Abstract
Recent advances in computer hardware and software have led to the development of increasingly successful molecular simulations of protein structural dynamics that are intrinsic to biological processes. These simulations have resulted in models that increasingly agree with experimental observations, suggest new experiments and provide insights into biological mechanisms. Used in combination with data obtained with sophisticated experimental techniques, simulations are helping us to understand biological complexity at the atomic and molecular levels and are giving promising insights into the genetic, thermodynamic and functional/mechanistic behaviour of biological processes. Here, we highlight some examples of such approaches that illustrate the current state and potential of the field of molecular simulation.
Keywords: molecular dynamics, function, catalysis, cellular, macromolecules
Introduction
These are remarkable times for the analysis of protein function. The fundamental mechanisms underlying an increasing range of biological processes are not only being revealed as a result of advances in experimental methods in the areas of crystallography, nuclear magnetic resonance (NMR), electron microscopy and single-molecule manipulations, but also because of an increase in computational power and the development of new theoretical methods. The desire to answer complex biological questions drives ever-increasing improvements in both experimental and computational methodologies. Two factors affect computational calculations: first, the attempt to associate motion with mechanism and second, the characterization of the detailed kinetic and thermodynamic parameters of biomolecular interactions by using site-directed mutagenesis, and biochemical, biophysical and bioinformatic techniques (Hammes-Schiffer & Benkovic, 2006). It is encouraging to see that the technology has evolved to a point where the observed and calculated values of chemical and physical parameters are increasingly concordant, as we explain below.
There are currently two main approaches to performing molecular simulations of protein motions. The first is molecular dynamics (MD), in which atoms and molecules are allowed to interact over time at a given temperature following the laws of classical mechanics and which provides a detailed description of atomic motion. The second approach is normal mode analysis (NMA), in which the simple harmonic motions of the molecule about a local energy minimum are calculated. Other commonly used techniques are continuum electrostatic calculations and Brownian dynamics (BD), a variation of MD in which the use of approximations makes possible long timescale calculations.
The pioneering NMR experiments of Sykes and colleagues (Snyder et al, 1975) and of Wuthrich & Wagner (1975), and the computations of Karplus and colleagues (McCammon et al, 1977) are now being followed most satisfactorily by the analyses of the close links between the intrinsic motions and dynamics of proteins and their functions (Karplus & Kuriyan, 2005; Dodson & Verma, 2006; Hammes-Schiffer & Benkovic, 2006; Sotomayor & Schulten, 2007). Significantly, both MD and NMA are now being used to investigate aspects of crystallography, NMR and electron microscopy (de Bakker et al, 2006; Zagrovic & van Gunsteren, 2006; Chen & Brooks, 2007).
It is now well understood that the structural changes, chemical pathways, and the thermodynamic character of cellular and physiological reactions cannot be fully characterized at the atomic level by experimentation alone. This is a serious limitation, which is particularly frustrating in view of the advances in molecular and cell biology, and biomedicine. However, MD simulations—with their unique property of probing the space and timescales simultaneously—are a promising avenue for the characterization of many biological reactions in significant detail (Tama & Brooks, 2006; Sottomayer & Schulten, 2007). Here, we look at how MD and associated simulation methods are elucidating the role of molecular motions in binding, enzymatic activity, signalling mechanisms and in macromolecular assemblies, all within relevant biological contexts. We initially highlight a few examples in which theory correlates with experimentation, followed by examples that link molecular motions to protein function, including enzyme mechanisms of action.Finally, we conclude by citing work in which computations have been extended to the larger scale of macromolecular assemblies.
Comparison of experimentation and theory
The following three studies have successfully compared experimental data and molecular calculations. Kuriyan and colleagues (Young et al, 2001) explored the links between conformational flexibility and long-range modulation of activity—a characteristic that is common in biology—in the Src family of tyrosine kinases. The kinase catalytic domain displays the consensus conformational changes associated with active-site activation and is linked to two peptide-binding domains, SH2 and SH3. Experimentation had shown that kinase activation and its subsequent interactions with the SH2 and SH3 domains are attenuated on phosphorylation of a carboxy-terminal tyrosine. However, the specific mechanism by which this interaction is communicated across the approximately 40 Å from the active site to the phosphorylation site remained a mystery. A series of MD studies now provide a testable mechanism for this long-range coupling: the motions of the SH2 and SH3 domains are strongly coupled (or uncoupled) when the tail is phosphorylated (or dephosphorylated). Furthermore, it was shown that the movement of the activation loop correlates with the motions of the contiguous SH3 domain and, surprisingly, is transmitted to the SH2 domain through the intervening linker peptide. To test the role of this linker, Kuriyan and colleagues mutated its linking amino acids to glycines and found that the correlated motions between the two domains were markedly reduced, mimicking the effects of C-terminal tyrosine dephosphorylation, as had been predicted by molecular modelling. This finding was further validated with an in vivo activation assay in Schizosaccharomyces pombe. This concordance between in silico predictions and in vivo experiments was an early illustration of the potential of MD.
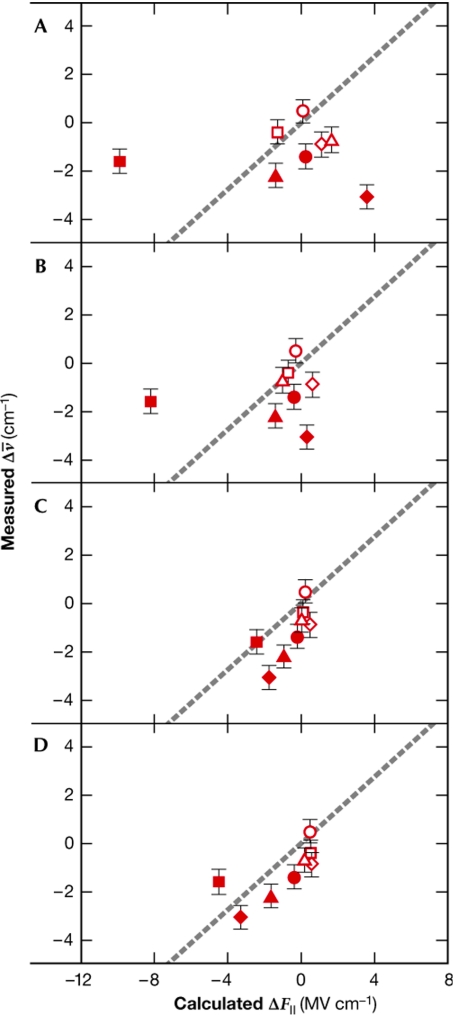
The second example concerns the human aldose reductase (ADH) in a complex with the nitrile-containing inhibitor IDD743. In this study, calculations were performed to evaluate the Stark effect—the influence of an electric field on quantum-mechanical states—at the enzyme active site of ADH (Suydam et al, 2006). The nitrile group is a sensitive reporter of electric field changes as its Stark frequencies are distinct from those of the protein. Electric fields measure the total collection of electric charges and di-electric properties of a given protein, and therefore provide a sensitive test for the widely used continuum electrostatic calculations (Baker, 2005). The electric fields calculated by using native and mutated ADH allowed the prediction of the vibrational changes of IDD743, which was used as the probe. Most interestingly, the correlation with experimental data improved markedly when MD-derived conformations were used (Fig 1), which is persuasive evidence that the structural behaviour of the protein was being accurately predicted by MD.
Figure 1.
Benchmarking computed values with experiments. Correlation, for several mutants of ADH, between the measured frequency changes and the calculated changes in electric field along the nitrile bond. Each symbol in the graph represents a mutant of ADH. (A) Electric field (F∥) calculated from initial models, interior dielectric set to 4. (B) F∥ calculated from energy-minimized models. (C) F/∥ calculated from the mean electric field generated from structures obtained by MD trajectories for protein dielectric 4. (D) F/∥ calculated from the mean electric field generated from structures obtained by MD trajectories for protein dielectric 2. From Suydam et al (2006) Science 313: 200–204. Reprinted with permission from AAAS. ADH, human aldose reductase; MD, molecular dynamics.
In the third example, syntaxin—a self-assembling protein—was investigated by Sieber et al (2007). It is known that proteins such as syntaxins occur in complex clusters that self-assemble through protein–protein interactions at specific motifs (SNARE motifs in the case of syntaxin). However, the physical basis underlying such supramolecular assemblies has yet to be characterized. A combination of sophisticated fluorescence microscopy and quantitative biochemistry revealed that syntaxins are in dynamic equilibrium between free and clustered states. It was observed that, in the presence of increasing amounts of syntaxin, there was an increase in the number of clusters, but not in cluster size. However, it was unclear whether the size and dynamics of these clusters was the result of simple physical interactions or of some other mechanism of biological regulation. BD simulations revealed that a simple balance between attractive and repulsive protein–protein interactions could account for the experimentally observed cluster sizes, and also that such dynamics were controlled by exchanges between freely diffusing syntaxin molecules and relatively immobile clusters. This consistency with experimental data prompted the characterization of syntaxin surfaces and their interactions, which led to the identification of the specific protein–protein contact sites. Although the attractive forces originated in the SNARE motifs, the origin of the repulsive forces proved to be more complex. A model of 75 syntaxin molecules oligomerizing through their SNARE motifs was constructed in which the amino-termini induced a half-closed conformation at the rim of the clusters (Fig 2). The calculations identified the diameter of the cluster, as well as epitopes of antibody binding, that are in agreement with experimental evidence. Thus, the molecular calculations defined the preferred stoichiometry of the cluster, thereby providing a molecular mechanism for the pattern of assembly and the dynamics of slow release of the inner molecules.
Figure 2.

Model of syntaxin clusters in the plasma membrane. Simulations of molecular dynamics were used to construct this model in which a balance between attractive interprotein interactions and steric repulsion result in clusters made up of approximately 75 syntaxins. Proteins within the cluster are immobile but can exchange with freely diffusing molecules. Image provided by B. Ramner and T. Lang.
Molecular motions and function
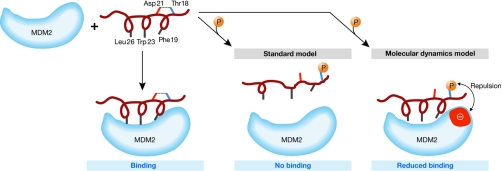
The role of MD simulations in understanding signalling pathways is described in the following three examples. The first example highlights the effects of phosphorylations on p53 binding (Lee et al, 2007). This protein is of exceptional importance for structural studies owing to its crucial role as a tumour suppressor (Vousden & Lane, 2007). The normal role of the p53 pathway is to respond to diverse stress signals by stimulating the transcriptional activation of genes that promote cell cycle arrest, senescence and apoptosis. The activity of p53 relies on its particular conformational properties and its ability to interact appropriately with its many cognate partners. One important partner is the ubiquitin E3-ligase MDM2, which interacts with the p53 N-terminal helical segment and modifies both p53 and itself. This interaction targets p53 for ubiquitination and degradation, thus regulating the levels of both p53 and MDM2. Of the many p53 post translational modifications, one crucial event is the phosphorylation of Thr 18, which releases p53 from its complex with MDM2. Structural analysis of the MDM2–p53 complex reveals that Thr 18 is hydrogen-bonded to Asp 21 (separated by one turn of the helix). It was assumed that this local interaction was decisive in stabilizing the p53 N-terminal helix by allowing the formation of a non-polar patch by residues Phe 19, Trp 23 and Leu 26, which are then embedded in the MDM2 binding pocket. Phosphorylation of Thr 18 was predicted to disrupt the formation of this helix and thus prevent the p53–MDM2 interaction. However, in agreement with NMR experiments, initial MD calculations showed that the p53 N-terminal peptide remains helical in solution. Further MD simulations showed that, in contrast to the prevailing theories, the helical structure is only marginally affected by the acquisition of a negatively charged phosphate group and that the affinity of p53 for MDM2 is only minimally decreased. If this marginal disturbance of the helical structure is not sufficient to cause dissociation of the phosphorylated complex, what causes it? An analysis of the MDM2 surface revealed a pair of acidic amino acids adjacent to the threonine docking site. Additional MD calculations revealed that, on phosphorylation, an electrostatic repulsion is generated between the phosphate group and acidic MDM2 residues that destabilizes the complex and probably releases the p53 helical peptide (Fig 3). This study emphasizes that there can be more than one route available to provide free energy for a molecular process and that any one or a combination of routes can contribute to the mechanism; the balance of these different contributions—helical disruption and charge repulsion—can now be estimated by using simulations. The effects of specific point mutations on p53 binding to MDM2 have been predicted and subsequently confirmed experimentally (Brown et al, 2008). This might be a mechanism by which overexpression of MDM2 isoforms overrides the phosphorylation-dependent upregulation of p53 in tumour cells.
Figure 3.
Schematic of the binding of p53 amino-terminal helical peptide to MDM2. The p53 hydrophobic residues, Phe 19, Trp 23 and Leu 26, interact with the MDM2 binding pocket. The standard model suggests that the hydrogen bond (dotted line) between Thr 18 and Asp 21 is responsible for the maintenance of the helical motif necessary to maximize the interactions with MDM2. On phosphorylation of Thr 18, the hydrogen bond with Asp 21 is broken and the helix is destabilized, resulting in disruption of the p53–MDM2 interaction. The simulations of molecular dynamics by Lee et al (2007) suggest that, although the hydrogen bond is broken on phosphorylation, the helix is marginally destabilized and the loss of binding also arises from repulsions generated between the negatively charged phosphate group and an anionic patch (red) on the surface of MDM2 in the vicinity of the phosphorylated Thr 18. MDM2, ubiquitin E3 ligase.
A second example is the analysis of how post-translational modifications of the nuclear factor of activated T cells (NFAT) modulate binding events. This has been studied by Shen et al (2005) using MD and combining detailed models (in which all atoms are explicitly represented) with coarse-grained models (in which each residue is represented by only three atoms) to investigate how phosphorylation leads to the nuclear localization of NFAT. Simulations show that when unphosphorylated, the helical bundle structure of NFAT is very flexible and its nuclear localization signal (NLS) remains exposed. Multiple phosphorylations lead to interactions of the negative charges of the phosphorylated groups with a series of exposed cationic residues, causing the charges to cluster. This brings the helices together, burying the NLS and preventing the transport of NFAT to the nucleus and the initiation of transcription. In this example, the combination of detailed and coarse-grained representations has been effective in modelling a complex multi-step process. These results still need experimental verification but we include them here because their validation is achievable and these theoretical experiments hold realistic promise that large, complex proteins and assemblies can be investigated by coupling such finely calculated chemical or structural local effects with global approximations.
As a third example, simulations have led Somani and colleagues to propose a new signalling mechanism (Somani et al, 2007) for the cytokine interleukin-1β (IL-1β). The role of IL-1β in inflammation is reasonably well understood and the IL-1 receptor is known. IL-1β is interesting both structurally and computationally as it contains a buried hydrophobic cavity that might or might not be occupied by water, an issue that has been controversial among crystallographers and NMR spectroscopists. NMR suggests that the cavity is hydrated, whereas crystallographers remain divided (Ernst et al, 1995; Yu et al, 1999; Quillin et al, 2006). This debate is a reminder of the technical limits to the structural methods we use. MD studies, in agreement with nuclear Overhauser effect data, suggest that the cavity is indeed hydrated. The fluctuations of the cavity waters, if present, are presumably too high for crystallographic detection, especially with data obtained at moderate resolutions such as 2.1 Å. The MD contribution to this debate became significant after the experimental characterization of a series of IL-1β mutants. The mutations that interfere with IL-1β signalling are located near the disputed cavity and between 8 Å and 20 Å from the receptor-binding surface. Molecular simulations showed a correlation between the motions at the mutated residues and the binding site residues, providing a simple explanation for the loss of signalling: changes in side-chain structures interfere with the natural fluctuations of the molecule, which present the appropriate interactions to the receptor. Strikingly, however, the ‘native' dynamics (signal transmissions) are observed only when there are two or three water molecules in the cavity. This example is a clear indication that MD simulations might assist in resolving debates originated by contradictory experimental observations.
Modelling enzyme mechanisms
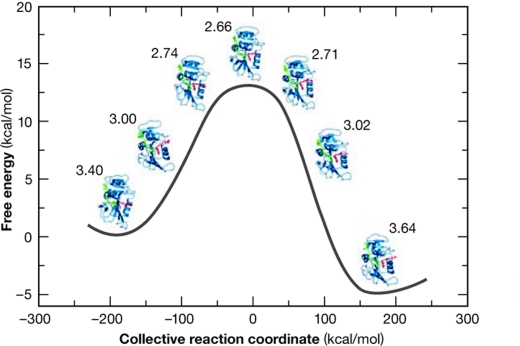
Our understanding of the precise chemical and structural processes underlying enzymatic activity is still incomplete. A pressing point in this regard is the need to incorporate quantum mechanics into the calculations. There has been a long history of research in this area, although significant progress has only been possible in recent years by using a combination of structural, kinetic and thermodynamic data, quantum mechanics and computer simulations for the study of enzymatic reactions (Garcia-Viloca et al, 2004). A crucial issue is whether intrinsic enzymatic motions correlate with and contribute to the energy requirements of a catalytic reaction (Hammes-Schiffer & Benkovic, 2006; Quaytman & Schwartz, 2007). By using extensive structural, mutagenic and computational analyses of the enzyme dihydrofolate reductase (DHFR) and of the intermediates in its reaction pathway, intrinsic atomic motions at the catalytic site were shown to correlate with those beyond the site (Fig 4; Sergi et al, 2006). This network of motions brings the substrate and cofactor close together, directs their chemical approaches appropriately and generates a favourable local electrostatic field for hydride transfer between cofactor and substrate. This emerging picture was further elaborated by detailed NMR measurements along the catalytic cycle (Boehr et al, 2006), making it possible to identify the kinetics between a series of ground states and their associated excited states. The timescales deduced support the view that motions over several timescales combine to modulate and/or orchestrate the functional dynamics of this enzyme. These experimental data agree with the calculations of Hammes-Schiffer & Benkovic (2006), and, by delineating the structural dynamics along the catalytic pathway, open the way for more detailed calculations. Similar behaviour was seen for the enzyme lactate dehydrogenase (LDH). Quaytman & Schwartz (2007) have used extensive computations to show that regions of LDH that are far from the active site contribute significantly to bring the reactants into close proximity to facilitate hydride transfer. Unexpectedly, a recent analysis of the enzyme aromatic amine dehydrogenase (AADH)—based on structural data from intermediates, stopped-flow kinetics, and on quantum mechanical and MD simulations—found that short-range motion modulates the AADH proton transfer mechanism, with no apparent role for the intrinsic large-scale or long-range coupled motions of AADH (Masgrau et al, 2006). These studies have established that picosecond and nanosecond vibrations are involved in modulating the dynamics of catalysis, which is a millisecond process, through conformational changes. Further studies will undoubtedly clarify the link between protein architecture, intrinsic protein motions and reactivity.
Figure 4.
Reaction profile and conformational dynamics of dihydrofolate reductase. The free energy profile of the hydride transfer reaction is shown as a function of a collective reaction coordinate, which includes motions of the enzyme, substrate and cofactor in the dihydrofolate-reductase-catalysed reaction. The donor–acceptor distance is shown next to each structure. Reprinted with permission from Sergi et al (2006). ©2006 American Chemical Society.
Biomolecular assemblies
The structures of impressive macromolecular complexes such as the F1ATPase, GroEL, virus particles, RNA polymerase and intact ribosomes have been solved. These structural data have underlined the need to address the relationship between the structural dynamics and the function of these macromolecular complexes (Gao et al, 2005; Tama & Brooks, 2006; Sotomayor & Schulten, 2007). Here, we highlight some studies that have made attempts in this direction.
The interactions between the ribosomal 30S subunit components during the assembly of the ribosome have been dissected by Hamacher et al (2006). The results were concordant with the relative binding affinities of various ribosome components for naked 16S ribosomal RNA that had been determined experimentally, establishing the validity of the theoretical approximations. This approach enabled a quantitative assessment of the inter-dependencies of the different components of the subunit and helped to establish the kinetics of assembly. The characterization of the relative importance of the various components makes it possible to determine the ‘Achilles heel' of such assemblies and could provide new therapeutic targets.
Proliferating cell nuclear antigen (PCNA) forms a circular belt that clamps around and slides along double-helical DNA during transcription. Dynamic requirements demand that it moves along the DNA rapidly, and that it appropriately interacts with the various enzymes responsible for the polymerase activity. Kazmirski et al (2005) have simulated the organization of this assembly. Although the PCNA crystal reveals a flat circular structure of two trimeric discs, there is evidence that the functional unit is a spiral. Such an organization is essential to clamp and release the DNA. MD simulations of the flat assembly performed on structures from yeast, humans and archaea showed that PCNA moved unambiguously into a right-handed spiral organization driven by the flexibility of the interfacial residues of the macromolecular ensemble. This organization was recently confirmed by electron microscopy (Johnson et al, 2006).
The protein-conducting channel (PCC) is a heterotrimeric integral membrane protein complex that assembles at a specific site on the endoplasmic reticulum and is responsible for translocating nascent proteins through or into the endoplasmic reticular membrane. The PCC structure of E. coli was built by homology-modelling using the crystal structure of the PCC heterotrimer of Methanococcus jannaschii as a template, and was fitted to images obtained by cryo-electron microscopy using a normal mode-based refinement (Mitra et al, 2005). A back-to-back organization of PCC did not correlate with the density on the micrographs at either of the observed binding sites. However, a much better correlation with experimental density was seen for a front-to-front arrangement. A thread of density entering the translocating PCC was interpreted as nascent protein chain. In addition, normal mode analysis of the two trimers arranged front-to-front revealed an organization and an opening mode appropriately placed for the exiting nascent protein, which were not apparent in the back-to-back arrangement. In a separate theoretical study, MD simulations showed that the fluctuations of M. jannaschii PCC heterotrimers embedded in membrane channels are large enough to enable proteins to translocate across or into the membrane (Tian & Andricioaei, 2006).
Conclusions
This necessarily selective review shows that there has been considerable progress in MD simulations in spite of the complexity of molecular behaviour. Technical advances in computation and simulation methods have been matched by the accurate measurement of kinetic and thermodynamic parameters. Comparison between experimental and calculated data have given confidence in the validity of simulations and have led to their successful application in a range of complex systems from which convincing insights and significant new information have been obtained.
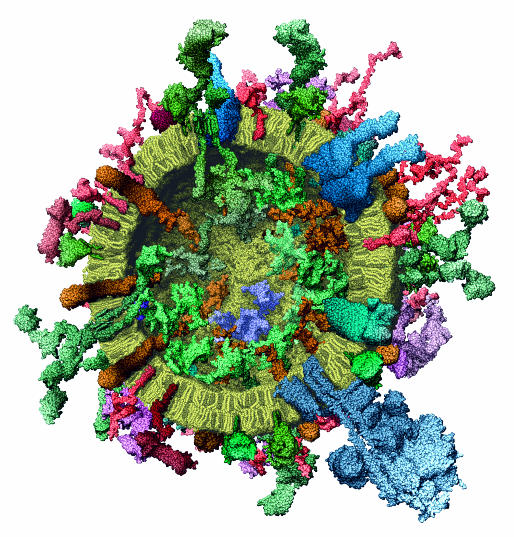
At present, simulations need to expand on three issues. The first is the need to improve the descriptions of interatomic forces to accurately incorporate features such as salt effects or pH to better mimic experimental conditions. This will rely on sophisticated quantum representations (Dal Peraro et al, 2007). The second is the need for longer timescale simulations to bridge the gap between the fast and slow motions (Henzler-Wildman et al, 2007), and associated improvements in quantifying thermodynamic parameters (Elber, 2005; Meirovitch, 2007). Finally, there is a need for the prediction and simulation of the interactions of very large and complex assemblies to shed light on the structural characterization of cellular pathways and networks (Chung et al, 2007; Elcock, 2006). This is dependent on the capacity to accurately compute the location, number and dynamics of macromolecules within the cell (Albeck et al, 2006; Sieber et al, 2007; Stein et al, 2007). Advances in labelling and in imaging are providing a framework to successfully integrate experimental data and have, for example, allowed the construction of a detailed molecular model of the synaptic vesicle (Fig 5; Takamori et al, 2006), which is a good illustration of the level of complexity that simulation methods will have to address in the future. We are confident that the simulation of complex assemblies, and the connection between quantum effects, atomic motions and macroscopic behaviour (Sanbonmatsu & Tung, 2007), will be areas of intense future research.
Figure 5.
Molecular model of a synaptic vesicle constructed on the basis of space-filling models of all its constituent macromolecules at near atomic resolution. The cross-section is shown to illustrate the complex nature of the assembly both inside and outside the cell. The structures were determined by X-ray analysis, nuclear magnetic resonance and single particle electron microscopy. Reprinted from Takamori et al (2006), with permission from Elsevier.
Note added in proof. After the completion of this manuscript, a complete atomic model of a bacterial photosynthetic membrane vesicle was reported (Sener et al, 2007). The particle was assembled by using structural data and was used to compute values for the kinetics and the efficiency of energy transfer that were found to be in agreement with experimental findings. This work illustrates, in a most encouraging manner, the potential of MD and associated computational approaches in probing molecular events in the cellular milieu.

Guy G. Dodson

David P. Lane

Chandra S. Verma
Acknowledgments
G.G.D. is funded by the Medical Research Council, UK. D.P.L. and C.S.V. are funded by the Agency for Science, Technology and Research (A*STAR), Singapore; D.P.L. is a Gibbs Fellow of Cancer Research UK.
References
- Albeck JG, MacBeath G, White FM, Sorger PK, Lauffenburger DA, Gaudet S (2006) Collecting and organizing systematic sets of protein data. Nat Rev Mol Cell Biol 7: 803–812 [DOI] [PubMed] [Google Scholar]
- Baker M (2005) Improving implicit solvent simulations: a Poisson-centric view. Curr Opin Struct Biol 15: 137–143 [DOI] [PubMed] [Google Scholar]
- Boehr DD, McElheny D, Dyson HJ, Wright PE (2006) The dynamic energy landscape of dihydrofolate reductase landscape. Science 313: 1638–1642 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Srinivasan D, Lee HJ, Coomber D, Verma CS, Lane DP (2008) The electrostatic surface of MDM2 modulates the specificity of its interaction with phosphorylated and unphosphorylated p53 peptides. Cell Cycle 7 (in press) [DOI] [PubMed] [Google Scholar]
- Chen J, Brooks CL (2007) Can molecular dynamics simulations provide high-resolution refinement of protein structure? Proteins 67: 922–930 [DOI] [PubMed] [Google Scholar]
- Chung JL, Wang W, Bourne PE (2007) High throughput identification of interacting protein–protein binding sites. BMC Bioinformatics 8: 223–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Peraro M, Ruggerone P, Raugei S, Gervasio FL, Carloni P (2007) Investigating biological systems using first principles Car-Parrinello molecular dynamics simulations. Curr Opin Struct Biol 17: 149–156 [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Furnham N, Blundell T, DePristo MA (2006) Conformer generation under restraints. Curr Opin Struct Biol 16: 160–165 [DOI] [PubMed] [Google Scholar]
- Dodson G, Verma CS (2006) Protein flexibility: its role in structure and mechanism revealed by molecular simulations. Cell Mol Life Sci 63: 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elber R (2005) Long-timescale simulation methods. Curr Opin Struct Biol 15: 151–156 [DOI] [PubMed] [Google Scholar]
- Elcock A (2006) Molecular simulations of cotranslational protein folding: fragment stabilities, folding cooperativity and trapping in the ribosome. PLoS Comput Biol 2: e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JA, Clubb RT, Zhou HX, Gronenborn AM, Clore GM (1995) Demonstration of positionally disordered water within a protein hydrophobic cavity by NMR. Science 267: 1813–1817 [DOI] [PubMed] [Google Scholar]
- Gao YQ, Yang W, Karplus M (2005) A structure-based model for the synthesis and hydrolysis of ATP by F1-ATPase. Cell 123: 195–205 [DOI] [PubMed] [Google Scholar]
- Garcia-Viloca M, Gao J, Karplus M, Truhlar DG (2004) How enzymes work: analysis by modern rate theory and computer simulations. Science 303: 186–195 [DOI] [PubMed] [Google Scholar]
- Hamacher K, Trylska J, McCammon JA (2006) Dependency map of proteins in the small ribosomal subunit. PLoS Comput Biol 2: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes-Schiffer S, Benkovic SJ (2006) Relating protein motions to catalysis. Ann Rev Biochem 75: 519–541 [DOI] [PubMed] [Google Scholar]
- Henzler-Wildman KA et al. (2007) Intrinsic motions along an enzymatic reaction trajectory. Nature 450: 838–844 [DOI] [PubMed] [Google Scholar]
- Johnson A, Yao NY, Bowman GD, Kuriyan J, O'Donnell M (2006) The replication factor C clamp loader requires arginine finger sensors to drive DNA binding and proliferating cell nuclear antigen loading. J Biol Chem 281: 35531–35543 [DOI] [PubMed] [Google Scholar]
- Karplus M, Kuriyan J (2005) Molecular dynamics and protein function. Proc Natl Acad Sci USA 102: 6679–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmirski SL, Zhao Y, Bowman GD, O'Donnell M, Kuriyan J (2005) Out-of-plane sliding motions in open sliding clamps: molecular dynamics simulations of eukaryotic and archaeal proliferating cell nuclear antigen. Proc Natl Acad Sci USA 102: 13801–13806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Srinivasan D, Coomber D, Lane DP, Verma CS (2007) Modulation of the p53–MDM2 interaction by phosphorylation of Thr 18: a computational study. Cell Cycle 6: 2604–2611 [DOI] [PubMed] [Google Scholar]
- McCammon JA, Gelin BR, Karplus M (1977) Dynamics of folded proteins. Nature 267: 585–590 [DOI] [PubMed] [Google Scholar]
- Masgrau L, Roujeinikova A, Johannissen LO, Hothi P, Basran J, Ranaghan KE, Mulholland AJ, Sutcliffe MJ, Scrutton NS, Leys D (2006) Atomic description of an enzyme reaction dominated by proton tunneling. Science 312: 237–241 [DOI] [PubMed] [Google Scholar]
- Meirovitch H (2007) Recent developments in methodologies for calculating the entropy and free energy of biological systems by computer simulation. Curr Opin Struct Biol 17: 181–186 [DOI] [PubMed] [Google Scholar]
- Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, Ban N, Frank J (2005) Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438: 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaytman SL, Schwartz SD (2007) Reaction coordinate of an enzymatic reaction revealed by transition path sampling. Proc Natl Acad Sci USA 104: 12253–12258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillin ML, Wingfield PT, Matthews BW (2006) Determination of solvent content in cavities in IL-1β using experimentally phased electron density. Proc Natl Acad Sci USA 103: 19749–19753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanbonmatsu KY, Tung CS (2007) High performance computing in biology: multimillion atom simulations of nanoscale systems. J Struct Biol 157: 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sener MK, Olsen JD, Hunter CN, Schulten K (2007) Atomic-level structural and functional model of a bacterial photosynthetic membrane vesicle. Proc Natl Acad Sci USA 104: 15723–15728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergi A, Watney JB, Wong KF, Hammes-Schiffer S (2006) Freezing a single distal motion in dihydrofolate reductase. J Phys Chem B 110: 2435–2441 [DOI] [PubMed] [Google Scholar]
- Shen T, Zong C, Hamelberg D, McCammon JA, Wolynes PG (2005) The folding energy landscape and phosphorylation: modeling the conformational switch of the NFAT regulatory domain. FASEB J 19: 1389–1395 [DOI] [PubMed] [Google Scholar]
- Sieber JJ et al. (2007) Anatomy and dynamics of a supramolecular membrane protein cluster. Science 317: 1072–1076 [DOI] [PubMed] [Google Scholar]
- Snyder GH, Rowan R 3rd, Karplus S, Sykes BD (1975) Complete tyrosine assignments in the high field 1H nuclear magnetic resonance spectrum of the bovine pancreatic trypsin inhibitor. Biochemistry 14: 3765–3777 [DOI] [PubMed] [Google Scholar]
- Somani S, Chng CP, Verma CS (2007) Hydration of a hydrophobic cavity and its functional role: a simulation study of human interleukin-1β. Proteins 67: 868–885 [DOI] [PubMed] [Google Scholar]
- Sotomayor M, Schulten K (2007) Single-molecule experiments in vitro and in silico. Science 316: 1144–1148 [DOI] [PubMed] [Google Scholar]
- Stein M, Gabdoulline RR, Wade RC (2007) Bridging from molecular simulation to biochemical networks. Curr Opin Struct Biol 17: 166–172 [DOI] [PubMed] [Google Scholar]
- Suydam IT, Snow CD, Pande VS, Boxer SG (2006) Electric fields at the active site of an enzyme: direct comparison of experiment with theory. Science 313: 200–204 [DOI] [PubMed] [Google Scholar]
- Takamori S et al. (2006) Molecular anatomy of a trafficking organelle. Cell 127: 671–673 [DOI] [PubMed] [Google Scholar]
- Tama F, Brooks CL (2006) Symmetry, form and shape: guiding principles for robustness in macromolecular machines. Annu Rev Biophys Biomol Struct 35: 115–133 [DOI] [PubMed] [Google Scholar]
- Tian P, Andricioaei I (2006) Size, motion and function of the SecY translocon revealed by molecular dynamics simulations with virtual probes. Biophys J 90: 2718–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Lane DP (2007) p53 in health and disease. Nat Rev Mol Cell Biol 8: 275–283 [DOI] [PubMed] [Google Scholar]
- Wuthrich K, Wagner G (1975) NMR investigations of the dynamics of the aromatic amino acid residues in the basic pancreatic trypsin inhibitor. FEBS Lett 50: 265–268 [DOI] [PubMed] [Google Scholar]
- Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J (2001) Dynamic coupling between the SH2 and SH3 domains of c-Src and Hck underlies their inactivation by C-terminal tyrosine phosphorylation. Cell 105: 115–126 [DOI] [PubMed] [Google Scholar]
- Yu B, Blaber M, Gronenborn AM, Clore GM, Caspar DL (1999) Disordered water within a hydrophobic protein cavity visualized by X-ray crystallography. Proc Natl Acad Sci USA 96: 103–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagrovic B, van Gunsteren WF (2006) Comparing atomistic simulation data with NMR experiment: how much can NOEs actually tell us. Proteins 63: 210–218 [DOI] [PubMed] [Google Scholar]