Abstract
During protein synthesis, the two elongation factors Tu and G alternately bind to the 50S ribosomal subunit at a site of which the protein L7/L12 is an essential component. L7/L12 is present in each 50S subunit in four copies organized as two dimers. Each dimer consists of distinct domains: a single N-terminal (“tail”) domain that is responsible for both dimerization and binding to the ribosome via interaction with the protein L10 and two independent globular C-terminal domains (“heads”) that are required for binding of elongation factors to ribosomes. The two heads are connected by flexible hinge sequences to the N-terminal domain. Important questions concerning the mechanism by which L7/L12 interacts with elongation factors are posed by us in response to the presence of two dimers, two heads per dimer, and their dynamic, mobile properties. In an attempt to answer these questions, we constructed a single-headed dimer of L7/L12 by using recombinant DNA techniques and chemical cross-linking. This chimeric molecule was added to inactive core particles lacking wild-type L7/L12 and shown to restore activity to a level approaching that of wild-type two-headed L7/L12.
The ribosomal protein L7/L12 is present on the ribosome in four copies as two dimers and is required for the binding of translational factors. The association between these ribosomal proteins and factors to produce GTP hydrolysis-derived energy for template-guided movement of the ribosome is of interest with respect to the mechanism of protein synthesis on the ribosome and as an example of a mechanochemical system (1), or molecular motor, and as an effector G protein system.
The unique quaternary structure of L7/L12 has been conserved in eubacteria, eukaryotes, and archea (2, 3). Protein L7/L12 has been studied in great detail because of the ease with which it can be removed selectively from the large ribosomal subunits. The removal of L7/L12 reduces the rate of protein synthesis by an order of magnitude and its accuracy (4, 5). Wild-type L7/L12 can be replaced easily by variant polypeptides created by chemical or proteolytic cleavage, chemical modification, or recombinant DNA techniques as long as the ribosome binding domain is intact.
Detailed structures of both organized domains [the globular C-terminal domain (residues 53–120) from x-ray crystallography (6) and the helical N-terminal dimerization domain (residues 1–37) by NMR (7)] have been determined. The two organized domains are connected by a flexible hinge sequence. Constructs comprised of the N-terminal dimerization domain bind to core particles and compete with the binding of wild-type L7/L12 but have no activity in protein synthesis because factor binding is lost (8, 9, and 10). Crystallographic structure analysis indicated two globular C-terminal domains packed such that there is a contiguous surface containing evolutionarily conserved residues from both domains. It was suggested that this surface was functionally important, a hypothesis that would imply both the essentialness of two C-terminal domains and association in a specific orientation. The latter implication first was questioned when L7/L12 variants with C-terminal domains cross-linked in different disparate orientations that precluded the formation of the conserved surface were constructed and found to be active (11). Moreover, the C-terminal domains are separated from each other by an average of 85 Å (12) and are independently mobile (13). The results imply that a preferred close orientation of C-terminal domains relative to each other is not important for L7/L12 activity, nor is it likely that two heads function while associated with each other. The results suggest that a dimer with a single-head domain might be active. To test this hypothesis, a chimeric L7/L12 variant that retained a dimeric N-terminal domain to facilitate binding but that contained only one C-terminal domain was constructed, reconstituted into ribosomes in vitro, and tested in a polyphenylalanine synthesis assay.
MATERIALS AND METHODS
Construction of L7/L12 Protein Variants.
The genetic construction, expression, purification, and characterization of the L7/L12Cys99 variant (full length L7/L12 Ser99 → Cys99) were performed as described (14, 15). The DNA construct for an N-terminal fragment of L7/L12 terminated at Cys52 (NTF-Cys52) was made by PCR amplification of the appropriate N-terminal part of the L7/L12 gene and introduction of a Cys codon at position 52 followed by a stop codon. Expression and purification were as described (15–17). The DNA construct for the C-terminal fragment (CTF) was prepared as follows. A pT7–6 vector was digested with EcoRI, filled with T4 DNA polymerase, and ligated with T4 DNA ligase to yield a vector lacking an EcoRI site. The 790-bp HindII–SalI fragment containing the intact rplL gene (15) was cloned into this pT7–6 (without EcoRI) vector. This construct was digested with HindII and EcoRI to remove the coding sequence for the amino acids 1–51 of L7/L12 and the ribosome binding site. A synthetic double-stranded oligonucleotide adapter encoding the ribosome binding site, the initiator Met codon, and the codons for residues 53 and 54 was inserted in place of the fragment removed. The resulting construct was coded for a C-terminal fragment starting with Met and containing residues 53–120 of L7/L12. Expression and purification were performed as described (15–17). All genetic constructs were confirmed by sequencing. The oligonucleotide sequences can be obtained on request.
Preparation of Single-Headed L7/L12 Dimer.
The NTF terminated by Cys52 (NTF:Cys52) was reduced by 2% β-mercaptoethanol, purified by two sequential Bio-Spin 6 column (Bio-Rad) centrifugations, and modified with dithionitrobenzoate as described (18, 19) to yield NTF:Cys52 thionitrobenzoate (TNB). Cys99 L7/L12 was reduced and purified by two sequential Bio-Spin 6 column centrifugations. NTF:Cys52TNB and Cys99 were mixed in a 2:1 molar ratio in the presence of 6 M urea and were incubated for 2 hr at room temperature. The resulting single-headed hybrid was purified by reverse-phase chromatography on an Altex C-4 column (4.6 × 250 mm) by using a gradient of acetonitrile. The single-headed dimer preparation was dissolved in 6 M urea and further purified by gel filtration on Tosohaas (Montgomeryville, PA) TSK-3000SW (7.5 × 300 mm) column in buffer A (20 mM Tris⋅HCl (pH 7.4), 10 mM MgCl2, and 100 mM NH4Cl). The purified protein was analyzed by electrophoresis in gels containing 20% acrylamide and SDS but without reducing agent (Pharmacia). Contamination of the preparation by full length Cys 99 L7/L12 was <3% as estimated by scanning the gel and by immunoblotting with an mAb to the N-terminal region of L7/L12 (20).
RESULTS AND DISCUSSION
There is a high rate of exchange of monomers among L7/L12 dimers in solution (12). Any hypothetical construct containing one full length monomer in association with a headless monomer would reorganize to a mixture of full length dimers, one-headed dimers, and N-terminal dimers. To avoid this exchange of monomer subunits, a strategy to cross-link the full length monomer to a monomeric N-terminal fragment that contained both the dimerization domain and the full hinge was designed. The location of the cross-link should be such that there is no major distortion in the attachment of the single “head” to the flexible hinge. Inspection of the crystallographic structure (6) showed that residue 99 is located in the exposed loop between the βB sheet and the αC helix and is within 5 Å of residue 53, which normally joins the organized C-terminal domain to hinge residue 52. Two-headed dimers cross-linked at Cys99 were shown to be active (see below). It was plausible that cross-linking at Cys99 of the full length monomer to the Cys52 introduced at the end of the hinge region of an N-terminal fragment by itself would not distort significantly the conformation of the dimer and that the chimeric molecule would provide a suitable test for the activity of a one-headed dimer.
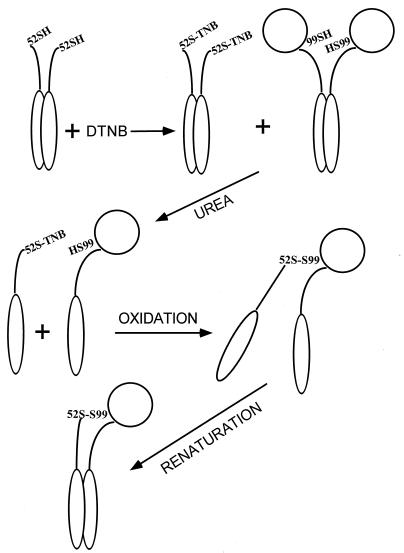
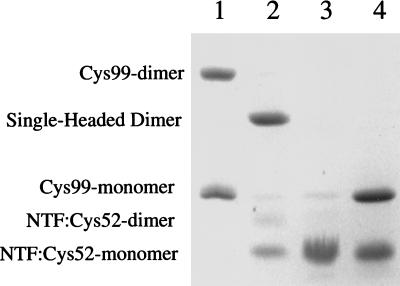
DNA constructs coding for a full length variant of L7/L12 with a Ser99 to Cys99 substitution (L7/L12:Cys99), an N-terminal fragment of L7/L12 terminated at position 52 by Cys (NTF:Cys52), and a C-terminal fragment (residues 53–120) were made by standard techniques described earlier (14, 15) and above. These variant proteins were over expressed and purified. L7/L12:Cys99, reduced or cross-linked by disulfide oxidation, was fully active in polyphenylalanine synthesis (Table 1) as had been shown earlier for Cys63 and Cys89 substituted proteins (11). The hybrid single-headed variant was constructed as outlined in Fig. 1. Fully reduced NTF:Cys52 was modified by reaction with dithionitrobenzoate. The fully modified product NTF:Cys52TNB was mixed in a 2-fold excess with L7/L12:Cys99 in buffer containing 6 M urea. Dissociation to form monomers of both species precludes intramolecular oxidation of L7/L12:Cys99. Disulfide cross-links between L7/L12:Cys99 and NTF:Cys52 were formed by displacement of the TNB residues by the Cys99 sulfhydryl groups. The rate of reaction of the TNB-activated cysteine with free sulfhydryl greatly exceeds the rate of disulfide bond formation in the absence of any oxidizing agent (18). The excess of NTF-Cys52TNB insures that the amount of residual nonreacted full length L7/L12:Cys99 will be minimized. Single-headed hybrid dimers were purified by reverse-phase and gel filtration HPLC. Renaturation led to reassociation of the N-terminal domains to form a functional ribosome binding domain. The purity of the resulting preparation is shown by SDS/PAGE under nonreducing conditions (Fig. 2). Fig. 2, lane 2 shows the mobility of the single-headed dimer to be intermediate between marker preparations of oxidized (disulfide-linked) and monomeric full length L7/L12:Cys99 (Fig. 2, lane 1). Fig. 2, lane 3 shows the mobility of marker monomeric NTF:Cys52TNB. Fig. 2, lane 4 shows the behavior of the hybrid dimer after reduction. Bands corresponding to monomeric L7/L12:Cys99 and to NTF:Cys52 are generated, confirming the composition of the one-headed construct.
Table 1.
Activity of 70S ribosomes reconstituted from Po cores with L7/L12:Cys99
| L7/L12 variant added | Activity |
|---|---|
| Po | 3.5 |
| Po + L7/L12:Cys 99 - reduced | 12.5 |
| Po + L7/L12:Cys 99 - oxidized | 12.9 |
Results represent Phe/70S particle/15 min in poly[U]-directed polyphenylalanine synthesis (11) (average of three different experiments; SD was <10%). Six equivalents of each indicated protein were added to the Po cores.
Figure 1.
Single-headed L7/L12 formation from genetically prepared constructs NTF:Cys52 and full length L7/L12:Cys99.
Figure 2.
Analysis of purified single-headed L7/L12 dimer by gel electrophoresis. The FAST Gel system (Pharmacia) with 20% SDS polyacrylamide was used. Lanes: 1, mixture of oxidized and reduced Cys99; 2, single-headed dimer preparation; 3, NTF:Cys52; 4, single-headed dimer preparation after reduction with 2% β-mercaptoethanol.
As seen in Fig. 2, lane 2, the single-headed hybrid preparation contains a significant amount of the NTF monomer, a small amount of NTF dimer, and even smaller amounts of the Cys99 monomer and a trace amount of Cys99 (oxidized) dimer. Contamination by NTF is difficult to avoid because of the similar chromatographic behavior of NTF and the single-headed dimer. The relative quantities of all of the species in the preparation were determined by scanning of the Coomassie blue-stained gel. The sum of monomeric Cys99 and dimeric oxidized Cys99 on SDS/PAGE was <3% of the single-headed dimer. Because both species contain tail and head domains, it is unlikely that putative differential staining can distort significantly the quantitative analysis.
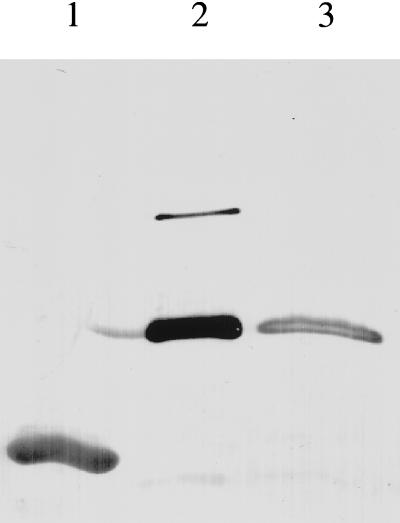
This construct, wild-type L7/L12, NTF:Cys52, and CTF were tested in polyphenylalanine synthesis in the absence of reducing agent as described (11). The results (Table 2) show that the single-headed dimer was active whereas neither NTF:Cys52 nor CTF was active. Fig. 3 demonstrates by immunoblotting of SDS/PAGE of the entire protein synthesis reaction mixture that the single-head dimer retained its disulfide cross-linked structure; there was no rearrangement producing a full-length dimer that could account for the activity. The amount of Cys99 dimer detected (see above) is far too low to account for the activity observed. The slightly reduced activity of single-head dimer preparation relative to the wild-type L7/L12 may be due to its contamination by an NTF dimer (Fig. 2, lane 2), which competes for the binding to the ribosome. It might also be due to the lowering of the local concentration of C-terminal domains on the ribosome (two per ribosome vs. four per ribosome). The new disulfide bond may in some manner strain the necessary orientation of the C-terminal domain. The experiments clearly demonstrate that a single C-terminal domain of L7/L12 dimer retains activity comparable to that of a wild-type two-headed protein. The result confirms the prediction that the Cys99–Cys52 cross-link would not itself result in loss of activity. The hypothetical opportunity for two single C-terminal domains from two single-headed dimers bound to the ribosome to interact with each other and organize the associated dimeric C-terminal structure seems unlikely in the light of the fact that C-terminal domains of two dimers have different locations on the ribosome so that one is protruding away from the body and the other is located on the body of the ribosome (21, 22).
Table 2.
Activity of 70S ribosomes reconstituted from Po cores with different L7/L12 variants
| L7/L12 variant added | Activity |
|---|---|
| Po | 3.9 |
| Po + WT | 12.3 |
| Po + CTF (53-120) | 3.9 |
| Po + NTF:Cys52 | 3.0 |
| Po + NTF:Cys52 + CTF | 3.0 |
| Po + Single-headed dimer | 10.0 |
See footnote to Table 1.
Figure 3.
Persistence of single-headed dimer after protein synthesis. Wild-type L7/L12 (lane 1), single-headed dimer (2 μg) (lane 2), and protein synthesis reaction mixture after 15 min of incubation containing 0.5 μg of single-headed dimer (lane 3) were separated by 16% SDS/PAGE, transferred to nitrocellulose and probed with monoclonal anti-NTF antibodies (20).
It was proposed early in the description of L7/L12 that it resembled a “mini myosin” (1, 23). It is now clear that single-headed myosin retains motor–protein activity (24) and that it occurs normally inside cells. In this case, a single myosin head, with or without attachment to the tail responsible for dimerization, retains the ability to interact with actin. There is no similar evidence that the C-terminal domain of L7/L12 in solution interacts with elongation factors. The effect of adding excess L7/L12 up to 32 equivalents of L7/L12 dimers present in the ribosome did not inhibit the ribosomal activity (data not shown). Neither the NTF nor the CTF separately or in combination had any effect on restoration of activity to the ribosomal cores deprived of L7/L12 (Table 2). That lack of effect implies that only the C-terminal domain, when retained on the ribosome by the rest of the molecule, is able to promote the binding of elongation factors, probably by complementing some additional ribosomal component(s) and organizing a specific structure with them. Previously, we demonstrated that an L7/L12 construct with a shortened hinge bound to ribosome core particles and restored binding of elongation factors; however, this interaction did not result in functional activity, either GTP hydrolysis or translocation (25). The flexible hinge itself must facilitate the functional interaction of one or more C-terminal domains with elongation factors. The single-headed chimeric construct having all the domain features of wild-type L7/L12, except the presence of a second “head,” functions well in the simple protein synthesis assay used here that requires only the binding of elongation factors. The result suggests that a single C-terminal domain of L7/L12 is sufficient for this molecule to support functional binding of the elongation factors. Whether there is any effect on fidelity or whether the single-headed dimer would function equally well in a more natural protein synthesis assay involving initiation and termination, in vitro or in vivo, remains an open question. Yeast protein P0, the protein equivalent to L10, has a C-terminal sequence homologous to proteins P1 and P2, the proteins equivalent to L7/L12. Strains lacking P1 and P2 but retaining P0 with its single C-terminal domain are viable but grow slowly with an altered pattern of protein synthesis (26).
Acknowledgments
This work was supported by the National Institutes of Health Grant GM 17924 (R.R.T).
ABBREVIATIONS
- NTF
N-terminal fragment construct of residues 1–52 of L7/L12
- CTF
C-terminal fragment construct of residues 53–120 of L7/L12
- TNB
thionitrobenzoate
References
- 1.Kischa K, Möller W, Stöffler G. Nat New Biol. 1971;233:62–63. doi: 10.1038/newbio233062a0. [DOI] [PubMed] [Google Scholar]
- 2.Uchiumi T, Wahba A J, Traut R R. Proc Natl Acad Sci USA. 1987;84:5580–5584. doi: 10.1073/pnas.84.16.5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casiano C, Matheson A T, Traut R R. J Biol Chem. 1990;265:18757–18761. [PubMed] [Google Scholar]
- 4.Hamel E, Koka M, Nakamoto T. J Biol Chem. 1972;10:805–814. [PubMed] [Google Scholar]
- 5.Pettersson I, Kurland C G. Proc Natl Acad Sci USA. 1980;77:4007–4010. doi: 10.1073/pnas.77.7.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leijonmarck M, Liljas A. J Mol Biol. 1987;195:555–581. doi: 10.1016/0022-2836(87)90183-5. [DOI] [PubMed] [Google Scholar]
- 7.Bocharov E V, Gudkov A T, Arseniev A S. FEBS Lett. 1996;379:291–294. doi: 10.1016/0014-5793(95)01531-0. [DOI] [PubMed] [Google Scholar]
- 8.Koteliansky V E, Domogatsky S P, Gudkov I T. Eur J Biochem. 1978;90:319–323. doi: 10.1111/j.1432-1033.1978.tb12607.x. [DOI] [PubMed] [Google Scholar]
- 9.Agthoven A, Maassen J A, Schrier P I, Möller W. Biochim Biophys Res Commun. 1975;64:1184–1189. doi: 10.1016/0006-291x(75)90818-9. [DOI] [PubMed] [Google Scholar]
- 10.Schop R N, Maassen J. Eur J Biochem. 1982;128:371–375. doi: 10.1111/j.1432-1033.1982.tb06974.x. [DOI] [PubMed] [Google Scholar]
- 11.Oleinikov A V, Jokhadze G G, Traut R R. Proc Natl Acad Sci USA. 1993;90:9828–9831. doi: 10.1073/pnas.90.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamman B D, Oleinikov A V, Jokhadze G G, Traut R R, Jameson D M. Biochemistry. 1996;51:16680–16686. doi: 10.1021/bi9624189. [DOI] [PubMed] [Google Scholar]
- 13.Hamman B D, Oleinikov A V, Jokhadze G G, Traut R R, Jameson D M. Biochemistry. 1996;51:16672–16679. doi: 10.1021/bi9615001. [DOI] [PubMed] [Google Scholar]
- 14.Hamman B D, Oleinikov A V, Jokhadze G G, Bochkariov D E, Traut R R, Jameson D M. J Biol Chem. 1996;271:7568–7573. doi: 10.1074/jbc.271.13.7568. [DOI] [PubMed] [Google Scholar]
- 15.Zecherle G N, Oleinikov A, Traut R R. Biochemistry. 1992;31:9526–9532. doi: 10.1021/bi00155a003. [DOI] [PubMed] [Google Scholar]
- 16.Zecherle G N, Oleinikov A, Traut R R. J Biol Chem. 1992;267:5889–5896. [PubMed] [Google Scholar]
- 17.Oleinikov A V, Perroud B, Wang B, Traut R R. J Biol Chem. 1993;268:917–922. [PubMed] [Google Scholar]
- 18.Milligan D L, Koshland D E., Jr Science. 1991;254:1651–1655. doi: 10.1126/science.1661030. (1991). [DOI] [PubMed] [Google Scholar]
- 19.Falke J F, Koshland D E. Science. 1987;237:1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- 20.Sommer A, Etchison JR, Gavino G, Zecherle N, Casiano C, Traut RR. J Biol Chem. 1985;260:6522–6527. [PubMed] [Google Scholar]
- 21.Theilen A, Maassen J A, Kriek J, Möller W. Biochemistry. 1984;23:3317–3322. [Google Scholar]
- 22.Olson H M, Sommer A, Tewari D S, Traut R R, Glitz D G. J Biol Chem. 1986;261:6924–6932. [PubMed] [Google Scholar]
- 23.Möller W. Biochimie (Paris) 1991;73:1093–1100. doi: 10.1016/0300-9084(91)90151-p. [DOI] [PubMed] [Google Scholar]
- 24.Albanesi J P, Fujisaki H, Hammer J A, III, Korn E D, Jones R, Sheetz M P. J Biol Chem. 1985;260:8649–8652. [PubMed] [Google Scholar]
- 25.Dey D, Oleinikov A V, Traut R R. Biochimie (Paris) 1995;77:925–930. doi: 10.1016/0300-9084(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 26.Remacha M, Jimenez–Diaz A, Santos C, Briones E, Zambrano R, Rodriguez–Gabriel M A, Guarinos E, Ballesta J P. Biochem Cell Biol. 1995;73:959–968. doi: 10.1139/o95-103. [DOI] [PubMed] [Google Scholar]