Abstract
Difficulties in determining composition and sequence of glycosaminoglycans, such as those related to heparin, have limited the investigation of these biologically important molecules. Here, we report methodology, based on matrix-assisted laser desorption ionization MS and capillary electrophoresis, to follow the time course of the enzymatic degradation of heparin-like glycosaminoglycans through the intermediate stages to the end products. MS allows the determination of the molecular weights of the sulfated carbohydrate intermediates and their approximate relative abundances at different time points of the experiment. Capillary electrophoresis subsequently is used to follow more accurately the abundance of the components and also to measure sulfated disaccharides for which MS is not well applicable. For those substrates that produce identical or isomeric intermediates, the reducing end of the carbohydrate chain was converted to the semicarbazone. This conversion increases the molecular weight of all products retaining the reducing terminus by the “mass tag” (in this case 56 Da) and thus distinguishes them from other products. A few picomoles of heparin-derived, sulfated hexa- to decasaccharides of known structure were subjected to heparinase I digestion and analyzed. The results indicate that the enzyme acts primarily exolytically and in a processive mode. The methodology described should be equally useful for other enzymes, including those modified by site-directed mutagenesis, and may lead to the development of an approach to the sequencing of complex glycosaminoglycans.
The growing interest in glycoprotein, glycolipid, and proteoglycan function in biology increases the demand for methods to characterize the structure of oligosaccharides (1). Heparan sulfate proteoglycans are probably the most complex glycoconjugates (2). One or more heparin-like glycosaminoglycan chains (HLGAGs)—heterogenous in length, composition, and sequence—are linked covalently to a single core protein and can account for 50–90% of the molecular weight of the proteoglycan (3). HLGAGs are linear chains of 20–100 disaccharide units comprised of a uronic acid (iduronic or glucuronic acid) and a glucosamine. Various disaccharide modifications are introduced by enzymatic deacetylation, epimerization, and sulfation resulting in chemically highly heterogeneous, mature HLGAG chains (3). Many proteins have been shown to bind HLGAGs with high affinity, but only in a few cases (e.g., antithrombin III and basic fibroblast growth factor) has the HLGAG sequence responsible for high affinity binding been elucidated (4, 5). This is, at least in part, because of the lack of effective and simple methods to isolate and characterize HLGAG oligosaccharides. Full length HLGAGs cannot be separated into individual components because of the large number of different HLGAG chains of closely related structure. Attempts at sequencing therefore have focused on oligosaccharide fragments obtained by HLGAG depolymerization. Such fragments are generated either chemically by nitrous acid, copper/peroxide, or base (6) or enzymatically by heparinases (7). The resulting mixtures of oligosaccharide fragments then are fractionated by size exclusion chromatography, HPLC anion exchange chromatography, or gel electrophoresis to obtain discrete components. Composition and sequence of the components then are deduced from enzymatic or chemical reactions in combination with electrophoresis, chromatography (5), NMR (8), or tandem MS (9). This approach can provide the complete sequence of small oligosaccharide fragments, but accuracy of sequence assignments decreases with HLGAG chain length. Furthermore, relatively large amounts of material (nano- to micromolar quantities) are required.
Recently, we developed a highly sensitive method for obtaining the molecular weight of femto- to picomolar amounts of HLGAG oligosaccharides by matrix-assisted laser desorption ionization MALDI MS (10). The composition can be deduced from the molecular weight alone (11), but structural information is limited because stereoisomers (iduronic acid vs. glucuronic acid) and positional isomers (e.g., sulfation of the 3- vs. 6-hydroxyl groups of glucosamine) cannot be distinguished.
For oligosaccharides derived from glycoproteins, MS has been combined with selective glycosidases for sequencing (12). Bacterial heparinases can be used to obtain composition and sequence of HLGAGs (13), but these enzymes primarily have been used to determine the disaccharide composition of HLGAGs after exhaustive digestion and information about the digested regions of the substrate is lost. Incomplete digestion, however, would generate oligosaccharide fragments of specific length and composition, which then could be analyzed to obtain sequence information. Here, we report the development of a methodology to study HLGAGs by a combination of MALDI MS, capillary electrophoresis (CE), and HLGAG- degrading enzymes (heparinases). We used these techniques to investigate the dynamics of heparinase I depolymerization of HLGAG oligosaccharides.
EXPERIMENTAL PROCEDURES
Materials.
Substrate H1 was a gift from D. Tyrrell of Glycomed (Alameda, CA) (14), and T5 was a gift of P. Østergaard of Novo-Nordisk (Gentofte, Denmark). Substrates O2 and D3 kindly were provided by R. J. Linhardt after repurification by R. Hileman of the University of Iowa (15). Oligosaccharides were dissolved in deionized water at concentrations of 10–35 μM. Sucrose octasulfate (SOS) was added as an internal standard where indicated (final concentration: 10 μM). Heparinase I was derived from Flavobacterium heparinum or in recombinant form from Escherichia coli cultures (16). Buffer A was 1 mM calcium acetate and 10 mM ethylenediamine/acetic acid (pH 7.0). Buffer B was 1 mM calcium acetate, 10 μM ovalbumin, 1 μM dextran sulfate, 50% glycerol, and 10 mM ethylenediamine/acetic acid (pH 7.0). Buffer C was 10 μM ovalbumin, 1 μM dextran sulfate, 50% glycerol, and 20 mM phosphoric acid/NaOH (pH 7.0). Enzyme solutions in buffer B and C were maintained at −15°C (Nalgene cryobox) and pipetted without prior warming.
Digests.
Enzyme reactions were performed by adding 1 μl of enzyme solution to 5 μl of substrate solution. The mass spectra shown in Figs. 1 and 3A were obtained by using 100 nM heparinase I in buffer A. All other digests were obtained by using a solution of 100 nM enzyme in buffer B (heparinase I) or buffer C (heparinase II). Digests were carried out at room temperature (22°C). Each time point was obtained by removing 0.5 μl of the reaction mixture and adding it to 4.5 μl of matrix solution.
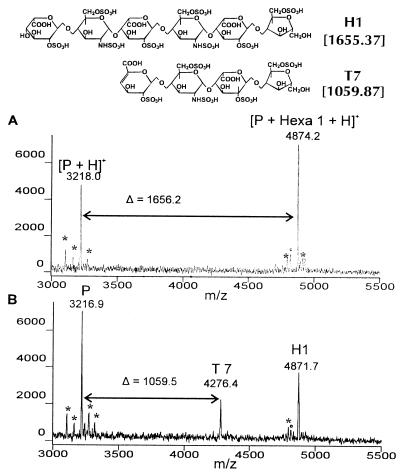
Figure 1.
MALDI mass spectrum of substrate H1 before (A) and after (B) digestion with heparinase I for 30 s. P, the protonated basic peptide (arg-gly)15; T7 (etc.), its protonated complex with HLGAG oligosaccharides. Peaks marked with an asterisk (∗) are peptide impurities (see text), and those marked with a circle (°) are -SO3 fragments of HLGAG oligosaccharides.
Figure 3.
(A) MALDI mass spectrum of a 3-min digest of O2 with heparinase I. (B) A 10-min digest with heparinase II.
Derivatizations.
Derivatizations were carried out by reacting 5 μl of oligosaccharide solution with 5 μl of 50 mM semicarbazide and 60 mM Tris/acetic acid (pH 7.0, prepared fresh daily) for 16 h at 30°C. Before digestion with enzyme, the pH was adjusted to 7.0 by the addition of 1 μl of 50 mM Tris.
MS.
Caffeic acid (≈12 mg/ml) in 70% acetonitrile/water was used as matrix solution. Seeded surfaces were prepared by the method of Xiang and Beavis (17). A 2-fold molar excess of basic peptide (arg-gly)15 was premixed with matrix before being added to the oligosaccharide solution (sample-to-matrix ratio was 1:10) (18). A 1-μl aliquot of sample/matrix mixture (1–3 pmol of oligosaccharide) was deposited on the seeded surface (stainless steel plate). After crystallization had occurred (typically within 60 s), excess liquid was rinsed off with water. MALDI MS spectra were acquired in the linear mode by using a PerSeptive Biosystems (Framingham, MA) Voyager Elite reflectron time-of-flight instrument fitted with a 337-nm nitrogen laser. Delayed extraction (19) was used to increase resolution (22 kV, grid at 93%, guide wire at 0.15%, pulse delay 150 ns, low mass gate at 1,000, 128 shots averaged). Mass spectra were calibrated externally by using the signals for protonated (arg-gly)15 and its complex with oligosaccharide H1.
CE.
CE was performed on a Hewlett–Packard 3DCE unit by using uncoated fused silica capillaries (i.d. 75 μm, o.d. 363 μm, ldet. 72.1 cm, and ltot 80.5 cm). Analytes were monitored by using UV detection at 230 nm (20) and an extended light path cell (Hewlett–Packard). The electrolyte was a solution of 10 μM dextran sulfate and 50 mM Tris/phosphoric acid (pH 2.5). Dextran sulfate was used to suppress nonspecific interactions of HLGAG oligosaccharides with the silica wall. Separations were carried out at 30 kV with the anode at the detector side (reversed polarity). A mixture of 1,5-naphtalenedisulfonic acid and 2-naphtalenesulfonic acid (10 μM each) was used as internal standard where indicated.
RESULTS
Enzymatic Depolymerization and Sample Preparation.
Conventional reaction and buffer conditions for heparinase I digestion of HLGAGs were modified to be compatible with MALDI MS analysis and to yield intermediate enzymatic reaction products after experimentally convenient incubation times. Conventional digestion buffers, such as 100 mM 3-(N-morpholino)propanesulfonic acid (Mops) and 5 mM calcium acetate (pH 7.0) (16), were used initially. However, the high concentrations of buffer ions and inorganic salt were incompatible with MALDI. By testing the activity of heparinase I in different buffer systems, a solution (buffer A) was found to combine low ionic strength with optimum enzyme activity. Digests were complete in 10 min for hexasaccharide H1 and in <1 min for larger substrates when using 500 nM heparinase I in buffer A. Under these conditions, it was difficult to detect enzymatic intermediates of the larger substrates because aliquots could not be taken fast enough. At a concentration of 100 nM, heparinase I activity was lost within 20 min. Hence, it was not practical to lower the enzyme concentrations to slow the reaction.
A modified buffer therefore was developed to ensure a low, but stable, enzyme activity. This new buffer B consisted of buffer A plus 10 μM ovalbumin, 50% glycerol, and 1 μM dextran sulfate. It was equally applicable for heparinase I derived from F. heparinum and for recombinant heparinase I derived from E. coli (16). Ovalbumin was chosen over the commonly used BSA because of its low ionization efficiency in MALDI MS. The facile ionization of the latter protein suppressed the signals of the oligosaccharide–peptide complexes. Glycerol prevented freezing of the solution, thus eliminating denaturation by repeated freeze-thaw cycles. Dextran sulfate played a dual role in the modulation of enzyme activity: time-dependent losses of enzyme activity were eliminated fully by dextran sulfate so that solutions as low as 1 nM heparinase I maintained full activity for 24 h and longer; furthermore, the absolute level of enzyme activity was lowered proportional to the concentration of dextran sulfate. Thus, the rate of depolymerization of HLGAG oligosaccharides now could be influenced in a reproducible manner by either lowering the concentration of enzyme or by increasing that of dextran sulfate. The conditions best suited for MALDI MS analysis of intermediates and end products were 100 nM heparinase I dissolved in buffer B.
The conventional “dried droplet” technique (using unseeded crystallization) of preparing MS samples failed in this case because nascent crystals did not form or adhere to the metal surface because of the presence of glycerol. Using surfaces “seeded” with crushed crystals eliminated this problem (17). Despite the presence of glycerol, polycrystalline films formed rapidly (in less than 60 s) on such surfaces. Caffeic acid was found to be the most reliable matrix and was used for all subsequent experiments. Another advantage is the improvement in quantitation because relative signal intensities are far more reproducible because of the uniform sample preparation.
Mass Spectrometric Analysis.
MS not only measures the abundance of each component in a mixture (albeit semiquantitatively) without prior separation but also permits the structural characterization of compounds without the need for well defined standards. Furthermore, MALDI MS is very sensitive and easy to use.
To overcome problems caused by the large number of negative charges, HLGAG oligosaccharides were detected as noncovalent complexes with the basic peptide (arg-gly)15 (10, 18). The molecular weight of HLGAG oligosaccharides then was calculated by subtracting the measured m/z value of the protonated peptide (calculated m/z 3217.62) from the measured m/z value of the protonated peptide/oligosaccharide complex. For substrate H1, a molecular weight of 1656.2 was obtained (Fig. 1A). A peak at m/z 4793.3 (marked o) was detected with an abundance of ≈10% of the peptide/substrate H1 complex (see Table 1 for correlation of molecular weight with m/z of protonated complexes). The calculated molecular weight is 1575.3 Da, which is 80.9 Da lower than that of substrate H1. This is a consequence of the loss of a sulfate group (in the form of SO3, Δ molecular weight = 80.06 Da) during ionization in the mass spectrometer and is not the result of sample inhomogeneity (18). Peaks marked with an asterisk (−58 Da, −114 Da, +54 Da, +98 Da) are impurities or adducts (−Gly, −Arg, +Fe, and +H3PO4) of the basic peptide used.
Table 1.
Molecular weights of HLGAG oligosaccharides and m/z values of their protonated complexes with (arg-gly)15
| Structure | MW | MW deriv. | m/z complex | m/z deriv. c. |
|---|---|---|---|---|
| H1 (Fig. 1) | 1655.37 | N.A. | 4872.99 | N.A. |
| O2 (Fig. 3) | 2229.85 | N.A. | 5447.47 | N.A. |
| D3 (Fig. 4) | 2887.40 | 2943.47 | 6105.02 | 6161.09 |
| T5 (Fig. 2) | 1154.96 | 1211.03 | 4372.58 | 4428.65 |
| Di6 (Fig. 5) | 557.48 | (633.55) | 3795.10 | (3851.17) |
| T7 (Fig. 1) | 1059.87 | N.A. | 4277.49 | N.A. |
| H8 (Fig. 3) | 1732.44 | 1788.51 | 4950.06 | 5006.13 |
| T9 (Fig. 3) | 1074.89 | N.A. | 4292.51 | N.A. |
| H10 (Fig. 3) | 1652.37 | N.A. | 4869.99 | N.A. |
| O11 (Fig. 4) | (2309.92) | (2365.99) | (5527.54) | (5583.61) |
The numbering is coordinated with the accompanying paper by Ernst et al. (23). Substrates are numbered 1–3 and products 5–11. Di, disaccharide; T, tetrasaccharide; H, hexasaccharide; O, octasaccharide; D, decasaccharide; MW, calculated molecular weight (average of isotope cluster) of neutral oligosaccharide; m/z, calculated values of protonated complex with (arg-gly)15; N.A., not applicable. Numbers in parentheses are oligosaccharides that were not observed.
After digestion with heparinase I, a new peak at m/z 4276.4 appeared and the abundance for the complex of substrate H1 with the basic peptide decreased (Fig. 1B). The calculated molecular weight of 1059.5 identifies this component as T7 (Fig. 1), a Δ4,5 unsaturated tetrasaccharide produced by eliminative cleavage (5) of H1. Previous experiments (21) using commercial heparinase I from Sigma instead of heparinase I purified to homogeneity from F. heparinum or E. coli had resulted in abundant peaks at −80 Da because of the exolytic activity of contaminating sulfatases. No increase in the signal at −80 Da was observed in this study, indicating the absence of sulfatases.
It should be noted that the number of chemical structures that can match a particular molecular weight is restricted by the compositions possible for HLGAG oligosaccharides. For natural heparin, only the residue masses (11) of uronic acid, glucosamine, and acetyl- and sulfate groups need to be taken into account. The smallest mass difference that can occur among HLGAG oligosaccharides is 4 Da, corresponding to the difference in mass between two acetyl groups (2 × 42 Da) and a sulfate group (80 Da). In the mass range of m/z 3,000–7,000, we obtained a mass accuracy of ±1 Da under the described experimental conditions. Hence, composition (including the number of acetyl and sulfate groups) can be deduced from molecular weight.
It was observed that peptide complexes of HLGAG oligosaccharides ionize with different efficiencies depending on the number of sulfate groups. In these and other experiments, disaccharides, which contain three or fewer sulfates, generally could not be observed. Their complexes with (arg-gly)15 appear to be too weak. Disaccharide Di6 was detected occasionally; however, its peak height did not reproducibly correlate with concentration. The presence of larger HLGAG oligosaccharides tends to suppress the signal of the Di6/peptide complex. The mass spectrum of an equimolar mixture of tetrasaccharide T7 and hexasaccharide H1 showed a ratio of peak heights of 1:2, indicating that hexasaccharides ionize with twice the efficiency of tetrasaccharides. Octasaccharides had an ionization efficiency approximately equal to hexasaccharides. HLGAGs with 12 or more sulfate groups (most decasaccharides) tended to ionize poorly. Therefore, the disappearance of a decasaccharide substrate could not be monitored directly.
The ratios of peak heights for well ionizing complexes in MALDI MS were reproducible to within ±10% when the sample was prepared as a polycrystalline film. Absolute peak heights were less reproducible. Therefore, an internal standard had to be used to allow quantitative comparisons between samples. SOS was chosen because it complexes well with (arg-gly)15 and is therefore required only in very low quantities; it is not cleaved by heparinase I (or any other heparinase); the protonated complex appears at m/z 4200.43 (molecular weight 982.81), lower than that of most HLGAG oligosaccharides; and it does not inhibit heparinase I activity. Enzyme digests were monitored by removing aliquots at several time points. Matrix solution (caffeic acid) then was added to each aliquot, and the resulting drop in pH stopped the digestion.
To compare the relative rates of degradation of substrates of different length, a mixture of compounds H1, T5, and SOS was digested and analyzed (Fig. 2) (22). The complex of (arg-gly)15 with T7 (molecular weight = 1059.9 Da), a digestion product of H1, appeared at m/z 4276.7, clearly separated from the complex of T5 at m/z 4371.5 (Fig. 2A). Because of the mass differences, each component of the mixture could be followed individually by MS. The peak height (relative to SOS) of hexasaccharide H1 declined more rapidly during the course of digestion than did that of tetrasaccharide T5 (Fig. 2B). Nonlinear least squares fitting of the data to a model for enzymatic degradation of H1, T5, and T7 showed that the apparent first-order rate constant for degradation of hexasaccharides is one order of magnitude larger than it is for tetrasaccharides. Octasaccharide O2 (Fig. 3) and decasaccharide D3 (Fig. 4) were digested under the same experimental conditions to explore the action of heparinase I on larger oligosaccharides. Tetrasaccharides T9 and T5 (peak ratio 10:1), but no hexasaccharide intermediates, were observed when O2 was digested (Fig. 3A). The hexasaccharide at m/z 4790.1 is a sample impurity resistant to heparinase I. Fig. 3B represents the spectrum obtained with heparinase II; it is described later. In the case of decasaccharide D3, T5 (at m/z 4371.6) was the dominant digestion product (Fig. 4A). Its peak height continuously increased over a time period of 2 h (Fig. 4B). A signal for hexasaccharide H8 appeared at m/z 4949.0 after 40 min and was significantly weaker than that of T5. The sensitivity of the mass spectrometer was too low to yield a detectable signal for H8 earlier in the reaction and permitted only poor quantitation because of the high signal-to-noise ratio. At no point during the digestion could an octasaccharide intermediate be detected.
Figure 2.
(A) MALDI mass spectrum of a mixture of substrates H1 and T5, as well as SOS, after a 40-min digestion with heparinase I. (B) Peak ratios of oligosaccharides H1, T5, and T7 relative to SOS during a 2-h heparinase I digest.
Figure 4.
(A) MALDI mass spectrum of a 60-min digest of D3 with heparinase I. (B) Peak ratios of T5 and H8 relative to sucrose octasulfate during the full 2-h digest. Parentheses indicate oligosaccharides that were expected but not observed.
CE Analysis.
For more rigorous quantitation of disaccharides and other reaction intermediates, CE was used. Heparinase digestion produces Δ4,5 unsaturated HLGAGs that have a uniform extinction coefficient at 230 nm (7), which allows the determination of their molar ratios by UV detection. Peak areas are more reproducible (better than ±3%) because they are less sensitive to separation conditions. Internal standards are nevertheless used to compensate for variations in injection volume (absolute quantitation) and migration times (caused by small variations in endoosmotic flow). Furthermore, disaccharides also can be observed (unlike in MS) so that the mass balance can be calculated. However, migration times give only limited information about the identity of a compound. The migration times of substrates and Di6, T5, and T9 could be determined by using pure standards. For all other components, data obtained by MS was used for identification.
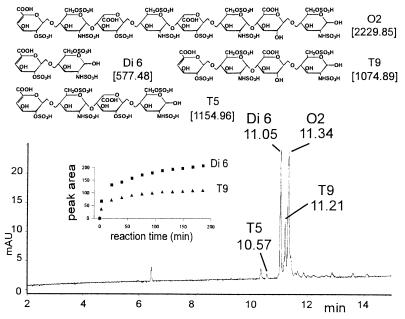
Octasaccharide O2 was chosen as a model compound because a precise quantitation of all its reaction products promised more detailed information about the mechanism of heparinase I. All components of the reaction mixture were separated based on shape and charge (Fig. 5). The peak areas of disaccharide Di6 and tetrasaccharide T9 steadily increased over a period of 3 h (Fig. 5 Inset), maintaining a ratio of 2:1. This ratio suggested that each substrate-enzyme encounter produced two molecules of Di6 and one molecule of T9 by a processive, exolytic mechanism (23). The peak area of T5 (produced by endolytic cleavage) was only 5% of that of T9. The peak area for substrate O2 did not decline to 0. By comparison with MS (Fig. 3A), it was deduced that an enzyme-resistant hexasaccharide impurity (H10-SO3) overlapped with O2.
Figure 5.
CE separation of a 3-min heparinase I digest of O2 and peak areas of oligosaccharides Di 6 and T9 over the full 4-h digestion period (Inset).
Mass Tags.
A number of enzymatic intermediates and end products were detected in the previous experiments. Surprisingly, fewer products were observed than expected based on the known specificity of heparinase I (7) and assuming random cleavage. Of the observed products, most were derived from the reducing end of the substrates. This interpretation is based on the characteristic masses of the observed products. Of the four uronic acids in O2, only one is unsulfated, and it is located at the reducing terminus of the molecule. Consequently, all fragments of O2 that include this group (e.g., T9) have a molecular weight 80.06 Da lower than those derived from the nonreducing terminus (e.g., T5). The unsulfated glucuronic acid in this experiment served as a “mass tag” for the reducing terminus. Hexasaccharide H1 has a mass tag that is a direct modification of the terminal glucosamine. The sugar residue had been transformed into a anhydromannitol by nitrous acid depolymerization followed by reduction (a change of 95.09 Da). This mass tag was used to identify the peak at m/z 4276.4 (T7, Fig. 1B) as a fragment retaining the reducing terminus. The chemical properties of the terminal anomeric center also can be used to create artificial mass tags. Reductive amination and reaction with hydrazine derivatives (24) have been used to generate UV absorbing carbohydrate derivatives. The same chemistry can be used to generate artificial mass tags. Semicarbazide was chosen as the derivatizing reagent because it is a small, neutral molecule expected to have only a minimal effect on the cleavage of HLGAG substrates by heparinase I. The resulting semicarbazone moiety does not absorb appreciably at 230 nm and therefore does not interfere with quantitation based on the absorption of unsaturated uronic acid at this wavelength. Furthermore, the ionization efficiency of the derivatized oligosaccharide should remain unchanged.
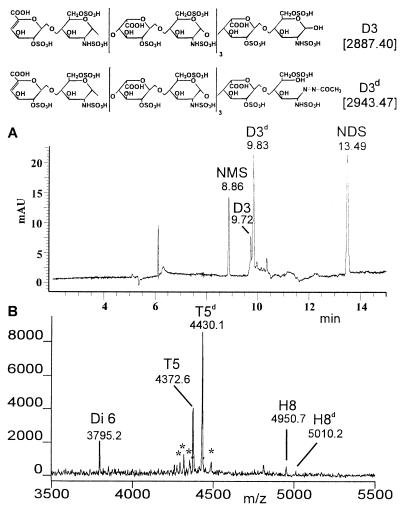
In the case of decasaccharide D3, oligosaccharides obtained from the reducing terminus have the same molecular weight as those derived from internal sequences or the nonreducing end of the molecule. Therefore, it was reacted with semicarbazide for 16 h to form the derivative D3d (increasing the molecular weight by 56.07 Da). The reaction yield was determined by CE because decasaccharides complexed with (arg-gly)15 have a poor ionization efficiency in MALDI MS. The yield was 76 ± 2% (Fig. 6A) assuming that the extinction coefficients for D3 and its derivative D3d are identical. It is noteworthy that CE resolves the native from the derivatized decasaccharide even for such a highly negatively charged molecule (15 sulfate groups). After digesting D3d for 60 min with heparinase I, the main products were observed to be T5 (molecular weight = 1154.96 Da) at m/z 4372.6 and its derivatized form T5d (molecular weight = 1211.03 Da) at m/z 4430.1 (Fig. 6B) with a peak ratio of 1:2. Hence, 66% of the total tetrasaccharide pool was derived from the reducing end (T5d) and 33% was derived from the nonreducing end or internal sequences. By contrast, only Di6, but not its derivatized form, was observed. Hence, the observed disaccharide originates exclusively from the nonreducing terminus or internal residues.
Figure 6.
(A) CE separation of D3 after derivatization with semicarbazide (D3d). (B) Mass spectrum after a 30-min heparinase I digest of the D3/D3d (1:3) mixture.
Other Enzymes.
Heparinase I predominantly cleaves the α-1,4 glycosidic linkage of N-sulfated glucosamine and sulfated iduronic acid (7). A secondary specificity for cleavage at 2-sulfated glucuronic (25) or unsulfated iduronic acid (26) also has been reported. For sequencing purposes, enzymes that can cleave other linkages as well and generate a larger number of intermediates through random endolytic cleavage are needed. Heparinase II was used as a second enzyme because of its broader specificity. Because it is inhibited by calcium, the digestion buffer had to be slightly modified (buffer C). Otherwise, the same experimental conditions were found to be applicable to generate intermediates in a controlled and reproducible manner. A larger number of fragments was observed when O2 was digested with heparinase II (Fig. 3B) instead of heparinase I (Fig. 3A). Hexasaccharide H8 (m/z 4949.0) was produced by cleavage of a glucuronic acid linkage that is resistant to heparinase I. Hexasaccharide H10 (m/z 4869.5) was observed in the digest with heparinase II but not with heparinase I (23). Also, the tetrasaccharide distribution for the two enzymes was markedly different. The observation of fragments derived exclusively from the reducing end and the absence (or low abundance) of hexasaccharides and octasaccharide fragments in heparinase I digests (Figs. 3A, 4A, and 6B) was hence not a result of methodological factors but was caused by the mode of action of heparinase I, indicating a processive mechanism for the enzyme (23).
DISCUSSION
By integrating MS and CE with enzymatic degradation by heparinases, we have shown in this study that one can investigate the depolymerization of HLGAG oligosaccharides in detail. MS is used to follow the enzymatic degradation of HLGAG oligosaccharides and to identify the fragments generated, and CE is applied as a complement to MS for quantitation of HLGAG fragments. Heparinase I and II are used in this methodology development as model enzymes for studying HLGAG oligosaccharides and to illustrate how it is possible to exploit the specificity of the enzymes to investigate HLGAG composition and sequence.
MALDI MS is a very sensitive method requiring only picomoles of material. It can be used for the identification and semiquantitative measurement of components in mixtures of substrates, intermediates, and end products. The simple operation of time-of-flight mass spectrometers and the easy interpretation of MALDI MS spectra make it an attractive technique to investigate glycosaminoglycans and related materials. Capillary electrophoresis is a rapid separation method for the rigorous quantitation of HLGAG oligosaccharides. It is capable of resolving even small changes, such as the difference between a decasaccharide and its semicarbazone derivative. Heparinase I and II generated different patterns that provided complementary sequence information. Other methods rely on parameters, such as viscosity (27) or total UV absorbance (16), that do not provide information about individual saccharide components.
The combination of CE and MS data can better define mechanisms by tracing reaction pathways directly from a defined substrate through various intermediates to the final end products. Each reaction component thus can be placed within the context of the original sequence. We also confirmed a reduced rate of tetrasaccharide cleavage by heparinase I. The pattern for this enzyme is predominantly exolytic, processive (23), and not endolytic as suggested previously (27). It might be possible to generate an exclusively exolytic or a nonprocessive heparinase I by protein engineering (28). Such an enzyme would be suited more ideally for sequencing. Nevertheless, combinations of heparinase I, heparinase II, or mutant heparinases and other enzymes can be used together with CE and MALDI MS to investigate and sequence HLGAG oligosaccharides. The methodology is also adaptable to other glycosaminoglycans and to enzymes that do not act through an eliminative mechanism. The absence of methods such as the one described here has been one of the fundamental barriers in exploring the function of HLGAG oligosaccharides in different biological systems. This methodology may be useful to investigate the biosynthesis, expression, and interactions of glycosaminoglycans. In particular, we would expect applications of the technique to the elucidation of protein-binding GAG structures.
Acknowledgments
We thank Drs. R. Hileman, R. J. Linhardt, P. Østergaard, and D. Tyrrell for the HLGAG oligosaccharides used as substrates. Funding was provided by the National Institutes of Health (GM05472 to K.B. and GM57073 to R.S.), the Fulbright Commission/Austrian-American Educational Commission (to A.R.), and the National Science Foundation (to SE through the Biotechnology Process Engineering Center at MIT).
ABBREVIATIONS
- HLGAG
heparin-like glycosaminoglycan
- MALDI
matrix-assisted laser desorption ionization
- CE
capillary electrophoresis
- SOS
sucrose octasulfate
References
- 1.Varki A. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kjellèn L, Lindahl U. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 3.Salmivirata M, Lidholt K, Lindahl U. FASEB J. 1996;10:1270–12794. doi: 10.1096/fasebj.10.11.8836040. [DOI] [PubMed] [Google Scholar]
- 4.Casu B, Oreste P, Torri G, Zoppetti G, Choay J, Cormeau J-C, Petitou M, Sinay P. Biochem J. 1981;197:599–609. doi: 10.1042/bj1970599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maccarana M, Casu B, Lindahl U. J Biol Chem. 1993;268:23898–23905. [PubMed] [Google Scholar]
- 6.Casu B. Adv Carbohydr Chem Biochem. 1985;43:51–134. doi: 10.1016/s0065-2318(08)60067-0. [DOI] [PubMed] [Google Scholar]
- 7.Ernst, S., Langer, R., Cooney, L. C. & Sasisekharan, R. CRC Crit. Rev. Biochem. Mol. Biol. 30, 387–444. [DOI] [PubMed]
- 8.Gettins P, Horne A P. Carbohydr Res. 1992;223:81–98. doi: 10.1016/0008-6215(92)80008-o. [DOI] [PubMed] [Google Scholar]
- 9.Ii T, Kubota M, Okuda S, Hirano T, Ohashi M. Glycoconj J. 1995;12:162–172. doi: 10.1007/BF00731361. [DOI] [PubMed] [Google Scholar]
- 10.Juhasz P, Biemann K. Proc Natl Acad Sci USA. 1994;91:4333–4337. doi: 10.1073/pnas.91.10.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhomberg, A. J. & Biemann, K. (19XX) in A Laboratory Guide to Glycoconjugate Analysis, eds. Jackson, P. & Gallagher, J. T. (Birkäuser, Basel), pp. 77–89.
- 12.Sutton C W, O’Neil J A, Cottrell J S. Anal Biochem. 1994;218:34–46. doi: 10.1006/abio.1994.1138. [DOI] [PubMed] [Google Scholar]
- 13.Linhardt R J, Wang H M, Ampofo S A. In: Heparin and Related Polysaccharides. Lane D A, Björk I, Lindahl U, editors. New York: Plenum; 1992. pp. 37–47. [Google Scholar]
- 14.Tyrrell D J, Ishihara M, Rao N, Horn A, Kiefer M C, Stauber G B, Lam L H, Stack R J. J Biol Chem. 1993;268:4684–4689. [PubMed] [Google Scholar]
- 15.Pervin A, Gallo C, Jandik K A, Han X, Linhardt R J. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 16.Ernst S, Venkataraman G, Winkler S, Godavarti R, Langer R, Cooney C L, Sasisekharan R. Biochem J. 1996;315:589–597. doi: 10.1042/bj3150589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang F, Beavis R C. Rapid Commun Mass Spectrom. 1994;8:199–204. [Google Scholar]
- 18.Juhasz P, Biemann K. Carbohydr Res. 1994;270:131–147. doi: 10.1016/0008-6215(94)00012-5. [DOI] [PubMed] [Google Scholar]
- 19.Vestal M L, Juhasz P, Martin S A. Rapid Commun Mass Spectrom. 1995;9:1044–1050. [Google Scholar]
- 20.Ampofo S A, Wang H M, Linhardt R J. Anal Biochem. 1991;195:68–73. doi: 10.1016/0003-2697(91)90296-6. [DOI] [PubMed] [Google Scholar]
- 21.Juhasz P, Biemann K. Proceedings of The 42nd ASMS Conference on MS and Allied Topics. Santa Fe, CA: ASMS; 1994. p. 890. [Google Scholar]
- 22.Rice K G, Linhardt R J. Carbohydr Res. 1989;190:219–233. doi: 10.1016/0008-6215(89)84127-8. [DOI] [PubMed] [Google Scholar]
- 23.Ernst S, Rhomberg A, Biemann K, Sasisekharan R. Proc Natl Acad Sci USA. 1998;95:4182–4187. doi: 10.1073/pnas.95.8.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hase S. In: Carbohydrate Analysis. El Rassi Z, editor. Amsterdam: Elsevier; 1995. pp. 555–575. [Google Scholar]
- 25.Yamada S, Murakami T, Tsuda H, Yoshida K, Sugahara K. J Biol Chem. 1995;270:8696–8705. [PubMed] [Google Scholar]
- 26.Desai R, Wang H, Linhardt R J. Arch Biochem Biophys. 1993;306:461–468. doi: 10.1006/abbi.1993.1538. [DOI] [PubMed] [Google Scholar]
- 27.Jandik K A, Gu K, Linhardt R J. Glycobiology. 1994;4:284–296. doi: 10.1093/glycob/4.3.289. [DOI] [PubMed] [Google Scholar]
- 28.Godavarti R, Sasisekharan R. J Biol Chem. 1998;273:248–255. doi: 10.1074/jbc.273.1.248. [DOI] [PubMed] [Google Scholar]