Abstract
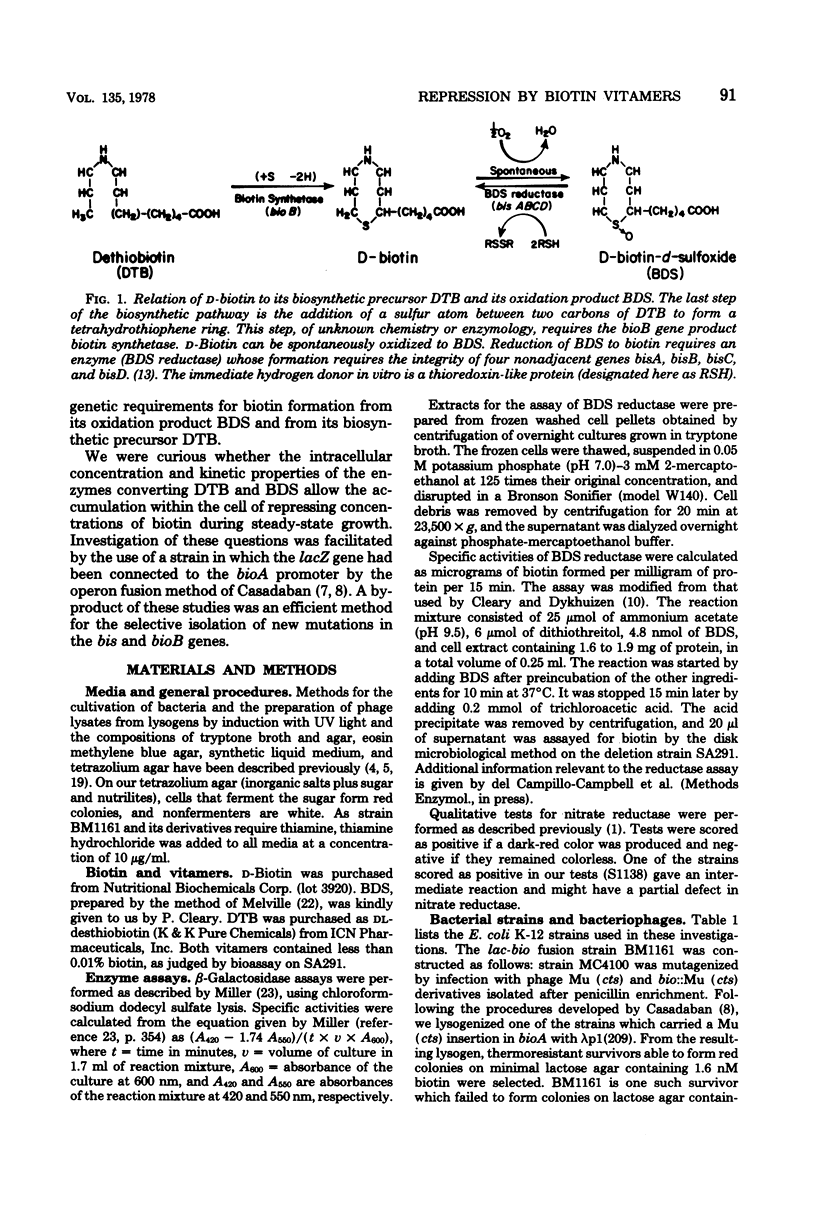
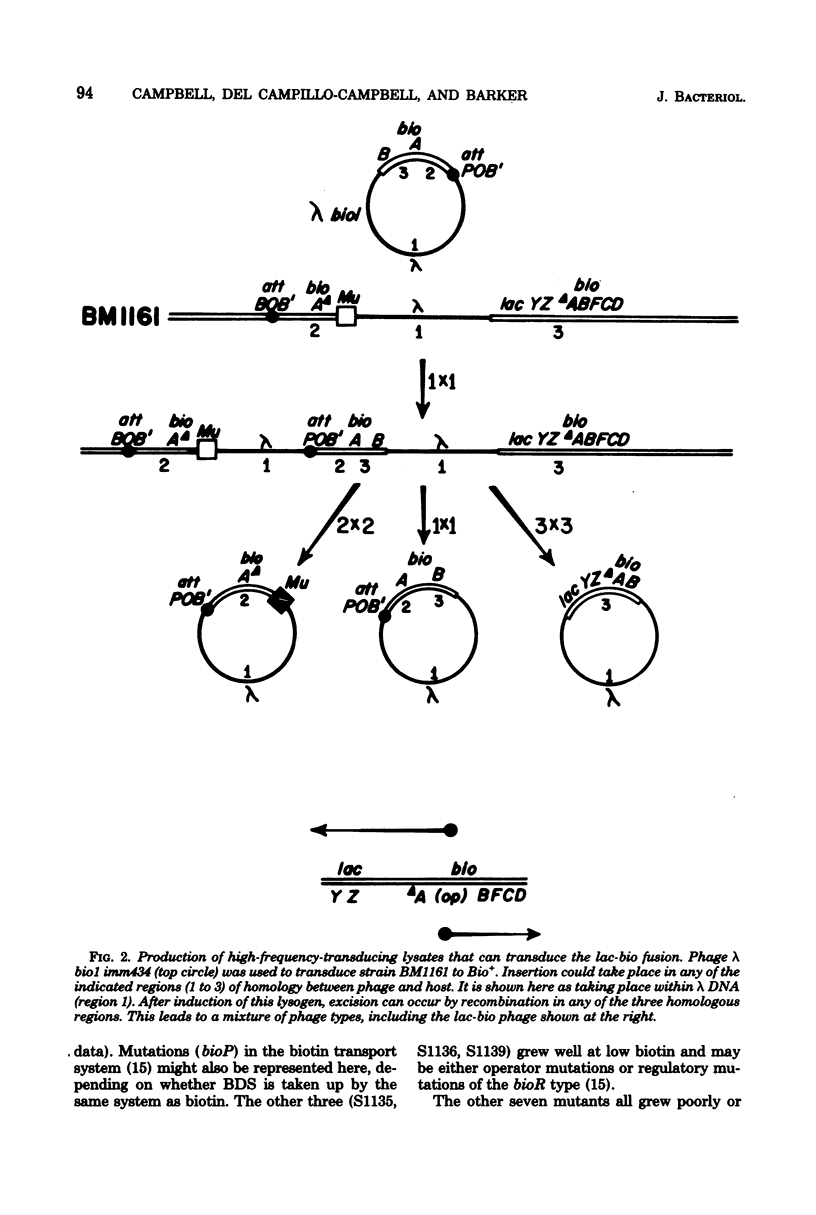
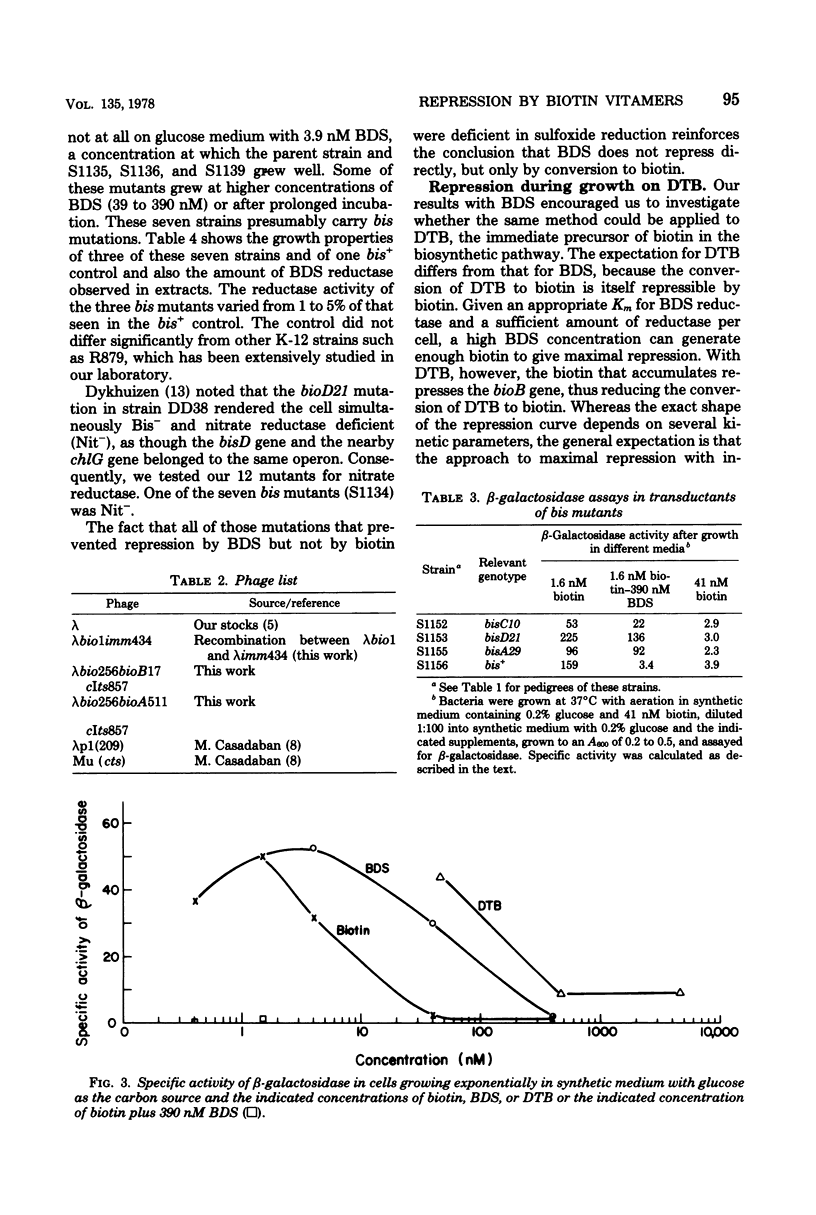
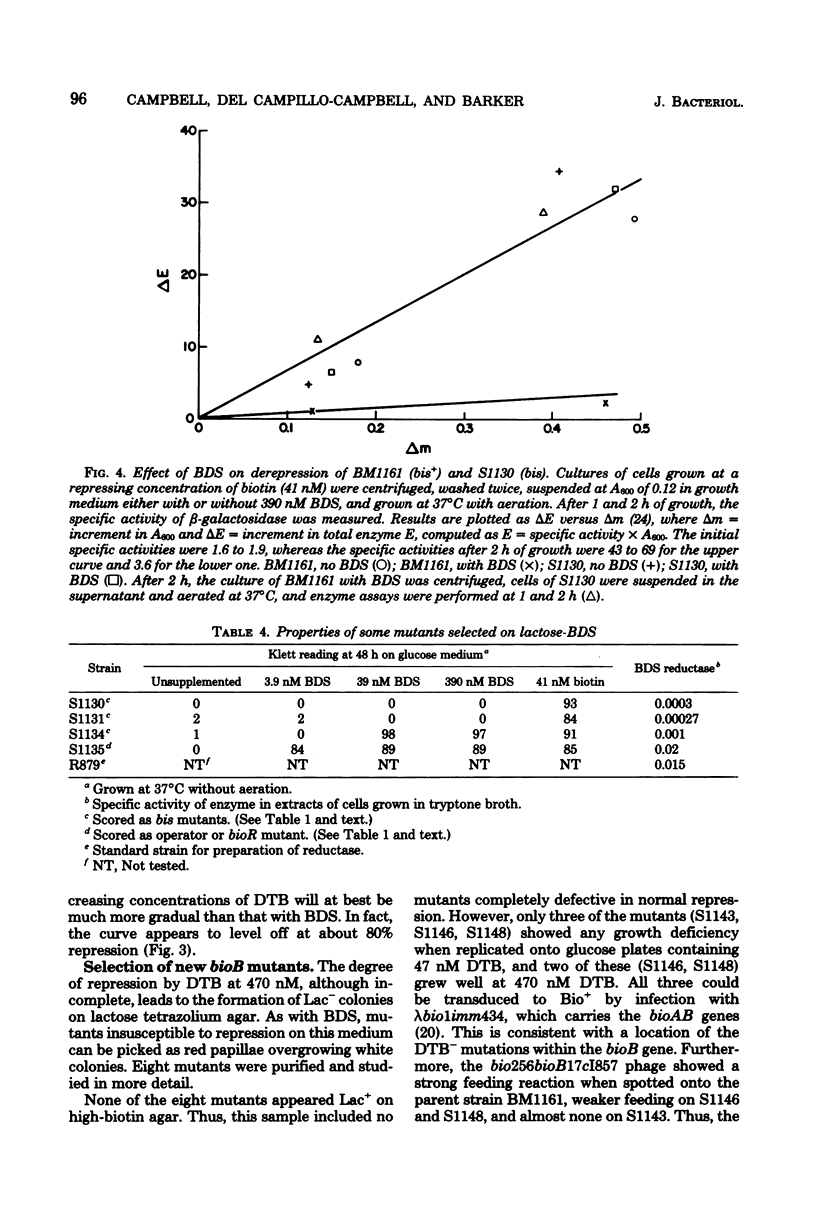
A strain of Escherichia coli in which the lacZ gene was fused to the bioA promoter was constructed. Colonies of this strain formed Lac+ colonies on low-biotin agar (1.6 to 4.1 nM) and Lac− colonies on high-biotin agar (41 nM). This lac-bio fusion strain was used to study the question of whether cells growing on the biotin vitamers d-biotin-d-sulfoxide (BDS) and dethiobiotin (DTB) generate enough biotin to give maximal repression of β-galactosidase synthesis. Repression by high concentrations (400 nM) of BDS was almost maximal (about 96%), whereas DTB repression reached a saturation level of about 80% with increasing DTB concentrations. The levels of repression obtained with both vitamers were sufficient to cause the colonies to appear Lac−. When the lac-bio fusion was transduced into lines carrying mutations (bis) that prevent reduction of BDS to biotin, the transductants were not repressed by added BDS. Repression by BDS is unlikely to result from accumulation of extracellular biotin-related substances because (i) washed bis+ cells were not detectably derepressed when transferred into medium containing BDS and (ii) washed bis cells were not detectably repressed when transferred into medium in which bis+ cells had grown. Lactose agar plates containing high concentrations of DTB or BDS comprise an efficient selective medium for bioB or bis mutants and were used to isolate spontaneous mutations of these genes. This method should be adaptable to the selection of mutations in any biosynthetic pathway subject to end-product repression.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Cleary P., Campbell A. A deletion analysis of prophage lambda and adjacent genetic regions. Proc Natl Acad Sci U S A. 1968 Nov;61(3):956–962. doi: 10.1073/pnas.61.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum J., Pai C. H., Lichstein H. C. Biosynthesis of biotin in microorganisms. V. Control of vitamer production. J Bacteriol. 1967 Dec;94(6):1846–1853. doi: 10.1128/jb.94.6.1846-1853.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAMPBELL A. Sensitive mutants of bacteriophage lambda. Virology. 1961 May;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- CAMPBELL A. Transduction and segregation in Escherichia coli K12. Virology. 1957 Oct;4(2):366–384. doi: 10.1016/0042-6822(57)90070-3. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J. Fusion of the Escherichia coli lac genes to the ara promoter: a general technique using bacteriophage Mu-1 insertions. Proc Natl Acad Sci U S A. 1975 Mar;72(3):809–813. doi: 10.1073/pnas.72.3.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Cleary P. P., Campbell A., Chang R. Location of promoter and operator sites in the biotin gene cluster of Escherichia coli. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2219–2223. doi: 10.1073/pnas.69.8.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary P. P., Campbell A. Deletion and complementation analysis of biotin gene cluster of Escherichia coli. J Bacteriol. 1972 Nov;112(2):830–839. doi: 10.1128/jb.112.2.830-839.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary P. P., Dykhuizen D. Enzymatic reduction of D-biotin-d-sulfoxide with cell-free extracts of Escherichia coli. Biochem Biophys Res Commun. 1974 Feb 4;56(3):629–634. doi: 10.1016/0006-291x(74)90651-2. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Genetic expression in bacteriophage lambda. 3. Inhibition of Escherichia coli nucleic acid and protein synthesis during lambda development. J Mol Biol. 1970 May 14;49(3):557–575. doi: 10.1016/0022-2836(70)90281-0. [DOI] [PubMed] [Google Scholar]
- Del Campillo-Campbell A., Kayajanian G., Campbell A., Adhya S. Biotin-requiring mutants of Escherichia coli K-12. J Bacteriol. 1967 Dec;94(6):2065–2066. doi: 10.1128/jb.94.6.2065-2066.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen D. Genetic analysis of the system that reduces biotin-d-sulfoxide in Escherichia coli. J Bacteriol. 1973 Aug;115(2):662–667. doi: 10.1128/jb.115.2.662-667.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg M. A. Biotin: biogenesis, transport, and their regulation. Adv Enzymol Relat Areas Mol Biol. 1973;38:317–372. doi: 10.1002/9780470122839.ch7. [DOI] [PubMed] [Google Scholar]
- Guha A. Divergent orientation of transcription from the biotin locus of Escherichia coli. J Mol Biol. 1971 Feb 28;56(1):53–62. doi: 10.1016/0022-2836(71)90083-0. [DOI] [PubMed] [Google Scholar]
- Hofnung M. Divergent operons and the genetic structure of the maltose B region in Escherichia coli K12. Genetics. 1974 Feb;76(2):169–184. doi: 10.1093/genetics/76.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G. A. Control of the argECBH cluster in Escherichia coli. Mol Gen Genet. 1972;117(4):337–348. doi: 10.1007/BF00333027. [DOI] [PubMed] [Google Scholar]
- Kayajanian G., Campbell A. The relationship between heritable physical and genetic properties of selected gal- and gal+ transducing lambda dg. Virology. 1966 Nov;30(3):482–492. doi: 10.1016/0042-6822(66)90124-3. [DOI] [PubMed] [Google Scholar]
- Ketner G., Campbel A. Operator and promoter mutations affecting divergent transcription in the bio gene cluster of Escherichia coli. J Mol Biol. 1975 Jul 25;96(1):13–27. doi: 10.1016/0022-2836(75)90179-5. [DOI] [PubMed] [Google Scholar]
- Lane M. D., Moss J., Polakis S. E. Acetyl coenzyme A carboxylase. Curr Top Cell Regul. 1974;8(0):139–195. [PubMed] [Google Scholar]
- MELVILLE D. B. Biotin sulfoxide. J Biol Chem. 1954 Jun;208(2):495–501. [PubMed] [Google Scholar]
- MONOD J., PAPPENHEIMER A. M., Jr, COHEN-BAZIRE G. La cinétique de la biosynthèse de la beta-galactosidase chez E. coli considérée comme fonction de la croissance. Biochim Biophys Acta. 1952 Dec;9(6):648–660. doi: 10.1016/0006-3002(52)90227-8. [DOI] [PubMed] [Google Scholar]
- Signer E. R., Manly K. F., Brunstetter M. A. Deletion mapping of the c-3-N region of bacteriophage. Virology. 1969 Sep;39(1):137–141. doi: 10.1016/0042-6822(69)90356-0. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Casadaban M. J., Shuman H. A., Beckwith J. R. Conversion of beta-galactosidase to a membrane-bound state by gene fusion. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3423–3427. doi: 10.1073/pnas.73.10.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]


