Abstract
In yeast, global genome nucleotide-excision repair (GG-NER) requires a protein complex containing Rad7 and Rad16. Rad16 is a member of the switch/sucrose nonfermentable superfamily, and it is presumed that chromatin remodelling is its primary function during repair. We show that RAD16 is required for ultraviolet-dependent hyperacetylation of histone H3 (Lys 9 and Lys 14) at the MFA2 promoter and throughout the genome. The yeast repressor complex Ssn6–Tup1 represses many genes including MFA2. TUP1 deletion results in constitutive hyperacetylation of histone H3, nucleosome disruption and derepression of gene transcription in Tup1-regulated genes. GG-NER in the MFA2 promoter proceeds more rapidly in tup1Δ α-cells compared with wild type, even when transcription is inhibited. We show that elevated histone H3 acetylation levels in the MFA2 promoter in tup1Δ α-cells result in Rad7- and Rad16-independent GG-NER, and that Rad16 mediates the ultraviolet-induced acetylation of histone H3, necessary for efficient GG-NER.
Keywords: chromatin, histone acetylation, MFA2, NER, yeast
Introduction
Nucleotide-excision repair (NER) is a fundamental cellular process. The excision of lesions from non-transcribed regions of the genome involves global genome NER (GG-NER), which requires a specific subset of NER proteins (Friedberg et al, 2005). Various studies have provided a detailed mechanistic understanding of some of the core enzymatic activities associated with NER, but a precise mechanistic understanding of how it functions in relation to chromatin structure is lacking.
In eukaryotes, DNA associates with histones to form nucleosomes. The nucleosomal arrays are compacted further into higher order chromatin structures. Chromatin influences how proteins functionally interact with regulatory regions in the DNA to facilitate events such as transcription, replication, recombination and repair (Wolffe, 2000; Ataian & Krebs, 2006). This is achieved primarily by the activity of two main processes: histone modification and chromatin remodelling. Both processes have been studied for their roles in gene regulation (Wu & Grunstein, 2000; Waterborg, 2002), and more recently in NER (Yu et al, 2005; Gong et al, 2006).
In Saccharomyces cerevisiae, Rad7 and Rad16 form a protein complex that is specifically required for GG-NER (Verhage et al, 1996; Reed et al, 1999; Li & Smerdon, 2004). Rad16 shares marked homology with Snf2, the catalytic subunit of the switch/sucrose nonfermentable (SWI/SNF) chromatin-remodelling complex (Bang et al, 1992). SWI/SNF superfamily proteins show ATPase activity that is stimulated by DNA or chromatin (Eisen et al, 1995; Whitehouse et al, 1999), and all SWI/SNF-like proteins generate superhelical tension in linear DNA fragments through a DNA translocase activity associated with their ATPase function (Havas et al, 2000; Van Komen et al, 2000). It is now established that the generation of superhelicity in DNA is a fundamental mechanistic paradigm common to SWI/SNF-like chromatin remodelling complexes for altering chromatin structure (Havas et al, 2000). We showed previously that generation of superhelical torsion in DNA by the Rad16 component of a GG-NER complex is needed to excise DNA damage during GG-NER in vitro (Yu et al, 2004). Although these experiments indicated a fundamental activity of the GG-NER complex during NER, they were unable to provide information on how the GG-NER complex functioned on chromatin, as the experiments were conducted on naked DNA.
Recently, we reported that the histone acetyl transferase Gcn5 facilitates efficient NER at MFA2 (Teng et al, 2002; Yu et al, 2005). Our study showed that histone H3 acetylation (H3Ac), previously linked only to the regulation of gene transcription, can also facilitate GG-NER at this locus after the exposure of cells to ultraviolet light (Yu et al, 2005). We also showed that histone H3 is hyperacetylated throughout the genome after ultraviolet irradiation, but this is not significantly reduced in the gcn5 histone acetyl transferase mutant strain, possibly because specific histone acetyl transferases affect H3Ac in certain regions of the genome following ultraviolet irradiation. Surprisingly, we found that ultraviolet irradiation-induced H3Ac occurred independently of functional NER. Here, we report that ultraviolet irradiation-induced H3Ac depends on Rad16. Furthermore, by examining events at the MFA2 gene, we show that it is possible to eliminate the requirement for Rad7 and Rad16 during GG-NER by constitutively elevating the levels of H3Ac in the MFA2 gene.
MFA2 is an a-mating-type-specific gene, which is transcriptionally active in a-mating-type cells and silent in α-mating-type cells. The transcriptional regulation of this gene involves the yeast general repressor complex Ssn6–Tup1 (Keleher et al, 1992). Previously, we identified four positioned nucleosomes at MFA2 in α-cells where the gene is repressed (Teng et al, 2001). In TUP1-regulated genes, including MFA2, deletion of TUP1 results in the disruption of positioned nucleosomes, derepression of gene transcription and elevated histone acetylation (Cooper et al, 1994; Bone & Roth, 2001; Malave & Dent, 2006). We show that Rad16 mediates ultraviolet irradiation-induced H3Ac, which results in the chromatin remodelling required for efficient GG-NER. We also discuss possible mechanisms relating to how this is achieved.
Results
UV-dependent histone H3 acetylation requires Rad16
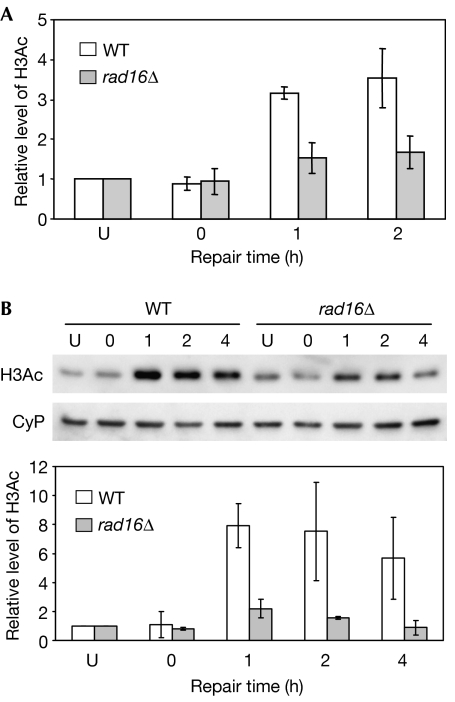
Ultraviolet irradiation stimulates histone H3 hyperacetylation in the regulatory region of MFA2 and throughout the genome in wild-type cells. This established that, in yeast, repressed chromatin is specifically hyperacetylated in response to ultraviolet irradiation. We showed that although ultraviolet irradiation-induced hyperacetylation of histone H3 is required for efficient NER, functional NER is not required to observe H3Ac following ultraviolet irradiation (Yu et al, 2005). Fig 1 shows that most ultraviolet irradiation-induced H3Ac (Lys 9 and Lys 14), at the regulatory region of the MFA2 gene (Fig 1A) and in the genome overall (Fig 1B), requires the GG-NER factor Rad16. We surmised that Rad16-dependent acetylation of histone H3 in response to ultraviolet irradiation could represent an important stage during GG-NER in chromatin. To investigate this, we measured GG-NER in the MFA2 gene of TUP1-deleted cells in which H3Ac is constitutively elevated, even in the absence of ultraviolet irradiation.
Figure 1.
Histone acetylation in wild-type and rad16Δ cells after ultraviolet irradiation. (A) Chromatin immunoprecipitation analysis of histone H3 acetylation (H3Ac) level in the MFA2 promoter using H3Ac (Lys 9 and Lys 14) antibodies. U: untreated samples; 0: cells received 150 J/m2 of ultraviolet without repair; 1, 2 or 4 cells being irradiated with ultraviolet and then were allowed to repair in YPD for these hours. (B) Western analysis of H3Ac (Lys 9 and Lys 14) level in whole-cell extracts. Cyclophilin A (CyP) was used as a control. Acetylation is presented as the fold increase relative to unirradiated cells. WT, wild type.
TUP1 deletion alters gene expression of MFA2
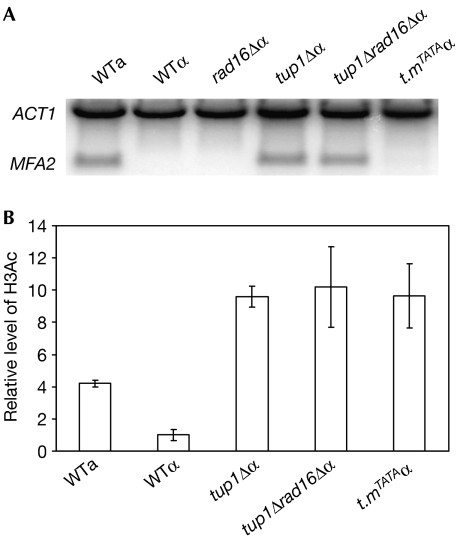
Fig 2A shows northern blots detecting the level of MFA2 messenger RNA in wild-type and mutant cells. MFA2 mRNA is detected in wild-type a-cells, but not in wild-type or rad16Δ α-cells. Deletion of TUP1 activates MFA2 transcription in α-cells to a level similar to that in wild-type a-cells. Deletion of RAD16 in a tup1Δ α-strain has no affect on the level of MFA2 mRNA. However, a TATA box mutation in the promoter of MFA2 abolishes the transcription induced by TUP1 deletion. These observations confirm that we can exclusively examine GG-NER in the promoter of MFA2 in subsequent experiments.
Figure 2.
Transcription and constitutive H3 acetylation (Lys 9 and Lys 14) level at MFA2. (A) MFA2 messenger RNA is detected by northern blots in wild-type (WT) a- and α-cells, rad16Δ, tup1Δ, tup1Δrad16Δ and the MFA2 TATA box mutant tup1Δmfa2TATA (t.mTATA) α-cells, using ACT1 as an internal control. (B) The level of H3Ac (Lys 9 and Lys 14) in the MFA2 promoter in wild-type a- and α-, tup1Δ, tup1Δrad16Δ and tup1Δmfa2TATA (t.mTATA) α-cells is presented as the fold increase relative to that in wild type α-cells, in which MFA2 is repressed. H3Ac, histone H3 acetylation.
TUP1 deletion elevates histone H3 acetylation at MFA2
We measured levels of H3Ac in the MFA2 promoter in the absence of ultraviolet light in wild-type a- and α-, tup1Δ α-, tup1Δrad16Δ α- and tup1Δ mfa2TATA α-cells (Fig 2B). The elevated level of H3Ac in the wild-type a-strain reflects the transcriptionally active status of MFA2. A tenfold elevation in the constitutive levels of H3Ac is seen in the tup1Δ α- and the tup1Δrad16Δ α-cells, and importantly in the transcription inhibited TATA box-mutated tup1Δmfa2TATA α-cells, even in the absence of ultraviolet light. Thus, absence of the Tup1 repressor results in constitutively higher levels of H3Ac in the MFA2 promoter, even when transcription is inhibited.
TUP1 deletion enhances CPD repair in MFA2
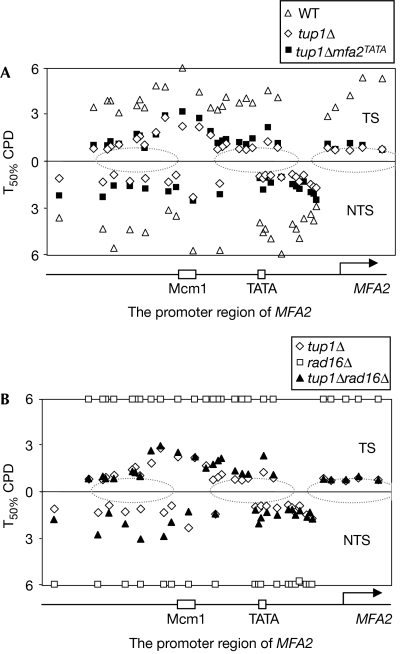
We examined the repair of cyclobutane pyrimidine dimers (CPDs) in the promoter region of MFA2 in wild-type, tup1Δ and TATA box-mutated tup1Δmfa2TATA α-cells. Typical gels to analyse this region are shown in supplementary Figs 1, 2 online. Repair was expressed as the time taken to remove 50% (T50%) of the CPDs at specific positions (Fig 3). NER in tup1Δ cells is enhanced for all detectable CPDs in both strands (Fig 3A). In wild-type α-cells, in which MFA2 is repressed, the overall repair rate of CPDs was T50% 3.3±0.2 h for the transcribed strand and T50% 3.7±0.3 h for the non-transcribed strand. In the tup1Δ α-strain, repair of the transcribed and non-transcribed strands had a T50% of 1.4±0.2 and 1.5±0.2 h, respectively.
Figure 3.
Removal of CPD from the promoter of MFA2. (A) Time to remove 50% of the initial CPDs (T50%) at the MFA2 promoter in wild-type (WT), tup1Δ and tup1Δmfa2TATA α-strains. T50% of a single CPD or a group of close CPDs with a similar repair rate was calculated or extrapolated as described by Teng et al (2002) from typical gels shown in supplementary Figs 1,2 online. The T50% of slowly repaired or unrepaired CPDs (T50%⩾6 h) is shown at the same level (6 h). The dotted ellipses represent the positioned nucleosomes. The diagram below the graph indicates the start of the MFA2 coding region (arrow) and its regulating elements (TATA box- and Mcm1-binding site). (B) Time to remove 50% of the initial CPDs (T50%) at the MFA2 promoter in tup1Δ, rad16Δ and tup1Δrad16Δ α-strains. See Fig 3A legend for labels. CPD, cyclobutane pyrimidine dimer; MCM, minichromosome maintenance protein; NTS, non-transcribed strand; TS, transcribed strand; WT, wild type.
GG-NER is unaltered when transcription is inhibited
The deletion of TUP1 results in histone H3 modification, chromatin structural changes and transcription activation in the MFA2 gene. As gene expression could influence NER in the region, we inhibited MFA2 transcription by mutating its TATA box to eliminate any effect of transcription on repair (Fig 2A). NER in the tup1Δmfa2TATA strain is not significantly altered compared with tup1Δ cells, and remains much faster than that in wild-type α-cells (Fig 3A; supplementary Fig 2 online). The average T50% for CPDs is 1.6±0.2 h for the transcribed strand and 1.7±0.2 h for the non-transcribed strand. Therefore, tup1Δ cells show enhanced lesion removal by GG-NER in the MFA2 promoter, and this is not affected following the inhibition of transcription.
Rad7- and Rad16-independent GG-NER in MFA2
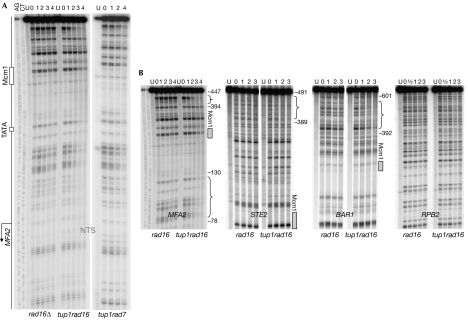
The Rad7–Rad16 complex is generally considered to be essential for GG-NER in S. cerevisiae (Verhage et al, 1994; Reed et al, 1999). Rad7 and Rad16 are required for CPD removal from the non-transcribed strand of MFA2 in both wild-type a- and α-cells (Teng et al, 1997). If the ultraviolet irradiation-induced Rad16-dependent H3Ac has a significant role in NER, one might predict Rad7- and Rad16-independent GG-NER in genomic regions where there are constitutively elevated levels of H3Ac, such as at MFA2 in tup1Δ α cells; Figs 3B, 4A show that this is indeed the case. In the rad7Δ and rad16Δ mutants, GG-NER is completely defective in the MFA2 promoter, whereas in the tup1Δrad7Δ and the tup1Δrad16Δ mutant, repair is restored even in the absence of functional transcription in the tup1Δrad16Δmfa2TATA and tup1Δrad7Δmfa2TATA strains (data not shown). Rad7 and Rad16 are no longer required for GG-NER in the region (Figs 3B,4A). We confirmed the absence of CPD repair in the tup1Δrad14Δ mutant in the region examined (data not shown), indicating that the enhanced CPD repair seen in the absence of Tup1 is unequivocally through Rad7- and Rad16-independent GG-NER.
Figure 4.
Typical sequencing gels showing the repair of ultraviolet-induced CPDs at nucleotide resolution. (A) Repair of ultraviolet-induced CPDs in the non-transcribed strand (NTS) of the HaeIII restriction fragment (−516 to +83) of MFA2 in rad16Δ, tup1Δrad16Δ and tup1Δrad7Δ α-strains. Reference sequences for the combined A and G or C and T are indicated. Lane 0 has DNA from cells receiving 150 J/m2 ultraviolet and extracted immediately after irradiation. Lanes numbered 1, 2, 3 and 4 contain DNA from cells that received ultraviolet light but that were incubated afterwards in medium for these hours before DNA extraction. MFA2 upstream activating sequences (Mcm1-binding site and TATA box) and the start of the coding region of MFA2 (arrow) are indicated. (B) Repair of ultraviolet-induced CPDs in the NTS of MFA2, STE2 and BAR1 promoters, and the NTS of the RPB2 gene in rad16Δ and tup1Δrad16Δ α-strains. See (A) for other labels. The brackets indicate the regions in which Rad16-independent GG-NER is observed. The numbers mark the relative nucleotide positions referring to the start of the coding region ATG as +1 for each related gene. Mcm1-binding sites in the promoter regions of MFA2, STE2 and BAR1 are indicated by grey boxes. CPD, cyclobutane pyrimidine dimers; GG-NER, global genome repair; MCM, minichromosome maintenance protein; U, untreated samples.
Rad16-independent GG-NER can occur in other genes
We examined NER in other Tup1-regulated genes (Fig 4B) to determine whether the effect of Tup1 deletion on GG-NER is unique to MFA2. In rad16Δ and tup1Δrad16Δ mutants, no CPD removal was detected up to 3 h after ultraviolet irradiation in the non-transcribed strand of RPB2, the transcription of which is not regulated by Tup1. However, for the Tup1-regulated genes, STE2 and BAR1, we detected GG-NER in regions of the promoters in the tup1Δrad16Δ strain, whereas GG-NER was absent in the rad16Δ mutant. In the regions where Rad7- and Rad16-independent GG-NER occurred in STE2 and BAR1 (see bracketed regions in Fig 4B), lesion removal occurred at a similar level to that observed in the MFA2 gene. Therefore, the suppression of Rad16 required for GG-NER extends to other Tup1-regulated genes.
Discussion
The Rad16 protein shares significant homology with the Snf2 protein—the catalytic subunit of the SWI/SNF chromatin remodelling complex (Bang et al, 1992)—and Snf2 contains conserved motifs found in a large family of ATPases thought to be involved in chromatin remodelling activities (Eisen et al, 1995). Studies with rad7Δ and rad16Δ mutants have indicated that they have almost identical, intermediate ultraviolet light sensitivity and the same molecular repair defect; failure to perform GG-NER. Given the homology of Rad16 to other chromatin remodelling proteins and the finding that GG-NER operates on transcriptionally repressed regions of the genome, it is not surprising that the role of Rad7 and Rad16 has long been thought to include a chromatin remodelling activity; however, to date none of the studies have suggested a role for these proteins in this process. Rad7 and Rad16 were not required in an in vitro-reconstituted GG-NER system; although, adding the purified Rad7–Rad16 complex moderately stimulated incision (Guzder et al, 1997). The same group also showed that the Rad7–Rad16 complex could bind to damaged DNA. Furthermore, the ATPase activity of Rad16 was attenuated at increasing levels of DNA damage (Guzder et al, 1998). The authors' interpretation of this result suggested a DNA scanning function for Rad7–Rad16, possibly using the ATPase motors of Rad16 during DNA-damage recognition. Our in vitro studies, using yeast whole-cell extracts (WCE), also indicated that Rad7 and Rad16 are not required in order to observe damage-dependent incision during global genome repair, but are required to detect efficient excision of the damaged DNA (Reed et al, 1998). We also showed that a complex containing Rad7–Rad16–Abf1 generates negative supercoiling in DNA through the activity of Rad16 (Yu et al, 2004). This showed that the supercoiling generated in the DNA by this complex is required for damage excision on a template that was not assembled into nucleosomes and that the core activity of the ATPases in Rad16 generates superhelical torsion in the DNA that facilitates excision. However, these studies were not able to determine whether or how Rad16 and the GG-NER complex function on chromatin.
Our results provide an insight into how Rad16 functions during NER in chromatin. We have shown, that although functional NER is not required to observe the ultraviolet irradiation-induced acetylation of histone H3, Rad16 is necessary. Significantly, neither Rad7 nor Rad16 is required for GG-NER in non-transcribed DNA at specific promoters in which constitutively high levels of H3Ac occur. The results indicate that Rad16 activity during GG-NER mediates H3Ac, which results in changes in chromatin structure. Below we describe a possible mechanism for this.
Speculation
We think it unlikely that Rad16 has a direct role in histone acetylation, either at MFA2 or globally. Indeed, we have shown previously that H3Ac at MFA2 is catalysed by the histone acetyl transferase Gcn5 (Teng et al, 2002; Yu et al, 2005). It is also evident that H3Ac is not stimulated by NER per se, as acetylation occurs in the absence of NER in rad4Δ or rad14Δ strains (Yu et al, 2005). Therefore, it is clear that the ultraviolet irradiation-induced H3Ac is specifically mediated by Rad16. We are now performing experiments to examine the effect of Rad16 on the ability of the Gcn5 histone acetyl transferase to bind to and acetylate histone H3 in the promoter of MFA2—and possibly other histone acetyl transferases in other regions of the genome—in response to ultraviolet irradiation. We speculate that Rad16 generates superhelical torsion in DNA through the activity of its ATPase motors, and that this mediates incremental changes in chromatin in response to ultraviolet irradiation. This might alter the ability of histone acetyl transferases such as Gcn5 to induce further changes in chromatin structure through histone acetylation necessary for efficient NER. Encoded within the RAD16 gene are two distinct biochemical functions: an ATPase activity able to generate superhelical torsion in DNA and an E3 ubiquitin ligase activity, the substrates of which include but might not be limited to the NER factor Rad4 (Gillette et al, 2006). In cells, both these activities are inactivated in Rad7-deleted cells. We showed that constitutively elevated H3Ac levels in the MFA2 promoter are sufficient to rescue GG-NER in both Rad7- and Rad16-deleted cells. Although this observation does not prove that the function of the GG-NER complex is necessary for H3Ac, our future work will focus on how the biochemical activities of Rad16 and other components of the GG-NER complex contribute to triggering histone acetyl transferase activity and chromatin remodelling in response to ultraviolet irradiation in yeast.
Methods
Details of the yeast strains and the methods are given in the supplementary information online.
Ultraviolet treatment of yeast cells, DNA isolation and high-resolution mapping of CPD sites. These were carried out as described by Teng et al (1997). In brief, exponential phase yeast cells were resuspended at 2 × 107 cells/ml in pre-chilled PBS and irradiated with 254 nm ultraviolet light and followed by incubation in YPD at 30°C for various repair times. DNA was isolated and digested with HaeIII, and cleaved specifically at CPD sites using T4 endonuclease V (T4 endo V; Epicentre Biotechnologies, Madison, WI, USA). The purified DNA fragments were end-labelled with [α-32P]dATP and resolved by electrophoresis on polyacrylamide gel.
RNA isolation and northern blot analysis. These were carried out as described previously (Teng et al, 2002; Yu et al, 2005).
Chromatin immunoprecipitation. This was carried out as described by Yu et al (2005). In brief, 100 ml of exponential phase yeast cells was crosslinked by formaldehyde. Washed cells were lysed by vortexing with glass beads. The lysate was sonicated to generate DNA fragments ranging from 200 to 800 bp in size. Histone H3 acetyl antibody (at Lys 9 and Lys 14; Upstate Biotechnology, Millipore, MA, USA) was used to immunoprecipitate sheared chromatin and input samples were used as controls. Quantitative PCR was carried out in real time to analyse the enrichment of DNA fragments after immunoprecipitation.
Western blot analysis. This was carried out according to as in Reed et al (1999). In brief, ultraviolet-treated and untreated cells were washed with 10 ml of dialysis buffer (20 mM HEPES-KOH, 10 mM MgSO4, 10 mM EGTA, 20% glycerol and 5 mM dithiothreitol) with 100 μl of 100 × protease inhibitor (100 mM phenylmethylsulphonyl fluoride, 30 mg/ml benzamidine, 100 μg/ml pepstatin, 100 μg/ml leupeptin, 100 μg/ml chymostatin and 100 μg/ml antipain) and resuspended in 0.5 ml of dialysis buffer with protease inhibitor in 2 ml eppendorf tubes. Cells were lysed by vortexing with glass beads. The supernatant phase was collected as the WCE for western blot. Protein electrophoresis on 6% polyacrylamide–SDS gel and western blotting were carried out according to the standard procedure.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Information
Acknowledgments
This work was supported by a Medical Research Council (MRC) programme award to R.W., an MRC Career Establishment Grant to S.H.R. and a Cancer Research Wales Award to Y.T.
References
- Ataian Y, Krebs JE (2006) Five repair pathways in one context: chromatin modification during DNA repair. Biochem Cell Biol 84: 490–504 [DOI] [PubMed] [Google Scholar]
- Bang DD, Verhage R, Goosen N, Brouwer J, van de Putte P (1992) Molecular cloning of RAD16, a gene involved in differential repair in Saccharomyces cerevisiae. Nucleic Acids Res 20: 3925–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone JR, Roth SY (2001) Recruitment of the yeast Tup1p-Ssn6p repressor is associated with localized decreases in histone acetylation. J Biol Chem 276: 1808–1813 [DOI] [PubMed] [Google Scholar]
- Cooper JP, Roth SY, Simpson RT (1994) The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev 8: 1400–1410 [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC (1995) Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res 23: 2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T (2005) DNA Repair Mutagenesis. Washington DC, USA: ASM Press [Google Scholar]
- Gillette TG, Yu S, Zhou Z, Waters R, Johnston SA, Reed SH (2006) Distinct functions of the ubiquitin-proteasome pathway influence nucleotide excision repair. EMBO J 25: 2529–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Fahy D, Smerdon MJ (2006) Rad4–Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol 13: 902–907 [DOI] [PubMed] [Google Scholar]
- Guzder SN, Sung P, Prakash L, Prakash S (1997) Yeast Rad7–Rad16 complex, specific for the nucleotide excision repair of the nontranscribed DNA strand, is an ATP-dependent DNA damage sensor. J Biol Chem 272: 21665–21668 [DOI] [PubMed] [Google Scholar]
- Guzder SN, Sung P, Prakash L, Prakash S (1998) Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J Biol Chem 273: 31541–31546 [DOI] [PubMed] [Google Scholar]
- Havas K, Flaus A, Phelan M, Kingston R, Wade PA, Lilley DM, Owen-Hughes T (2000) Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103: 1133–1142 [DOI] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD (1992) Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68: 709–719 [DOI] [PubMed] [Google Scholar]
- Li S, Smerdon MJ (2004) Dissecting transcription-coupled and global genomic repair in the chromatin of yeast GAL1–10 genes. J Biol Chem 279: 14418–14426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malave TM, Dent SY (2006) Transcriptional repression by Tup1-Ssn6. Biochem Cell Biol 84: 437–443 [DOI] [PubMed] [Google Scholar]
- Reed SH, You Z, Friedberg EC (1998) The yeast RAD7 and RAD16 genes are required for postincision events during nucleotide excision repair. In vitro and in vivo studies with rad7 and rad16 mutants and purification of a Rad7/Rad16-containing protein complex. J Biol Chem 273: 29481–29488 [DOI] [PubMed] [Google Scholar]
- Reed SH, Akiyama M, Stillman B, Friedberg EC (1999) Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev 13: 3052–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Li S, Waters R, Reed SH (1997) Excision repair at the level of the nucleotide in the Saccharomyces cerevisiae MFA2 gene: mapping of where enhanced repair in the transcribed strand begins or ends and identification of only a partial Rad16 requisite for repairing upstream control sequences. J Mol Biol 267: 324–337 [DOI] [PubMed] [Google Scholar]
- Teng Y, Yu S, Waters R (2001) The mapping of nucleosomes and regulatory protein binding sites at the Saccharomyces cerevisiae MFA2 gene: a high resolution approach. Nucleic Acids Res 29: E64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Yu Y, Waters R (2002) The Saccharomyces cerevisiae histone acetyltransferase Gcn5 has a role in the photoreactivation and nucleotide excision repair of UV-induced cyclobutane pyrimidine dimers in the MFA2 gene. J Mol Biol 316: 489–499 [DOI] [PubMed] [Google Scholar]
- Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P (2000) Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell 6: 563–572 [DOI] [PubMed] [Google Scholar]
- Verhage R, Zeeman AM, de Groot N, Gleig F, Bang DD, van de Putte P, Brouwer J (1994) The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol Cell Biol 14: 6135–6142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage RA, van Gool AJ, de Groot N, Hoeijmakers JH, van de Putte P, Brouwer J (1996) Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol Cell Biol 16: 496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterborg JH (2002) Dynamics of histone acetylation in vivo. A function for acetylation turnover?. Biochem Cell Biol 80: 363–378 [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T (1999) Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400: 784–787 [DOI] [PubMed] [Google Scholar]
- Wolffe AP (2000) Chromatin Structure and Function. San Diego, CA, USA: Academic [Google Scholar]
- Wu J, Grunstein M (2000) 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci 25: 619–623 [DOI] [PubMed] [Google Scholar]
- Yu S, Owen-Hughes T, Friedberg EC, Waters R, Reed SH (2004) The yeast Rad7–Rad16–Abf1 complex generates superhelical torsion in DNA that is required for nucleotide excision repair. DNA Repair 3: 277–287 [DOI] [PubMed] [Google Scholar]
- Yu Y, Teng Y, Liu H, Reed SH, Waters R (2005) UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc Natl Acad Sci USA 102: 8650–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Information