures in the 3′-proximal region of the BMV 3a ORF. Fourteen-base sequences in SL-I, SL-II, and SL-III and in the sequence between SL-II and SL-III (bases are shaded), were replaced with synthetic oligonucleotides of identical length (bases bracketed in a box) to produce SL1 M, SL2 M, SL3 M, and CM cDNA clones, respectively.
SL1 M, SL2 M, SL3 M, and CM cDNA clones (see Fig. 4) were also constructed by PCR-based in vitro mutagenesis in which each SL was replaced with a synthetic oligonucleotide of identical length. The amplified cDNA fragments were digested with ClaI and Aor51HI, and these fragments were used to replace the corresponding regions in pBTF3WSS5R25 to create pBTF3WSS5R25SL1 M, pBTF3WSS5R25SL2 M, pBTF3WSS5R25SL3 M, and pBTF3WSS5R25 CM.
The plasmid pT7B3Cmp is a chimeric BMV RNA3 with the 3a movement protein gene replacing that of cucumber mosaic virus (CMV) (27). This plasmid was used to create B3Cmp derivatives containing the 3′-proximal region of the BMV 3a gene (nt 866 to 1003). A PstI site was created by introducing four bases between nt 835 and 836 in the CMV 3a gene in pT7B3Cmp and by introducing six bases between nt 865 and 866 in the BMV 3a gene of pBTF3WSS5R25 using the appropriate primers with recombinant PCR, both of which were carried out to produce pT7B3CmpPST and pBTF3WSS5R25PST, respectively. The PstI-to-BglII fragment of pT7B3Cmp was substituted for that of pBTF3WSS5R25 to create pT7B3CmpBR3SL, in which the 3′-proximal region of the CMV 3a gene (nt 835 to 931 in B3Cmp) was substituted for that of the BMV 3a gene (nt 866 to 1003 in BMV RNA3). To disrupt stem-loop structure II in B3CmpBR3SL, a PstI site was created in pBTF3WSS5R25SL2 M in the same manner as described for the construction of pBTF3WSS5R25PST. The PstI-to-BglII fragment was substituted for the corresponding fragment in pB3CmpBR3SL to create pB3CmpBR3SL2 M. A plasmid was constructed to express another derivative of B3Cmp, with the 3′-proximal region of the BMV 3a gene (nt 866 to 1003) between the end of the CMV 3a gene and the intercistronic region of BMV RNA3, by using a method similar to that described for the construction of pT7B3CmpBR3SL, i.e., by introducing a PstI site (6 nt) immediately after the stop codon of the CMV 3a gene in pT7B3Cmp.
CP frameshift mutants, B3RD(CP-fs)s, SLM(CP-fs)s, CM(CP-fs), B3Cmp(CP-fs), and B3CmpBR3SL(CP-fs), were constructed as described previously (9).
All plasmids were linearized with EcoRI, and RNA transcripts were synthesized in vitro with T7 RNA polymerase as described previously (26).
Protoplast inoculation, RNA extraction, and analysis of viral RNA.
Isolation and inoculation of barley protoplasts (Hordeum vulgare cv. Gose-shikoku) and extraction of total and virion fraction RNAs were performed as described previously (8). RNAs were subjected to Northern blot analysis essentially as described previously (8, 9). RNA accumulation was quantified as described previously (9). The quantified values for RNA3, its derivatives, and RNA4 relative to the value for the internal standards, RNAs 1 and 2, in each sample were used to calculate the mean values (with standard deviations) from at least three independent experiments.
Prediction of RNA structures.
RNA structure was predicted by using a computer-assisted RNA folding analysis (24).
RESULTS
A 69-nt sequence in the 3′-proximal region of the 3a ORF is required for efficient packaging of RNA3.
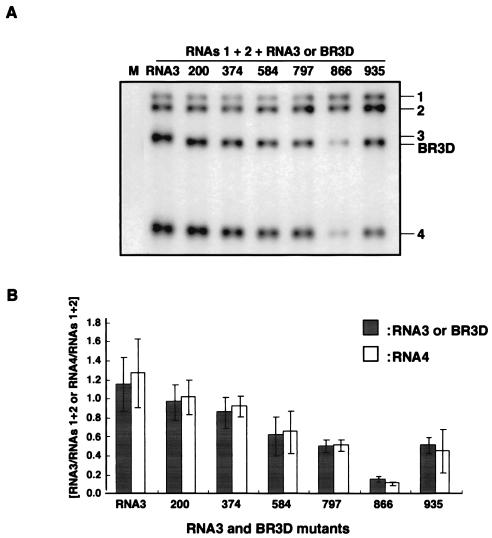
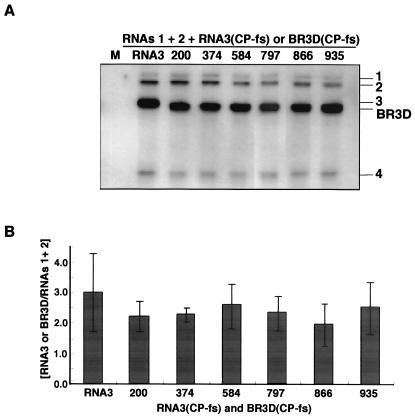
Our previous study suggested that the 3′-proximal region of the 3a ORF, encompassing nt 866 to the end of the ORF (nt 1003), contained nucleotide sequences required for the efficient packaging of BMV RNA3 (9). To delimit and to reevaluate nucleotide sequences in the 3a ORF required for BMV RNA3 packaging, we constructed six BMV RNA3 mutants (BR3Ds) with a small deletion of 66 or 69 nucleotides—two with a deletion in the 3′-proximal region of the 3a ORF and four with a deletion in other regions of the 3a ORF (Fig. 1). These BR3D mutants were tested for their packaging ability in barley protoplasts that had been coinoculated with RNAs 1 and 2. Virion fraction RNAs were extracted at 24 h after inoculation and were analyzed by the Northern blot method using a probe complementary to the 3′-terminal conserved sequence of BMV RNAs. The packaging efficiency of BR3Ds and RNA3 was assessed by calculating their accumulation levels relative to those of RNAs 1 and 2. BR3D-200 and BR3D-374 accumulated to a level similar to that of wild-type (wt) RNA3, whereas the accumulations of BR3D-584, BR3D-797, BR3D-866, and BR3D-935 were low compared with that of wt RNA3. In particular, the level of accumulation of BR3D-866 was less than 10% of that of wt RNA3 (Fig. 2). These results suggest that the deletion of 69 nucleotides introduced into the 3′-proximal half of the BMV 3a ORF, especially the deletion from nt 866 to 934, decreased the packaging efficiency of BMV RNA3. However, these deletions might affect the replication or stability of BMV RNA3. Northern blot results using the total RNA fraction showed an RNA accumulation pattern basically similar to that of the virion RNA fraction; the RNA accumulation level of BR3D-866 relative to that of RNAs 1 and 2 was approximately 40% of that of wt RNA3 (data not shown). Northern blot results from the total RNA fraction may reflect RNA packaging efficiency as well as replication competence or stability. Therefore, to test RNA replication or stability independently of packaging factors, a series of BR3D derivatives with a frameshift mutation in the CP gene (9) were constructed and viral RNA accumulation was compared among these mutants. All BR3D(CP-fs) mutants accumulated to a level similar to that of wt RNA3(CP-fs) (Fig. 3). This indicated that small deletions introduced into the BMV 3a ORF did not affect the replication and stability of the BR3D mutants. Together, these results indicate that the region from nt 866 to 934 in the 3a ORF plays an important role in the packaging of RNA3. We designated this region a “packaging element” of BMV RNA3 (BR3PE).
FIG. 2.
Northern blot analysis of the virion RNA fraction extracted from barley protoplasts at 24 h after inoculation with a mixture of in vitro transcripts of BMV RNAs 1, 2, and 3 or BR3D-RNAs or wt RNA3. The virion RNA fraction was prepared from 1.25 × 104 protoplasts. (A) Northern blot results from an experiment. The complete set of inocula is shown above the Northern blot photograph with individual BR3D-RNAs and RNA3 designated lane by lane. Lane M, inoculated with water only. Positions of RNAs 1, 2, 3, and 4 and BR3D are indicated on the right. (B) Relative values for the accumulation of RNA3s. RNAs 1 and 2 (RNAs 1+2) was used as an internal standard. The mean values for RNA3 and its derivatives (shaded columns) and for RNA4 (white columns) with standard deviations (thin vertical lines) were calculated from three independent experiments.
FIG. 3.
Northern blot analysis of total RNA extracted from barley protoplasts at 24 h after inoculation with a mixture of in vitro transcripts of BMV RNAs 1 and 2 and BR3D(CPfs) RNAs. Total RNA was extracted from 3.0 × 104 protoplasts. (A) Northern blot results from an experiment. (B) Relative values for the accumulation. For others, refer to the Fig. 2 legend.
It is noteworthy that the accumulation of RNA4 relative to that of RNAs 1 and 2 in the virion RNA fraction also differed in protoplasts infected with BR3Ds and that its accumulation paralleled those of BR3Ds (compare the shaded and white columns in Fig. 2B). This supports the previous conclusion that RNA3 and RNA4 are copackaged in BMV-infected cells (21).
In addition, as we reported and discussed previously (9), it was noted that the accumulation levels of wt RNA3 or RNA3 derivatives relative to those of RNAs 1 and 2 were higher in the absence of CP (Fig. 3, 5B, and 6E) than those in the presence of CP (Fig. 2, 5A, and 6C). The absence of functional CP stimulates expression of BMV 1a protein in barley protoplasts (34), and the BMV 1a protein stabilizes BMV RNA3 in yeast cells (16). The stabilization of RNA3(CP-fs) and its derivative RNA3(CP-fs)s by the 1a protein may lead to their higher accumulation in the absence of CP. Lower accumulation of RNA4 than that of its parental RNA3 suggests that a four-base insertion into the CP ORF in RNA3(CP-fs)s or lack of CP affects synthesis of subgenomic RNA4.
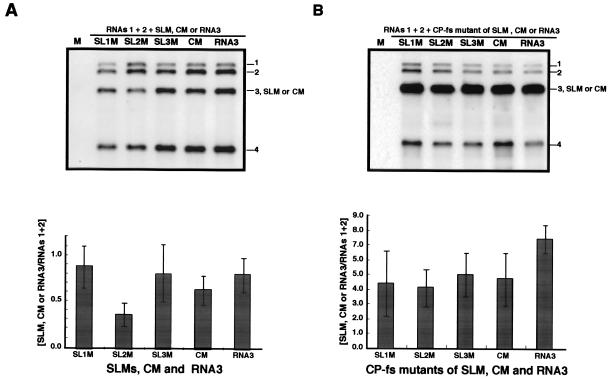
FIG. 5.
Northern blot analysis of the virion RNA or total RNA fraction extracted from barley protoplasts at 24 h after inoculation with a mixture of in vitro transcripts of BMV RNAs 1 and 2 and RNA3 mutants of SLMs and CM or their CP frameshift derivatives. Virion RNA fraction was extracted from 1.25 × 104 protoplasts and total RNA was extracted from 3.0 × 104 protoplasts. (A) Northern blot results of the virion RNA fraction from an experiment using SLMs, CM, or wt RNA3; (B) Northern blot results of the total RNA fraction from an experiment using CP frameshift mutants. The complete set of inocula is shown at the top. Positions of RNAs 1, 2, 3, and 4 and RNA3 mutants are indicated on the right. Relative values for the accumulation of RNA3s are presented below Northern blot photographs. Coinoculated RNA1 + RNA2 was used as an internal standard. The mean values (shaded columns) and standard deviations (thin vertical lines) were calculated from three independent experiments.
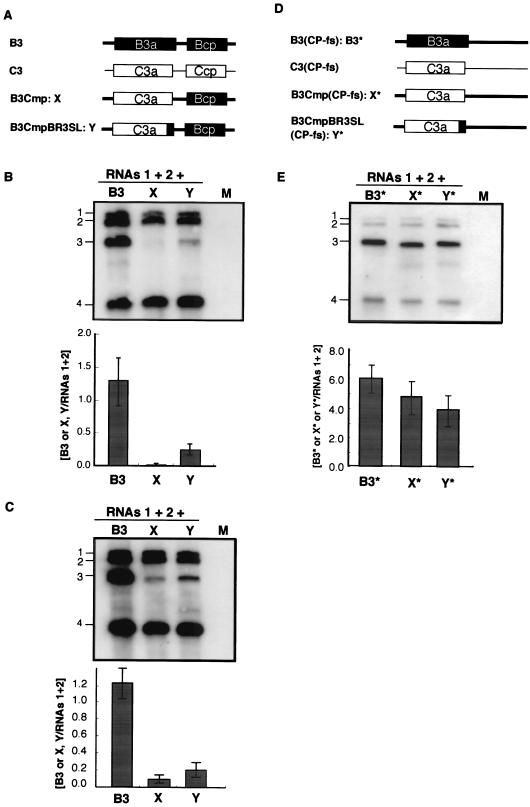
FIG. 6.
Accumulation of chimeric BMV RNA3s and their CP frameshift mutants in virion or total RNA fraction from infected barley protoplasts. Schematic representations of chimeric BMV RNA3s (A) and their CP frameshift mutants (D) with the corresponding abbreviations. Wild-type BMV RNA3 (B3) and CMV RNA3 (C3) are also shown. The ORFs encoded by B3 and C3 are shown as solid and open boxes, respectively, and the noncoding regions of BMV and CMV are represented as thick and thin lines, respectively. Northern blot results from the virion RNA fraction (B) and from the total RNA fraction (C) extracted from barley protoplasts inoculated with chimeric BMV RNA3s or wt BMV RNA3 together with RNAs 1 and 2. Northern blot results from total RNA fraction (E) extracted from barley protoplasts inoculated with CP frameshift mutants of chimeric BMV RNA3s or wt BMV RNA3 together with RNAs 1 and 2. The complete set of inocula is shown above the photograph, with the designations of individual chimeric RNAs given lane by lane. Relative values for the accumulation of RNA3s are presented below the Northern blot photographs. For others, refer to the legend of Fig. 5.
Role of stem-loop structures in RNA3 packaging.
Computer-assisted RNA folding analysis (24) predicted three SL structures in the 3′-proximal region from nt 866 to the end of the BMV 3a ORF (Fig. 4). These SL structures are referred to as SL-I, SL-II, and SL-III. SL-I and SL-II occur in BR3PE. SL-I has a four-base loop and an 8-bp stem that includes a mismatched G-G base pair. SL-II has a four-base loop and a 12-bp stem with two bulged U residues. The 3′-proximal SL-III has a seven-base loop and an 11-bp stem interrupted by eight and three bulged residues.
To examine the role of these SL structures in RNA3 packaging, we constructed three SL mutants (SL1 M, SL2 M, and SL3 M) by substituting 14 bases for synthetic oligonucleotides of an identical size to specifically disrupt each SL structure. We also constructed one CM with a 14-base substitution in the region between SL-II and SL-III that did not disrupt any SL structure in the region (Fig. 4). These RNA structures in the mutants were predicted again by computer-based structure analysis (24), and the results confirmed that the RNA structures were the expected ones described above. These RNA3 mutants were inoculated into barley protoplasts together with BMV RNAs 1 and 2, and the virion fraction RNA was analyzed. SL1 M, SL3 M, and CM accumulated to levels similar to that of wt RNA3, whereas SL2 M accumulated to less than 50% of wt RNA3 levels or those of the other mutants (Fig. 5A). It is possible that base substitutions affect replication or stability of the mutant RNAs. To test this, CP frameshift mutations were introduced into all SLM and CM RNA3 mutants to produce SLM(CP-fs)s and CM(CP-fs) and these CP-fs mutants were tested for the ability to accumulate virus RNA. Northern blot results of total RNA from barley protoplasts coinoculated with RNAs 1 and 2 showed that these CP-fs mutants accumulated RNA3 derivatives to similar levels, although their accumulation was slightly low compared with that of RNA3(CP-fs) (Fig. 5B), suggesting that the low accumulation of SL2 M was due to a deficit in RNA packaging rather than to low-level RNA replication or stability.
The 3′-proximal region of the BMV 3a ORF can function in chimeric BMV RNA3.
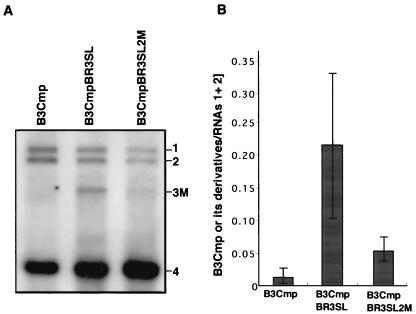
To investigate whether the 3′-proximal region of the BMV 3a ORF can function in a context other than the BMV 3a gene sequence as a cis-acting element for RNA packaging, we used a chimeric BMV RNA3 with the BMV 3a ORF precisely replaced with the CMV 3a ORF (B3Cmp) (27). This chimeric RNA3 was chosen because the accumulation of B3Cmp was extremely low compared with that of wt BMV RNA3 in the virion RNA fraction from barley protoplasts coinoculated with BMV RNAs 1 and 2 (H. Nagano and T. Okuno, unpublished data; Fig. 6). We created B3CmpBR3SL, in which the 3′-proximal region of CMV 3a ORF (nt 835 to 931) of B3Cmp was replaced with the 3′-proximal region of the BMV 3a ORF (nt 866 to 1003) (Fig. 6A). These chimeric RNAs were inoculated into barley protoplasts together with BMV RNAs 1 and 2, and the total and virion RNA fractions were analyzed by Northern blotting. In the virion RNA fraction, B3Cmp accumulated to a negligible level compared to B3CmpBR3SL (Fig. 6B), whereas the accumulation level of B3Cmp was nearly 40% of that of B3CmpBR3SL in the total RNA fraction (Fig. 6C). We also created another B3Cmp derivative, which contained the 3′-proximal region of the BMV 3a gene (nt 866 to 1003) between the end of the CMV 3a gene and the intercistronic region of BMV RNA3 in B3Cmp, and we tested its packaging as described above. This B3Cmp derivative was packaged as efficiently as B3CmpBR3SL (data not shown). To investigate the stability and replication competence of B3Cmp and B3CmpBR3SL independent of packaging factors, the accumulation of the corresponding RNA3 mutants containing CP frameshift mutations (Fig. 6D) was tested in barley protoplasts. Northern blot results from total RNA fraction showed that B3Cmp(CP-fs) and B3CmpBR3SL(CP-fs) accumulated to levels similar and nearly similar to that of B3(CP-fs), respectively (Fig. 6E). These results suggest that the 3′-proximal 138 nucleotides of the BMV 3a ORF, which includes BR3PE, functions as a cis-acting element for RNA packaging in nucleotide sequences partially dissimilar to that of BMV RNA3. To further investigate the role of stem-loop structure II in RNA packaging in B3CmpBR3SL, we created B3CmpBR3SL2 M, in which the SL-II structure was disrupted, and we compared its RNA packaging efficiency with that of B3CmpBR3SL. B3CmpBR3SL2 M accumulated much less than B3CmpBR3SL in the virion RNA fraction (Fig. 7). This suggests that disruption of the SL-II structure in B3CmpBR3SL strongly reduced the efficiency of RNA packaging, thus confirming that the SL-II structure plays an important role in BMV RNA3 packaging.
FIG. 7.
Effect of disruption of stem-loop structure II in chimeric BMV RNA3 (B3CmpBR3SL) on RNA-packaging efficiency. Virion RNA fractions were extracted from barley protoplasts at 24 h after inoculation with a mixture of in vitro transcripts of BMV RNAs 1 and 2 and chimeric RNAs. B3CmpBR3SLSL2 M is a derivative of B3CmpBR3SL in which SL-II is disrupted. The names of the RNA3 mutants are shown above the photograph. Positions of RNAs 1, 2, 4, and chimeric RNA3s (3 M) are indicated on the right. Relative values for the accumulation of RNA3s are presented on the right of the Northern blot photograph. For others, refer to the legend of Fig. 5.
Surprisingly, despite negligible accumulation of B3Cmp in the virion RNA fraction, the accumulation of RNA4 in the fraction was similar to or even higher than that of wt RNA3 relative to that of RNAs 1 and 2 (Fig. 6B). This not only indicates that RNA4 can be packaged into virions without RNA3 derivatives but also suggests that nucleotide sequences in the 3a ORF of BMV RNA3 are involved in the regulation of the copackaging of RNA3 and RNA4, as discussed below.
DISCUSSION
Our present study shows that the nucleotide sequence from nt 866 to 934 (B3PE) in the 3′-proximal region of the BMV 3a ORF, especially a stem-loop structure predicted between nt 904 and 933 (SL-II), plays an important role in the packaging of BMV RNA3 in vivo. The involvement of stem-loop structures in specific RNA packaging in vivo has been demonstrated in small icosahedral RNA viruses, including flock house virus (39), human immunodeficiency virus (14), and turnip crinkle virus (32), as well as in rod-shaped tobacco mosaic virus (36). Selective disruption of the SL-II structure in B3PE of BMV RNA3 did not completely abolish RNA packaging but did reduce packaging efficiency to 30 to 40% of that of wt RNA3 and B3CmpB3SL (Fig. 5 and 7). This suggests that the SL-II structure in BMV RNA3 functions as a cofactor for RNA packaging and also functions in concert with other cis-acting elements that may be present in BMV RNA3 regions other than the 3a ORF or within the 3a ORF. In BMV, the 3′-terminal TLS that is conserved in all BMV RNAs also plays a crucial role in RNA packaging in vitro (6). TLS and SL-II in B3PE may cooperate in RNA3 packaging in vivo through direct or indirect interactions between these regions. Alternatively, TLS and SL-II in B3PE may act independently to facilitate specific RNA3 packaging by BMV CP. In either case, the RNA structure, especially in the 3′-proximal region of the BMV 3a ORF, seems to be important for the efficient packaging of RNA3 because deletions on either side of B3PE also significantly reduced packaging efficiency compared to that of wt RNA3 without affecting the stability or replication competence of RNA3 mutants (compare BR3-D797 and BR3-D935 with wt RNA3 in Fig. 2 and 3). In contrast, deletions in the 5′-proximal region of the 3a ORF had no or mild effects on the packaging efficiency of RNA3 mutants (Fig. 2).
BMV CP expressed from a chimeric CMV RNA3, in which the CP gene was replaced with the BMV CP gene, is unable to package the chimeric CMV RNA, suggesting that BMV CP is highly specific for viral RNA packaging in vivo (30) and in vitro (5, 7). Our present study also shows that a chimeric BMV RNA3 (B3Cmp) was very poorly packaged (Fig. 6B), indicating that BMV TLS alone does not function as a cis-acting element for packaging BMV RNA3 in vivo, and this study confirms the requirement for other elements, including SL-II in B3PE. Although the structure of RNA3 is important for its efficient packaging, as discussed above, the significant increase and decrease in packaging efficiency observed in B3CmpBR3SL and B3CmpBR3SL2 M, respectively (Fig. 6 and 7), strongly indicate a crucial role for SL-II in B3PE in the packaging of BMV RNA3.
Physical and biochemical data suggest that BMV RNA3 is packaged into virions together with RNA4 in BMV-infected plant cells (12, 21). This is confirmed by the parallel accumulation of RNA3 and RNA4 in the virion RNA fraction in the experiments described here, using a series of RNA3 deletion mutants (Fig. 2). These results imply the presence of a mechanism that regulates the copackaging of RNA3 and RNA4 in vivo. Interestingly, in contrast to the parallel accumulation of RNA3 and RNA4, we found that the amount of RNA4 in the virion RNA fraction was similar to that in barley protoplasts infected with wt RNA3 and B3Cmp, whereas B3Cmp was packaged negligibly into virions (Fig. 6B). This suggests that B3Cmp not only lacks the element required for its own encapsidation (possibly B3PE) but that it also lacks the element(s) required to regulate the copackaging of RNA3 and RNA4. The latter element seems to suppress the packaging of RNA4 alone and to regulate the copackaging of RNA3 and RNA4 in BMV-infected cells (Fig. 6B). The regulatory element must occur in the 3a ORF of BMV RNA3, which is lacking in B3Cmp, and in regions other than those deleted from the RNA3 mutants used in this study (Fig. 1). We have observed that the level of RNA4 is consistently and markedly high in the virion RNA fraction when barley protoplasts are infected by some defective BMV RNA3s, with an approximately 500-nt deletion, relative to infections by wt RNA3 (reference 9; T. A. Damayanti and T. Okuno, unpublished data). All these defective RNA3s lack nucleotide sequences in the 3a ORF, from at least nt 95 to 302 (for a review, see reference 9), suggesting that the element occurs in the 5′-proximal region of the 3a ORF.
Another important message from the results shown in Fig. 6 is that RNA4 alone can be packaged into virions in vivo when RNA3 does not have a putative cis-acting element(s) involved in regulation of copackaging with RNA4. This suggests that BMV RNA4 has its own packaging elements. The packaging element is not necessarily TLS, because BMV CP can package an engineered BMV CP mRNA that lacks TLS into an unusual form of 120-CP-subunit virions in yeast cells (20). In BMV-infected cells, however, BMV virions composed of RNA4 without RNA3 are unlikely to be formed, because wt RNA3 encoding the 3a ORF that contains a putative element(s) involved in regulation of copackaging with RNA4 appears to allow only copackaging of RNA4 with RNA3 and to restrict the formation of virions containing RNA4 alone (Fig. 2 and 6). The mechanism underlying the copackaging of RNA3 and RNA4 is unknown. However, RNA3 and RNA4 must directly or indirectly interact with each other to be copackaged. In retroviruses, genomic RNA dimerization through a typical loop-loop “kissing” interaction regulates several steps in the viral life cycle, including encapsidation (31). It is noteworthy that the N-terminal domain of CP is involved in the selective packaging of BMV RNA4 (5).
Acknowledgments
This work was supported in part by a grant-in-aid for Scientific Research (A) (13306005) from the Japan Society for the Promotion of Science and in part by a grant-in-aid for Scientific Research on Priority Area (A) Molecular Mechanisms of Plant-Microbe Interaction toward Production of Disease-Resistant Plants from the Ministry of Education, Culture, Sports, Science, and Technology, Tokyo, Japan.
T.A.D. is a foreign fellowship scholar for the postdoctoral program from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Ahlquist, P. 1992. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 2:71-76. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 3.Callaway, A., D. Giesman-Cookmeyer, E. T. Gillock, T. L. Sit, and S. A. Lommel. 2001. The multifunctional capsid protein of plant RNA viruses. Annu. Rev. Phytopathol. 39:419-460. [DOI] [PubMed] [Google Scholar]
- 4.Choi, Y. G., G. L. Grantham, and A. L. N. Rao. 2000. Molecular studies on bromovirus capsid protein. VI. Contributions of the N-terminal arginine-rich motif of BMV capsid protein to virion stability and RNA packaging. Virology 270:377-385. [DOI] [PubMed] [Google Scholar]
- 5.Choi, Y. G., and A. L. N. Rao. 2000. Molecular studies on bromovirus capsid protein. VII. Selective packaging of BMV RNA4 by specific N-terminal arginine residues. Virology 275:207-217. [DOI] [PubMed] [Google Scholar]
- 6.Choi, Y. G., T. W. Dreher, and A. L. N. Rao. 2002. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl. Acad. Sci. USA 99:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuillel, M., M. Herzog, and L. Hirth. 1979. Specificity of in vitro reconstruction of bromegrass mosaic virus. Virology 95:146-153. [DOI] [PubMed] [Google Scholar]
- 8.Damayanti, T. A., H. Nagano, K. Mise, I. Furusawa, and T. Okuno. 1999. Brome mosaic virus defective RNAs generated during infection of barley plants. J. Gen. Virol. 80:2511-2518. [DOI] [PubMed] [Google Scholar]
- 9.Damayanti, T. A., H. Nagano, K. Mise, I. Furusawa, and T. Okuno. 2002. Positional effect of deletions on viability, especially on encapsidation, of Brome mosaic virus D-RNA in barley protoplasts. Virology 293:314-319. [DOI] [PubMed] [Google Scholar]
- 10.Dolja, V. V., R. Haldeman, N. L. Robertson, W. G. Dougherty, and J. C. Carrington. 1994. Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. EMBO J. 13:1482-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duggal, R., and T. C. Hall. 1993. Identification of domains in brome mosaic virus RNA-1 and coat protein necessary for specific interaction and encapsidation. J. Virol. 67:6406-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, J. M., J. E. Johnson, and M. J. Young. 1994. RNA/protein interactions in icosahedral virus assembly. Semin. Virol. 5:51-60. [Google Scholar]
- 13.French, R., M. Janda, and P. Ahlquist. 1986. Bacterial gene inserted in an engineered RNA virus: efficient expression in monocotyledonous plant cells. Science 231:1294-1297. [DOI] [PubMed] [Google Scholar]
- 14.Harrison, G. P., G. Mieli, E. Hunter, and A. M. L. Lever. 1998. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 77:5886-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, W., H. Ishiguro, and Y. Kurosawa. 1991. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene 102:67-70. [DOI] [PubMed] [Google Scholar]
- 16.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA3 replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Junker-Niepmann, M., R. Bartenschlager, and H. Schaller. 1990. A short cis-acting sequence is required for hepatitis B virus pregenome encapsidation and sufficient for packaging of foreign RNA. EMBO J. 9:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao, C. C., and K. Sivakumaran. 2000. Brome mosaic virus, good for an RNA virologist's basic need. Mol. Plant Pathol. 1:91-97. [DOI] [PubMed] [Google Scholar]
- 19.Kibertis, P. A., L. C. Loesch-Fries, and T. C. Hall. 1981. Viral protein synthesis in barley protoplasts inoculated with native and fractionated brome mosaic virus RNA. Virology 112:804-808. [DOI] [PubMed] [Google Scholar]
- 20.Krol, M. A., N. H. Olson, J. Tate, J. E. Johnson, T. S. Baker, and P. Ahlquist. 1999. RNA-controlled polymorphism in the in vivo assembly of 180-subunit and 120-subunit virions from a single capsid protein. Proc. Natl. Acad. Sci. USA 96:13650-13655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane, L. C., and P. Kaesberg. 1971. Multiple genetic components in bromegrass mosaic virus. Nat. New Biol. 232:40-43. [DOI] [PubMed] [Google Scholar]
- 22.Lever, A. M. 2000. HIV RNA packaging and lentivirus-based vectors. Adv. Pharmacol. 48:1-28. [DOI] [PubMed] [Google Scholar]
- 23.Lucas, R. W., S. B. Larson, and A. McPherson. 2002. The crystallographic structure of brome mosaic virus. J. Mol. Biol. 317:95-108. [DOI] [PubMed] [Google Scholar]
- 24.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 25.Miller, W. A., T. W. Dreher, and T. C. Hall. 1985. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature 313:68-72. [DOI] [PubMed] [Google Scholar]
- 26.Mori, M., K. Mise, K. Kobayashi, T. Okuno, and I. Furusawa. 1991. Infectivity of plasmids containing brome mosaic virus cDNA linked to the cauliflower mosaic virus 35S RNA promoter. J. Gen. Virol. 72:243-246. [DOI] [PubMed] [Google Scholar]
- 27.Nagano, H., K. Mise, T. Okuno, and I. Furusawa. 1999. The cognate coat protein is required for cell-to-cell movement of a chimeric brome mosaic virus mediated by the cucumber mosaic virus movement protein. Virology 265:226-234. [DOI] [PubMed] [Google Scholar]
- 28.Neeleman, L., R. C. Olsthoorn, H. J. Linthorst, and J. F. Bol. 2001. Translation of a nonpolyadenylated viral RNA is enhanced by binding of coat protein or polyadenylation of the RNA. Proc. Natl. Acad. Sci. USA 98:14286-14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okinaka, Y., K. Mise, E. Suzuki, T. Okuno, and I. Furusawa. 2001. The C terminus of brome mosaic virus coat protein controls viral cell-to-cell and long-distance movement. J. Virol. 75:5385-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osman, F., Y. F. Choi, G. L. Grantham, and A. L. N. Rao. 1998. Molecular studies on bromovirus capsid protein. V. Evidence for the specificity of brome mosaic virus encapsidation using RNA3 chimera of brome mosaic and cucumber mosaic viruses expressing heterologous coat proteins. Virology 251:438-448. [DOI] [PubMed] [Google Scholar]
- 31.Paillart, J. C., R. Marquet, E. Skripkin, C. Ehresmann, and B. Ehresmann. 1996. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie 78:639-653. [DOI] [PubMed] [Google Scholar]
- 32.Qu, F., and T. J. Morris. 1997. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J. Virol. 71:128-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao, A. L. N., and G. L. Grantham. 1996. Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement, and pathology. Virology 226:294-305. [DOI] [PubMed] [Google Scholar]
- 34.Sacher, R., and P. Ahlquist. 1989. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J. Virol. 63:4545-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmitz, I., and A. L. N. Rao. 1996. Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell movement-defective RNA3 variants of brome mosaic virus. Virology 226:281-293. [DOI] [PubMed] [Google Scholar]
- 36.Turner, D. R., L. E. Joyce, and P. J. G. Butler. 1988. The tobacco mosaic virus assembly origin RNA: functional characteristics defined by directed mutagenesis. J. Mol. Biol. 203:531-547. [DOI] [PubMed] [Google Scholar]
- 37.Weiss, B., U. Geigenmuller-Gnirke, and S. Schlesinger. 1994. Interactions (sic) between Sindbis virus RNAs and a 68 amino acid derivative of the viral capsid protein further defines the capsid binding site. Nucleic Acids Res. 22:780-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Witherell, G. W., J. M. Gott, and O. C. Uhlenbech. 1991. Specific interaction between RNA phage coat protein and RNA. Prog. Nucleic Acid Res. Mol. Biol. 40:185-220. [DOI] [PubMed] [Google Scholar]
- 39.Zhong, W., R. Dasgupta, and R. Rueckert. 1992. Evidence that the packaging signal for nodaviral RNA2 is bulged stem-loop. Proc. Natl. Acad. Sci. USA 89:11146-11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmern, D. 1977. The nucleotide sequences at the origin for assembly on tobacco mosaic virus RNA. Cell 11:463-482. [DOI] [PubMed] [Google Scholar]