Abstract
We have developed a reporting replicon of West Nile virus (WNV) that could be used to quantitatively distinguish viral translation and RNA replication. A Renilla luciferase (Rluc) gene was fused in-frame with the open reading frame of a subgenomic replicon in the position where the viral structural region was deleted, resulting in RlucRep. Transfection of BHK cells with RlucRep RNA yielded two distinctive Rluc signal peaks, one between 2 and 10 h and the other after 26 h posttransfection. By contrast, only the 2- to 10-h Rluc signal peak was observed in cells transfected with a mutant replicon containing an inactivated viral polymerase NS5 (RlucRep-NS5mt). Immunofluorescence and real-time reverse transcriptase PCR assays showed that the levels of viral protein expression and RNA replication increased in cells transfected with the RlucRep but not in those transfected with the RlucRep-NS5mt. These results suggest that the Rluc signal that occurred at 2 to 10 h posttransfection reflects viral translation of the input replicon, while the Rluc activity after 26 h posttransfection represents RNA replication. Using this system, we showed that mutations of conserved sequence (CS) elements within the 3′ untranslated region of the mosquito-borne flaviviruses did not significantly affect WNV translation but severely diminished or completely abolished RNA replication. Mutations of CS1 that blocked the potential base pairing with a conserved sequence in the 5′ region of the capsid gene (5′CS) abolished RNA replication. Restoration of the 5′CS-CS1 interaction rescued viral replication. Replicons containing individual deletions of CS2, repeated CS2 (RCS2), CS3, or RCS3 were viable, but their RNA replication was dramatically compromised. These results demonstrate that genome cyclization through the 5′CS-CS1 interaction is essential for WNV RNA replication, whereas CS2, RCS2, CS3, and RCS3 facilitate, but are dispensable for, WNV replication.
West Nile virus (WNV) belongs to the genus Flavivirus, which includes significant human pathogens such as dengue virus (DEN), yellow fever virus (YF), the tick-borne encephalitis virus complex, Japanese encephalitis virus (JE), Murray Valley encephalitis virus, and WNV. WNV is widely distributed throughout Africa, the Middle East, parts of Europe, Russia, India, Indonesia, and most recently, North America (6). Since the introduction of WNV into the United States in 1999, the virus has caused more than 4,156 known human cases and at least 284 deaths (for updates, see http://www.cdc.gov/ncidod/dvbid/westnile/surv&controlCaseCount03.htm). The WNV epidemic in the United States in 2002 represents the largest meningoencephalitis outbreak in the Western hemisphere and the largest WNV outbreak ever reported (7). Prevention and treatment of WNV infection have become public health priorities.
Flavivirus virions contain a single plus-sense RNA genome of approximately 11 kb in length, which consists of a 5′ untranslated region (UTR), a single long open reading frame (ORF), and a 3′ UTR (24). The 5′ and 3′ UTRs are approximately 100 nt and 400 to 700 nt in length, respectively. The encoded polyprotein is cotranslationally and posttranslationally processed by viral and cellular proteases into three structural proteins (capsid [C], premembrane [prM] or membrane [M], and envelope [E]) and seven nonstructural proteins (the glycoproteins NS1 and NS2a, the protease cofactor NS2b, the protease and helicase NS3, NS4a, and NS4b; and the methyltransferase and RNA-dependent RNA polymerase [RdRp] NS5) (8). During the flavivirus replication cycle, the synthesis of plus- and minus-sense RNAs is asymmetric; plus-sense RNA is produced in 10- to 100-fold excess over minus-sense RNA (11, 23, 28).
The terminal nucleotides of the 3′ UTR of flaviviruses can form highly conserved secondary and tertiary structures (4, 5, 24, 26, 31). Upstream of the 3′ stem-loop structure, conserved sequences (CS) CS1, CS2, and repeated CS2 (RCS2) are retained among mosquito-borne flaviviruses. The JE subgroup and YF also have a distinct CS3 and repeated CS3 (RCS3). There is limited information available in regard to the functions of these CS elements. CS1 has the potential to base pair with a sequence within the conserved sequence in the 5′ region of the capsid gene (5′CS) (13). Recent studies using DEN (33), Kunjin (16), and YF (10, 22) replicons have shown that base pairing between 5′CS and CS1 is essential for flavivirus replication. For CS2, RCS2, CS3, and RCS3 elements, studies using a Kunjin replicon showed that deletion of all 3′ UTR elements upstream of RCS2 is almost lethal (17). In contrast, results from a DEN type 4 infectious clone suggested that the region upstream of CS1 is dispensable for replication (21). However, the DEN 3′ UTR does not contain CS3 and RCS3 elements. The molecular details of whether and how each individual CS element affects viral translation and RNA replication remain to be elucidated.
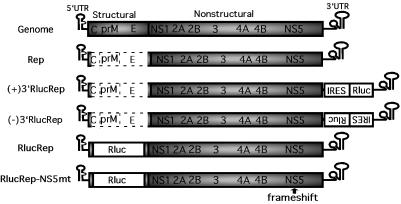
The functions of the CS elements in WNV have not been analyzed due to the lack of genetic systems. Yamshchikov and coworkers reported an infectious clone for lineage II WNV (32), which is largely maintained in enzootic cycles (3, 20). We recently developed an infectious clone (29) and a subgenomic replicon (28) for lineage I WNV, which is frequently involved in human and equine viral outbreaks (3, 20). Our original replicon (Fig. 1) contained large in-frame deletions (>92%) of the C-prM-E structural region yet still replicated efficiently in BHK cells (28). In this paper, we describe a reporting replicon that can be used to quantitatively distinguish viral translation and RNA replication. Functional analysis using such replicons has showed that mutations or deletions of the CS elements within the 3′ UTR of WNV do not significantly affect viral translation but severely or completely diminish RNA replication.
FIG. 1.
WNV genome and replicons with or without reporters constructed in this study. Compared with full-length WNV genome, the original replicon (Rep) contained an in-frame deletion of the structural region (dotted open box) from nt 190 to 2379. The N-terminal 31 amino acids from the capsid protein and the C-terminal 30 amino acids from the envelope protein were retained to maintain the essential 5′CS element and to ensure correct processing of the polyprotein, respectively. For construction of reporter replicons, Rluc driven by an IRES was inserted into the upstream end of the 3′ UTR of the Rep in either plus- or minus-sense orientation, resulting in (+)3′RlucRep and (−)3′GFPRep, respectively. Alternatively, Rluc reporter was fused in-frame with the ORF of the Rep in the position where the structural region was deleted, resulting in RlucRep. RlucRep-NS5mt contains a single-nucleotide frameshift upstream of the active site of the RdRp domain of NS5. The nucleotide positions were numbered according to the sequence of WNV strain 3356 (GenBank accession number AF404756). The drawing is not to scale.
MATERIALS AND METHODS
Plasmid constructions.
Recombinant DNA and cloning procedures were performed by standard methods (25) with modifications as previously described (28, 29). Replicon cDNAs of all constructs were driven by a T7 promoter for in vitro transcription. Four types of replicon plasmids were constructed in this study. Primers used for construction of the various replicons are listed in Table 1.
TABLE 1.
Oligonucleotidesa used for construction of WNV reporter replicons
| Primer | Primer sequence | Replicon name |
|---|---|---|
| F-Pst-IRES | ataattCTGCAGATCCGCCCCTCTCCCTC (PstI) | (+) or (−)3′RlucRep |
| R-Rluc-IRES | ACACCTTGGAAGCCATGGTTGTGGCCATATTATC | (+) or (−)3′RlucRep |
| F-IRES-Rluc | GGCCACAACCATGGCTTCCAAGGTGTAC | (+) or (−)3′RlucRep |
| R-Pst-Rluc | acagacCTGCAGTTACTGCTCGTTCTTCAGCAC (PstI) | (+) or (−)3′RlucRep |
| F-Afl-Rluc | tacactCTTAAGATGGCTTCCAAGGTGTACGA (AflII) | RlucRep |
| R-Afl-Rluc | cacaagCTTAAGCTGCTCGTTCTTCAGCACG (AflII) | RlucRep |
The oligonucleotides were named according to the polarity (F, forward sense primer; R, reverse antisense primer), restriction sites for cloning (underlined and indicated by Pst or Afl for PstI or AflII, respectively), and finally the IRES or Rluc sequences (indicated by italicized upper case or roman upper case, respectively).
Type 1 replicons contained an insertion of encephalomyocarditis virus internal ribosome entry site (IRES)-Rluc fragment at the upstream end of the 3′ UTR of the replicon in either plus or minus sense, resulting in (+)3′RlucRep and (−)3′RlucRep, respectively (Fig. 1). The IRES-Rluc DNA fragment was prepared through fusion PCR and inserted into a unique NsiI site of the original replicon (Rep). Individual fragments of IRES and Rluc were first PCR amplified from plasmid pIRES2-EGFP (Clontech, Palo Alto, Calif.) and phRL-SV40 (Promega, Madison, Wis.) using primers F-PstI-RES and R-Rluc-IRES and F-IRES-Rluc and R-Pst-Rluc, respectively. Fragments of IRES and Rluc were then fused to yield IRES-Rluc through an overlapping PCR. Because the Rluc gene contained an NsiI site, PstI restriction sites, containing cohesive ends compatible with the NsiI site, were engineered at both ends of the IRES-Rluc fragment by using primers F-Pst-IRES and R-Pst-Rluc. The resulting IRES-Rluc was digested with PstI and cloned into the NsiI site of the 3′ UTR of the Rep (28). The orientation of the insert was verified by restriction enzyme digestion followed by DNA sequencing.
Type 2 replicons contained the Rluc reporter fused in-frame with the ORF in the position where the viral structural region was deleted, resulting in RlucRep (Fig. 1). The Rluc fragment was amplified from plasmid phRL-SV40 (Promega), using primers F-Afl-Rluc and R-Afl-Rluc, and cloned into the AflII site of the original Rep (28). A mutant RlucRep, RlucRep-NS5mt, was prepared by insertion of a nucleotide U between nt positions 8027 and 8028, resulting in a frameshift upstream of the active site of the RdRp domain of the NS5 gene (Fig. 1). The nucleotide insertion was achieved through replacement of the SpeI (nt 8022)-NsiI (nt 10432) fragment from the RlucRep with the mutant counterpart.
Type 3 replicons were built on RlucRep and contained various CS mutations within the 3′ UTR or a 5′CS mutation. To facilitate cloning, the NsiI site at nt position 10432 from the RlucRep was changed to a unique MluI site by PCR mutagenesis. Rluc-CS1* and Rluc-5′CS* contained six nucleotide changes within the CS1 and 5′CS, respectively, which blocked the potential 5′CS-CS1 base pairing. Rluc-CS2-del, Rluc-RCS2-del, Rluc-CS3-del, and Rluc-RCS3-del contained deletions of individual CS elements. All mutants were constructed via site-directed PCR mutagenesis. Rluc-RCS3/RCS2-del contained a large truncation of the 3′UTR, effectively removing RCS3, CS3, and RCS2, but retaining CS2, CS1, and the 3′ terminal stem-loop. The Rluc-RCS3/RCS2-del mutant was constructed by digestion of the RlucRep with MluI and SacII, followed by filling in the 5′ overhang with deoxynucleoside triphophates and by removal of the 3′ overhang with the T4 DNA polymerase. The resulting plasmid was then blunt-end ligated using T4 DNA ligase.
Type 4 replicons were identical to type 3 replicons, except that each mutation was engineered into the original Rep without a reporter, resulting in CS1*, CS2-del, RCS2-del, CS3-del, RCS3-del, or 5′CS*. In addition, a compensatory mutant, 5′CS*/CS1*, which contained six complementary nucleotide mutations within both 5′CS and CS1, was constructed. These mutants were also generated by PCR-mediated mutagenesis. All plasmid constructs were verified by DNA sequencing.
RNA transcription, transfection, and immunofluorescence assay.
Procedures for RNA transcription and transfection were reported previously with modifications (28, 29). Plasmids (10 μg each) were linearized with XbaI, followed by Mung bean nuclease treatment at 30°C for 30 min to remove the nucleotides of the single-stranded overhang. The resulting plasmids were phenol-chloroform extracted twice, ethanol precipitated, and resuspended in 10 μl of RNase-free water. A mMESSAGE mMACHINE kit (Ambion, Austin, Tex.) was used for in vitro transcription in a 20-μl reaction with an extra 0.75 μl of GTP solution added after 5 min of incubation. The transcription reaction was incubated at 37°C for 2 h, followed by removal of DNA template by DNase I digestion at 37°C for 15 min. RNA was precipitated using lithium chloride, washed with 70% ethanol, resuspended in RNase-free water, quantified by spectrophotometry, and stored at −20°C in aliquots.
For RNA transfection, approximately 8 × 106 BHK cells suspended in 0.8 ml of ice-cold phosphate-buffered saline (PBS) were electroporated with 10 μg of RNA, along with 2 μg of mRNA containing a firefly luciferase gene for normalization of transfection. The plasmid, which was used for in vitro transcription of the firefly luciferase-coding RNA, was generously provided by Guangxiang Luo of the University of Kentucky. Electroporation was performed in 0.4-cm cuvettes using the GenePulser II apparatus (Bio-Rad), pulsed three times with a no-pulse controller, at settings of 0.85 kV and 25 μF. Transfected cells were resuspended in 25 ml of Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, and 3 ml of the cell resuspensions was seeded onto six-well plates. Indirect immunofluorescence assays (IFA) were performed as previously described (28, 29).
Renilla and firefly luciferase assays.
A dual-luciferase assay kit was used to quantify activities for both Renilla and firefly luciferases (Promega). The following procedures adhered strictly to the manufacturer's instructions unless stated otherwise. For harvesting of lysates at early time points (2 and 4 h posttransfection), six-well dishes were spun at 700 × g for 5 min in a desktop GS-6R centrifuge (Beckman, Fullerton, Calif.) to ensure adherence of transfected cells. The medium was then removed, and the cells were washed twice with PBS and spun for an additional 5 min at 700 × g between the washes. For samples collected at later time points (≥6 h posttransfection), cells had adhered well to the plates and were washed twice with PBS without centrifugation. After the PBS washing, cells were lysed with 500 μl of lysis buffer on a shaker for 25 min. Cell lysates were recovered, homogenized by brief vortexing, and stored at −80°C. Luciferase activity was assayed by mixing 20 μl of lysate with 100 μl of assay reagent and was measured with a Lumat luminometer (EG & G, Berthold, Australia).
RNA extraction and standard RT-PCR.
Cells were harvested by trypsinization at various time points posttransfection. Replicon RNA was extracted using RNeasy kits (Qiagen), quantified by spectrophotometry, and detected with the Onestep reverse transcriptase PCR (RT-PCR) kit (Qiagen). The resulting 388-bp DNA product, representing nt 8706 to 9093 within NS5, was analyzed on a 1.75% agarose gel stained with ethidium bromide (28, 29).
Quantification and stability assays of replicon RNAs using 5′ nuclease real-time RT-PCR.
A 5′ nuclease real-time RT-PCR assay was performed to assess the stability of various replicon RNAs and to quantify the level of RNA replication (27, 28). The assay was performed on an ABI Prism 7700 Sequence Detector using TaqMan One-Step RT-PCR master mix (Applied Biosystems, Foster City, Calif.). The primers and probe were targeted to amplify nt 3111 to 3239 in the NS1 gene (27, 28). RNA was quantified by reference to RNA extracted from a virus stock with a known titer (measured in PFU) or by reference to in vitro-transcribed RNA.
A Megashortscript T7 kit (Ambion) was used to prepare a 489-nt RNA spanning from nt 2937 to 3426 in the NS1 coding sequence. The DNA template was prepared by PCR with a primer containing a T7 promoter upstream of the NS1 sequence. After in vitro transcription, the DNA template was completely removed by three separate treatments with DNase I at 37°C for 30 min. The RNA was phenol-chloroform extracted, precipitated by ammonium acetate and ethanol, resuspended in RNase-free water, quantified by UV spectrophotometry, and calculated as copy numbers for the real-time RT-PCR.
For monitoring RNA replication, replicon RNAs extracted at various time points posttransfection were quantified. For testing of RNA stability, an equal amount of RNA (10 μg) for each replicon RNA was transfected into BHK cells under identical conditions. Replicon RNAs were extracted and quantified at 3, 5, 8, and 12 h posttransfection.
RESULTS
Strategies to engineer a Renilla luciferase (Rluc) reporter into WNV replicons.
We selected Rluc as a reporter for the development of a reporting replicon system due to its relatively small size (936 bp) and its robust enzymatic activity. Two approaches were taken to engineer the Rluc reporter into our original Rep (Fig. 1). The first approach was based on previous results (17, 28), which showed that a reporter protein driven by an IRES could be inserted into the upstream end of the 3′ UTR of flavivirus replicons. We demonstrated that such dicistronic replicons of WNV could express a high level of green fluorescent protein (GFP) when the IRES-GFP cassette was positioned in a plus-sense orientation (28). However, the GFP reporter is not as robust as enzyme-based reporters and therefore could not sensitively quantify the replication level of the replicon RNA. To overcome this limitation, we developed an Rluc version of the dicistronic replicons (Fig. 1). The IRES-Rluc was engineered in either plus- or minus-sense orientation, resulting in (+)3′RlucRep or (−)3′RlucRep, respectively (Fig. 1). Alternatively, the Rluc gene could be fused in-frame with the ORF of the original Rep in the position where a majority of the structural region was deleted, resulting in RlucRep. The Rluc coding sequence was placed between the N-terminal portion of the 5′ C coding sequence (nt 97 to 189) retained to preserve the 5′CS element and the C-terminal region of the E gene (nt 2380 to 2469) required for downstream polyprotein processing. Therefore, the Rluc protein expressed from such a monocistronic replicon would have an extra N-terminal 31 and C-terminal 30 amino acids derived from the C and E proteins, respectively (Fig. 1).
Rluc protein can be expressed only when positioned in the plus-sense orientation in the dicistronic replicon.
Initially, the (+)3′RlucRep was designed to quantify the plus-sense RNA synthesis. In contrast, the (−)3′RlucRep was designed to test, during WNV replication, whether minus-sense RNA would be accessible to translation, and if so, whether the Rluc signal could be used to quantify minus-sense RNA synthesis. BHK cells were transfected with either (+)3′RlucRep or (−)3′RlucRep RNA. Rluc assays and IFA were performed to indicate the RNA synthesis and WNV protein expression at 34, 48, and 72 h posttransfection. Upon transfection of cells with (+)3′RlucRep, the Rluc signals increased dramatically from 34 to 72 h posttransfection (data not shown). The Rluc activity was extremely robust, with levels up to 1.9 × 107 light units at 72 h posttransfection. The increasing Rluc activity correlated with the increasing number of IFA-positive cells (data not shown). These results indicated that the (+)3′RlucRep replicated efficiently in transfected cells and that its replication level could be monitored by the Rluc activity. In contrast, the level of Rluc activity measured from the (−)3′RlucRep-transfected cells was not different from that of mock-transfected cells. However, the (−)3′RlucRep was not replication deficient, as increasing numbers of IFA-positive cells were observed in the population of transfected cells (data not shown). To ensure that the lack of Rluc expression in the (−)3′RlucRep-transfected cells was not due to mutations in IRES-Rluc that were generated during the cloning procedure, we sequenced the entire (−)3′RlucRep construct and found no mutations. These results clearly show that the (−)3′RlucRep replicates efficiently in cells; however, no detectable level of Rluc can be translated from the minus-sense RNA.
In-frame fusion of the Rluc gene into the ORF of the replicon allows for quantification and differentiation of initial translation and RNA replication.
Although the Rluc signal could be used to indicate the replication level of the (+)3′RlucRep, it could not be utilized to directly measure viral translation of the replicon RNA. To overcome this limitation, we constructed a RlucRep that contained the Rluc gene fused in-frame with the ORF of the replicon (Fig. 1). The RlucRep allowed for cap-dependent translation of the Rluc gene along with downstream viral proteins, making it congruent with the normal process of viral translation during natural infection. As a negative control, we constructed a mutant counterpart of RlucRep that contained a frameshift mutation upstream of the active site GDD motif of the RdRp of NS5, resulting in RlucRep-NS5mt (Fig. 1).
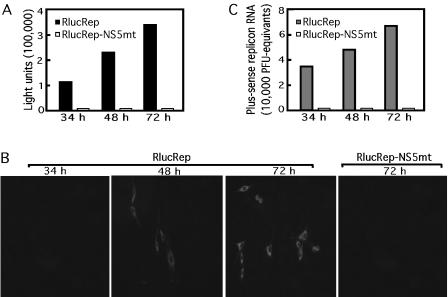
We first tested whether the Rluc activity could indicate the replication level of the RlucRep. Both RlucRep and RlucRep-NS5mt RNAs were transfected into BHK cells, and the Rluc activity was assayed from the cell lysates collected at 34, 48, and 72 h posttransfection. Figure 2A shows that, upon transfection of RlucRep RNA, Rluc activity increased dramatically between 34 and 72 h. Despite the fact that the Rluc protein has extra amino acids fused to both termini, robust Rluc signal was obtained. In contrast, transfection of RlucRep-NS5mt RNA yielded no Rluc activity (Fig. 2A). We then performed IFA to monitor viral protein expression in the transfected cells. As shown in Fig. 2B, no IFA signal was detected at 34 h posttransfection; however, increasing IFA signals were observed from 48 to 72 h in cells transfected with the RlucRep RNA. No IFA-positive signal was detected in cells transfected with the RlucRep-NS5mt RNA. Finally, we performed real-time RT-PCR to quantify the replicon RNA posttransfection. As shown in Fig. 2C, increasing amounts of RlucRep RNA were detected from 34 to 72 h posttransfection. On the other hand, during the same time period, only a background level of RlucRep-NS5mt RNA was observed. The lack of signals of Rluc, IFA, and real-time RT-PCR from the RlucRepNS-5mt-transfected cells was expected because of the inactivation of the viral polymerase of the replicon. Taken together, these results demonstrate that the replication level of the RlucRep can be monitored through the expression level of the Rluc reporter.
FIG. 2.
An Rluc reporter can be used to monitor viral RNA replication. (A) Rluc activities of cell lysates prepared at 34, 48, and 72 h after transfection with RlucRep or RlucRep-NS5mt; (B) IFA of BHK cells transfected with RlucRep or RlucRep-NS5mt at the indicated time points; (C) real-time RT-PCR quantification of RlucRep or RlucRep-NS5mt. RNA was quantified by reference to RNA extracted from a virus stock of a known titer (measured in PFU). Results of one of two representative experiments are shown.
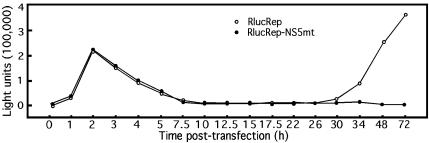
To further characterize the RlucRep system, we transfected BHK cells with an equal amount of RlucRep or RlucRep-NS5mt RNA and monitored Rluc activity within the first 34 h posttransfection. For each replicon transfection, we cotransfected cells with an mRNA transcript encoding a firefly luciferase. Since firefly luciferase and Renilla luciferase have completely different substrates, the activity of the firefly luciferase allowed us to normalize the transfection efficiencies of the two replicons. As shown in Fig. 3, almost identical curves of Rluc activity were obtained from cells transfected with either RlucRep or RlucRep-NS5mt up to 26 h posttransfection. Specifically, the Rluc signal peaked at 2 to 3 h, dropped substantially from 2 to 10 h, and remained at a low level from 10 to 26 h. Remarkably, after 26 h posttransfection, the Rluc signal from cells transfected with the RlucRep RNA rebounded and increased exponentially, while the Rluc signal from cells transfected with the RlucRep-NS5mt RNA remained at a background level. These results indicated that, up to 2 h posttransfection, the observed Rluc activity was derived from the translation of the input RNA. After the 2-h time point, the input RlucRep-NS5mt RNA and the initially translated Rluc protein (with a half-life of approximately 5 h) were degraded, resulting in the decrease of the Rluc signal. Similarly, the Rluc protein initially translated from the input RlucRep RNA was also degraded. However, a certain amount of the transfected RlucRep RNA underwent replication, and the newly synthesized RNA was released and subsequently translated, leading to a second Rluc signal increase after the 26-h posttransfection time point. In contrast, no second Rluc increase was observed in cells transfected with the RlucRep-NS5mt RNA because of its replication deficiency. These results suggest that Rluc signals at 2 to 3 h and at time points later than 26 h posttransfection can be used to quantify the levels of viral translation and RNA replication, respectively.
FIG. 3.
A time course of the Rluc activity in BHK cells transfected with RlucRep or RlucRep-NS5mt RNA. Rluc activity was normalized with respect to a cotransfected mRNA encoding a firefly luciferase. Similar results were obtained in two independent experiments.
Mutations of the CS elements within the 3′ UTR and mutations of the 5′CS do not significantly affect viral translation, but they can severely or completely diminish RNA replication.
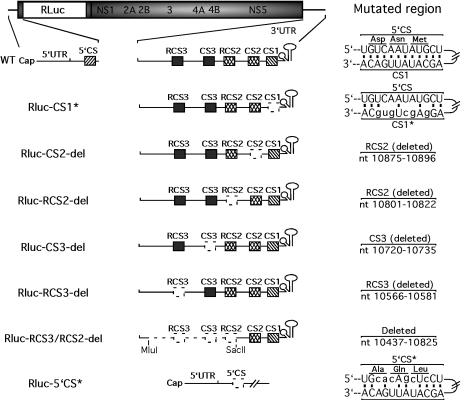
We applied the RlucRep system to examine the mutational effects of the CS elements within the 3′ UTR on WNV translation and RNA replication. A series of RlucRep variants were prepared (Fig. 4). Rluc-CS1* contained six nucleotide changes (indicated in lowercase type in Fig. 4) in the CS1, which disrupted the potential base pairing with the 5′CS element in the capsid-coding sequence. Rluc-CS2-del, Rluc-RCS2-del, Rluc-CS3-del, and Rluc-RCS3-del contained deletions of individual CS elements as indicated. Rluc-RCS3/RCS2-del contained a large truncation from nt 10437 to 10825 within the 3′ UTR, effectively removing RCS3, CS3, and RCS2 but retaining CS2, CS1, and the 3′ terminal stem-loop structure (Fig. 4).
FIG. 4.
Reporting replicons of WNV containing mutations or deletions of the CS elements. The terminal regions of the wild-type (WT) RlucRep have been enlarged to display the 5′CS within the capsid coding sequence and the CS1, CS2, RCS2, CS3, and RCS3 within the 3′ UTR. The potential base pairing between the 5′CS and CS1 is depicted. Rluc-CS1* and Rluc-5′CS* contained six nucleotide changes (indicated in lower case) in the CS1 and 5′CS, respectively, which disrupted the potential 5′CS/CS1 interaction. Rluc-CS2-del, Rluc-RCS2-del, Rluc-CS3-del, and Rluc-RCS3-del contained deletions of individual CS elements as indicated. Rluc-RCS3/RCS2-del contained a large truncation between the MluI and SacII sites. Mutated regions are indicated by dotted lines and are specified in the right column.
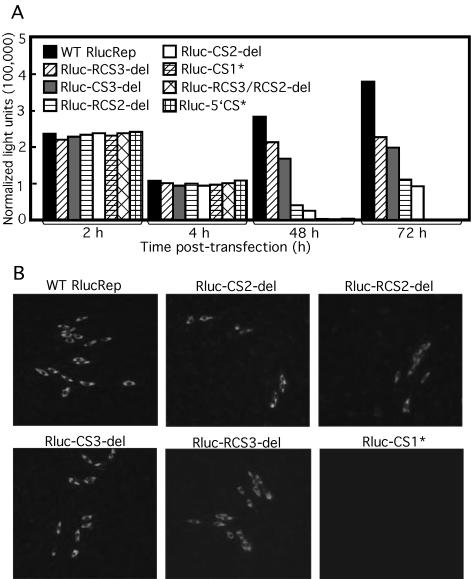
Equal amounts of wild-type and mutant replicon RNAs were transfected into BHK cells. Rluc activities at various time points posttransfection were quantified (Fig. 5A): Rluc signals at 2 and 4 h posttransfection were measured for estimation of the level of viral translation, while Rluc activities at 48 and 72 h posttransfection were quantified for assessment of the level of RNA replication. At 2 and 4 h posttransfection, Rluc signals derived from each mutant were only slightly lower than the signal from the wild type, indicating that none of the mutations significantly affected viral translation. By contrast, at 48 and 72 h posttransfection, Rluc signals were only detected from cells transfected with Rluc-CS2-del, Rluc-RCS2-del, Rluc-CS3-del, Rluc-RCS3-del, and wild-type RlucRep, but not from cells transfected with Rluc-CS1* or Rluc-RCS3/RCS2-del RNA. The Rluc signal disparity at 72 h was more dramatic than that detected at 48 h posttransfection. At the 72-h time point, the Rluc signals derived from the cells transfected with Rluc-CS2-del, Rluc-RCS2-del, Rluc-CS3-del, and Rluc-RCS3-del were approximately 24, 28, 51, and 57%, respectively, of the signal from the cells transfected with the wild-type RlucRep (Fig. 5A).
FIG. 5.
Mutational analyses of the CS elements using the Rluc-containing replicons. (A) Rluc activities in BHK cells transfected with wild-type (WT) or mutant RlucReps. Rluc activities at 2 and 4 h and at 48 and 72 h posttransfection were used to monitor viral translation and RNA replication, respectively. The transfection efficiency of the Rluc-containing replicons was normalized by cotransfection of an mRNA containing a firefly luciferase gene. Results of one of three representative experiments are shown. (B) IFA of BHK cells at 72 h posttransfection with WT or mutant RlucReps.
We also performed IFA to monitor viral protein expression upon transfection of the various replicons. The IFA results correlated well with the Rluc pattern. Positive IFA signals were observed in cells transfected with wild type, Rluc-CS2-del, Rluc-RCS2-del, Rluc-CS3-del, or Rluc-RCS3-del but not in cells transfected with Rluc-CS1* (Fig. 5B) or Rluc-RCS3/RCS2-del (data not shown).
To analyze the role of 5′CS on viral translation or RNA replication, we prepared an Rluc-5′CS* replicon that contained six nucleotide changes (indicated in lowercase type in Fig. 4) in the 5′CS, resulting in partial disruption of the 5′CS-CS1 base pairing. Since the 5′CS resides in the capsid-coding region, mutations within the 5′CS altered three amino acid residues (Fig. 4). Transfection of BHK cells with Rluc-5′CS* exhibited Rluc activities similar to those transfected with wild-type RlucRep at 2 and 4 h posttransfection (Fig. 5A). However, no Rluc signal was detected from cells at 48 and 72 h posttransfection (Fig. 5A). No positive IFA signal was observed from cells at 72 h after transfection with the Rluc-5′CS* (data not shown).
Finally, we recovered and sequenced the replicons. RT-PCR targeting of the complete 3′ UTR of WNV was performed on RNAs extracted from cells at 72 h posttransfection. DNA products of the expected sizes were obtained from cells transfected with wild-type RlucRep, Rluc-CS2-del, Rluc-RCS2-del, Rluc-CS3-del, and Rluc-RCS3-del, but not from cells transfected with Rluc-CS1* or Rluc-RCS3/RCS2-del (data not shown). These results were in agreement with the Rluc (Fig. 5A) and IFA (Fig. 5B) data. DNA sequencing of the RT-PCR products confirmed that each of the recovered replicons retained the engineered mutations. Overall, the results suggest that mutations or deletions of the CS elements within the 3′ UTR as well as the 5′CS mutation do not significantly affect viral translation but can severely diminish or completely abolish RNA replication.
The difference in RNA replication among various replicons is not due to differences in RNA stability.
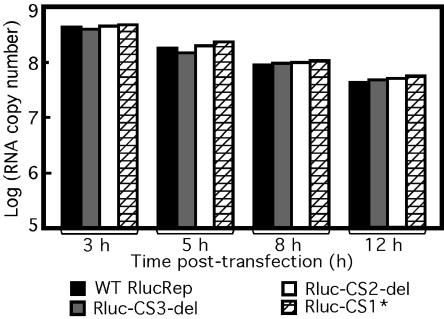
To exclude the possibility that the observed differences in RNA replication among various replicons were due to differences in RNA stability, we compared the RNA stability of wild-type RlucRep, Rluc-CS1*, Rluc-CS2-del, and Rluc-CS3-del in transfected cells. Equal amounts of RNA for each construct were transfected into BHK cells, and viral RNAs were extracted and quantified by real-time RT-PCR at 3, 5, 8, and 12 h posttransfection (Fig. 6). A dramatic decrease of replicon RNA was detected from 3 to 12 h posttransfection. Approximately 20% of RNA present at 3 h posttransfection was retained at 12 h posttransfection. However, no dramatic copy number differences were observed among various replicons at the same time points posttransfection. These results suggest that the observed replication differences of mutant replicons are not due to differences in RNA stability.
FIG. 6.
RNA stability of WT and mutant replicons in BHK cells. Equal amounts of WT RlucRep, Rluc-CS1*, Rluc-CS2-del, and Rluc-CS3-del were transfected into BHK cells as described in Materials and Methods. Replicon RNAs were extracted and quantified by real-time RT-PCT at 3, 5, 8, or 12 h posttransfection. The amount of RNA at each time point was estimated by reference to in vitro-transcribed RNA in copy number.
Rluc coding sequence is not responsible for changes in RNA replication for the mutant replicons.
To ensure that the mutational effects observed in the reporter replicon system were not substantially influenced by the addition of the Rluc coding sequence, we engineered individual mutations or deletions into the original Rep without the Rluc reporter. Six mutant counterparts were constructed: CS1*, CS2-del, RCS2-del, CS3-del, RCS3-del, and 5′CS*. The replication efficiency of each mutant, along with that of the wild-type replicon, was monitored by standard RT-PCR using RNA extracted from cells at 72 h posttransfection (Fig. 7). The resulting 388-bp DNA product, representing nt 8706 to 9093 within the NS5 gene, was analyzed on an agarose gel. Compared with the wild-type replicon, decreasing amounts of RT-PCR products were detected from the mutants, in this order: RCS3-del, CS3-del, RCS2-del, and CS2-del. No DNA product was observed from cells transfected with CS1* and 5′CS* (Fig. 7). These data correlated well with the levels of Rluc activity (Fig. 5A) and with the IFA results (Fig. 5B) derived from the Rluc-containing replicons, suggesting that the Rluc signals authentically reflect the level of replication of individual replicons.
FIG. 7.
Mutational analysis of the CS elements in replicons without a reporter. The original replicon without the Rluc reporter (Rep, Fig. 1) was used to introduce mutations or deletions as depicted in Fig. 4. The replication efficiency of the resulting replicons, CS1*, CS2-del, RCS2-del, CS3-del, and RCS3-del, along with that of the WT Rep, was monitored by standard RT-PCR using RNA extracted from cells at 72 h posttransfection. The resulting 388-bp DNA fragments were detected on an agarose gel (A). CS1* and 5′CS* contained six nucleotide changes (indicated in lowercase) within the CS1 and 5′CS element, respectively, which blocked the potential 5′CS-CS1 base pairing. 5′CS*/CS1* contained mutations in both 5′CS and CS1 with complementary sequences, restoring the 5′CS-CS1 interaction (B).
Restoration of the potential 5′CS-CS1 base pairing rescues the replication of WNV replicon.
As shown in Fig. 5 and 7, mutations in CS1 and 5′CS that partially blocked the 5′CS-CS1 interaction abolished RNA replication. To test whether restoration of the 5′CS-CS1 base pairing could rescue RNA replication, we constructed 5′CS*/CS1* using the original Rep containing no reporter. The 5′CS*/CS1* construct contained six nucleotide mutations within both 5′CS and CS1. However, the potential complementarity between 5′CS and CS1 was restored (indicated in lowercase type in Fig. 7B). Standard RT-PCR was used to monitor the replication of the replicon variants. Wild-type Rep yielded high levels of RT-PCR product of expected size, while CS1* and 5′CS* mutants generated no product (Fig. 7B). On the other hand, a substantial amount of RT-PCR product was obtained from the 5′CS*/CS1*-transfected cells. These results suggest that the potential base pairing between 5′CS and CS1 is essential for the replication of WNV RNA.
DISCUSSION
We have developed a reporting replicon of WNV that can differentiate between viral translation and RNA replication. As far as we are aware, this is the first report of such a system developed for flaviviruses. This replicon contains an Rluc gene fused in-frame with the viral ORF in the position where the structural genes were deleted. Using the reporting system, we initiated functional analyses of the CS elements of mosquito-borne flavivirus 3′ UTRs. Our results showed that mutations or deletions of the CS elements within the WNV 3′ UTR and 5′ CS do not significantly affect viral translation but can severely diminish or completely abolish RNA replication.
Mutations within the CS1 that partially blocked its base pairing with the 5′CS abolished the replication of the WNV replicon; restoration of the 5′CS-CS1 interaction with nonviral sequences rescued the replication. These results are in agreement with the emerging theme that plus-strand RNA viruses utilize interaction between the 5′ and 3′ termini of viral RNAs as a common strategy to regulate gene expression and RNA replication. Recent studies of poliovirus replication suggest that the 5′ and 3′ RNA termini coordinately mediate minus-strand RNA synthesis through protein-protein interactions (2, 14). Bacteriophage Qβ replication requires genome cyclization through RNA-RNA interactions (18). The essential role of interaction between 5′ and 3′ termini of RNA has also been indicated for Brome mosaic virus (12, 19) and coronavirus (15, 30). Among the flaviviruses, an essential role of the 5′CS-CS1 interaction in viral replication was recently demonstrated in replicons of Kunjin virus (16) and YF virus (10, 22). A minimum of 18 bp of duplex RNA formed between the 5′CS and CS1 was required for YF replication (10). Using cytoplasmic extracts from DEN viral-infected cells and recombinant RdRp, Ackermann and Padmanabhan showed that cyclization of DEN RNA template was essential for polymerase activity in vitro (1, 33). The latter results suggested that the 5′CS-CS1 duplex contains intrinsic signals for modulation of RdRp activities. Our results indicate that the 5′CS-CS1 interaction is essential for RNA replication but does not significantly regulate viral translation. Further studies are required to determine whether RNA cyclization functions at the stage of plus-strand RNA synthesis, minus-strand RNA synthesis, or both.
Deletions of individual CS2, RCS2, CS3, and RCS3 from WNV replicons significantly reduced the RNA replication but not viral translation. The deletions of these CS elements may affect the gross conformation of the RNA template, making it less optimal for RNA replication. The results suggest that CS2, RCS2, CS3, and RCS3 elements facilitate, but are not essential for, RNA replication. Previous studies showed that DEN virus type 4 mutants containing deletions of the 3′ UTR upstream of the CS1 were viable (21). Compared with the wild-type DEN virus, the viable deletion mutants exhibited a range of growth restriction in cultured cells as well as altered viremia pattern and immunogenicity in rhesus monkeys (21). It is noteworthy that the 3′ UTR of DEN virus lacks the CS3 and RCS3, which exist only in the YF and JE subgroup of flaviviruses, including WNV. Studies using a Kunjin virus replicon showed that deletion of the 3′ UTR upstream of RCS2 is almost lethal (17). Our results showed that further deletion of the RCS2, but retention of the CS2, CS1, and the 3′ terminal stem-loop (i.e., construct Rlu-RCS3/RCS2-del), completely abolishes WNV replication. These results indicate that attenuated WNV can be obtained through deletions of individual or combinatory CS elements within the 3′ UTR. Since the CS elements are conserved among the mosquito-borne flaviviruses, experiments are in progress to compare the replication differences between the wild-type and CS mutant WNV in both mammalian and mosquito cells. Further studies to compare the in vivo transmission capability by mosquitoes of the wild-type and the CS mutant viruses will provide more insights on the biological functions of the CS elements in WNV.
We also showed that Rluc reporter driven by an IRES can be engineered in the upstream region of the 3′ UTR. Compared with the in-frame fused RlucRep, the Rluc activity derived from the cells transfected with the (+)3′RlucRep was much stronger, e.g., a difference of approximately 60-fold was detected at 72 h posttransfection. The difference in intensity of the reporting signal between the (+)3′RLucRep and RlucRep could be due to one or more of the following. (i) In the (+)3′RlucRep, Rluc was expressed as an intact protein, while the Rluc expressed from the RlucRep was fused with the N-terminal 31 amino acids of the capsid gene and the C-terminal 30 amino acids of the E gene. The fusion partners may reduce the luciferase activity. (ii) The efficiency of an IRES-driven translation [in (+)3′RlucRep] may be more robust than that of the cap-scanning translation (in RlucRep). (iii) Insertion of the Rluc in the structural gene coding region (in RlucRep) may have a stronger negative effect on viral replication than does its insertion in the 3′ UTR (in (+)3′RlucRep).
Using the (+)3′RlucRep and (−)3′RlucRep, we found that although (−)3′RlucRep replicates efficiently in transfected cells, no detectable Rluc activity was observed, whereas a high level of Rluc was detected in the (+)3′RlucRep-transfected cells. Similar results were obtained for the GFP-containing replicons (28). One simple explanation is that, since the amount of minus-sense RNA synthesized was much less than that of plus-sense RNA, insufficient minus-sense RNA was synthesized to express a detectable level of Rluc from the (−)3′RlucRep. The extreme robustness of the Rluc signal in our system argues against this explanation. We previously estimated that the ratio of plus-sense to minus-sense RNA increased from several hundred- to 1,000-fold during culturing of the cells transfected with replicons (28). If the minus-sense RNA were accessible for translation, a detectable Rluc activity would have been observed. Therefore, we postulate that the lack of Rluc expression from the (−)3′RlucRep is due to the inaccessibility of the minus-sense RNA to the translational machinery, presumably because of its association with the plus-sense RNA in the form of replication forms or replication intermediates, according to the current model of flavivirus RNA synthesis (9).
The reporting replicons described in this paper will be useful tools to quickly screen for elements involved in viral translation and RNA replication. However, caution should be taken when interpreting the results derived from such replicons, as should be done for all reporting systems. A potential complication is that the nonviral reporter protein and its coding sequence could interfere with the viral elements of interest. However, when combined with other genetic tools such as full-length infectious clones, the reporting replicon system should greatly facilitate studies of many aspects of WNV replication.
Acknowledgments
Michael K. Lo and Mark Tilgner contributed equally to this work.
We thank Paul Masters for helpful discussions during the course of this work. We also thank the Molecular Genetics Core and the Cell Culture Facility at the Wadsworth Center for sequencing and oligonucleotide synthesis and for maintenance of BHK cells, respectively.
The work was funded in part by the National Institute of Allergy and Infectious Disease, National Institutes of Health under contract no. N01-AI-25490. M.K.L. was supported by the Emerging Infectious Diseases Fellowship Program funded by the New York State Department of Health.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ Cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthet, F. X., H. G. Zeller, M. T. Drouet, J. Rauzier, J. P. Digoutte, and V. Deubel. 1997. Extensive nucleotide changes and deletions within the envelope glycoprotein gene of Euro-African West Nile viruses. J. Gen. Virol. 78:2293-2297. [DOI] [PubMed] [Google Scholar]
- 4.Brinton, M. A., and J. H. Dispoto. 1988. Sequence and secondary structure analysis of the 5′-terminal region of flavivirus genome RNA. Virology 162:290-299. [DOI] [PubMed] [Google Scholar]
- 5.Brinton, M. A., A. V. Fernandez, and J. H. Dispoto. 1986. The 3′-nucleotides of flavivirus genomic RNA form a conserved secondary structure. Virology 153:113-121. [DOI] [PubMed] [Google Scholar]
- 6.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1126. In D. M. Knipe, and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 7.Centers for Disease Control and Prevention. 2002. Provisional surveillance summary of the West Nile virus epidemic—United States, January-November 2002. Morb. Mortal. Wkly. Rep. 51:1129-1133. [PubMed] [Google Scholar]
- 8.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 9.Chu, P. W., and E. G. Westaway. 1987. Characterization of Kunjin virus RNA-dependent RNA polymerase: reinitiation of synthesis in vitro. Virology 157:330-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corver, J., E. Lenches, K. Smith, R. Robison, T. Sando, E. Strauss, and J. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 77:2265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diamond, M. S., D. Edgil, T. G. Roberts, B. Lu, and E. Harris. 2000. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J. Virol. 74:7814-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 14.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, P., and M. M. C. Lai. 2001. Heterogeneous nuclear ribonucleoprotein A1 binds to the 3′-untranslated region and mediates potential 5′-3′-end cross talks of mouse hepatitis virus RNA. J. Virol. 75:5009-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klovins, J., and J. van Duin. 1999. A long-range pseudoknot in Qbeta RNA is essential for replication. J. Mol. Biol. 294:875-884. [DOI] [PubMed] [Google Scholar]
- 19.Lahser, F. C., L. E. Marsh, and T. C. Hall. 1993. Contributions of the brome mosaic virus RNA-3 3′-nontranslated region to replication and translation. J. Virol. 67:3295-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanciotti, R. S., J. T. Roehrig, V. Deubel, J. Smith, M. Parker, K. Steele, B. Crise, K. E. Volpe, M. B. Crabtree, J. H. Scherret, R. A. Hall, J. S. MacKenzie, C. B. Cropp, B. Panigrahy, E. Ostlund, B. Schmitt, M. Malkinson, C. Banet, J. Weissman, N. Komar, H. M. Savage, W. Stone, T. McNamara, and D. J. Gubler. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333-2337. [DOI] [PubMed] [Google Scholar]
- 21.Men, R., M. Bray, D. Clark, R. M. Chanock, and C. J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molenkamp, R., E. Kooi, M. Lucassen, S. Greve, J. Thijssen, W. Spaan, and P. Bredenbeek. 2003. Yellow fever virus replicons as an expression system for hepatitis C virus structural proteins. J. Virol. 77:1644-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muylaert, I. R., T. J. Chambers, R. Galler, and C. M. Rice. 1996. Mutagenesis of the N-linked glycosylation sites of the yellow fever virus NS1 protein: effects on virus replication and mouse neurovirulence. Virology 222:159-168. [DOI] [PubMed] [Google Scholar]
- 24.Rice, C. M., E. M. Lenches, S. R. Eddy, S. J. Shin, R. L. Sheets, and J. H. Strauss. 1985. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science 229:726-733. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Shi, P. Y., M. A. Brinton, J. M. Veal, Y. Y. Zhong, and W. D. Wilson. 1996. Evidence for the existence of a pseudoknot structure at the 3′ terminus of the flavivirus genomic RNA. Biochemistry 35:4222-4230. [DOI] [PubMed] [Google Scholar]
- 27.Shi, P. Y., E. B. Kauffman, P. Ren, A. Felton, J. H. Tai, A. P. Dupuis II, S. A. Jones, K. A. Ngo, D. C. Nicholas, J. Maffei, G. D. Ebel, K. A. Bernard, and L. D. Kramer. 2001. High-throughput detection of West Nile virus RNA. J. Clin. Microbiol. 39:1264-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 29.Shi, P. Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 76:5847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spagnolo, J. F., and B. G. Hogue. 2000. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 74:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallner, G., C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. The flavivirus 3′-noncoding region: extensive size heterogeneity independent of evolutionary relationships among strains of tick-borne encephalitis virus. Virology 213:169-178. [DOI] [PubMed] [Google Scholar]
- 32.Yamshchikov, V. F., G. Wengler, A. A. Perelygin, M. A. Brinton, and R. W. Compans. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294-304. [DOI] [PubMed] [Google Scholar]
- 33.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′ end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]