Abstract
Background
Xp10 and OP1 are phages of Xanthomonas oryzae pv. oryzae (Xoo), the causative agent of bacterial leaf blight in rice plants, which were isolated in 1967 in Taiwan and in 1954 in Japan, respectively. We recently isolated the Xoo phage Xop411.
Results
The linear Xop411 genome (44,520 bp, 58 ORFs) sequenced here is 147 bp longer than that of Xp10 (60 ORFs) and 735 bp longer than that of OP1 (59 ORFs). The G+C contents of OP1 (51%) and Xop411 and Xp10 (52% each) are less than that of the host (65%). The 9-bp 3'-overhangs (5'-GGACAGTCT-3') in Xop411 and Xp10 are absent from OP1. More of the deduced Xop411 proteins share higher degrees of identity with Xp10 than with OP1 proteins, while the right end of the genomes of Xp10 and OP1, containing all predicted promoters, share stronger homology. Xop411, Xp10, and OP1 contain 8, 7, and 6 freestanding HNH endonuclease genes, respectively. These genes can be classified into five groups depending on their possession of the HNH domain (HNN or HNH type) and/or AP2 domain in intact or truncated forms. While the HNN-AP2 type endonuclease genes dispersed in the genome, the HNH type endonuclease genes, each with a unique copy, were located within the same genome context. Mass spectrometry and N-terminal sequencing showed nine Xop411 coat proteins, among which three were identified, six were assigned as coat proteins (4) and conserved phage proteins (2) in Xp10. The major coat protein, in which only the N-terminal methionine is removed, appears to exist in oligomeric forms containing 2 to 6 subunits. The three phages exhibit different patterns of domain duplication in the N-terminus of the tail fiber, which are involved in determination of the host range. Many short repeated sequences are present in and around the duplicated domains.
Conclusion
Geographical separation may have confined lateral gene transfer among the Xoo phages. The HNN-AP2 type endonucleases were more likely to transfer their genes randomly in the genome and may degenerate after successful transmission. Some repeated sequences may be involved in duplication/loss of the domains in the tail fiber genes.
Background
Xanthomonas oryzae pv. oryzae (Xoo) is a gram-negative plant pathogenic bacterium that causes leaf blight in rice plants, thus having a serious effect on rice production in Taiwan, China, Japan, India, and South America [1]. Agrochemicals have been somewhat effective for disease control, although biological control using bacteriophages has been considered [2]. In addition, phages that specifically infect Xoo have been used to type Xoo hosts in the field [3,4].
Among the Xoo phages are the lytic phages Xp10, Xp12, OP1, and OP2, and the filamentous phages Xf and phiXo [2-8]. Recently, the genomic sequences of Xp10 (44,373 bp, 60 ORFs), OP1 (43,785 bp, 59 ORFs), and OP2 (46,643 bp, 62 ORFs) were determined. Xp10 and OP1 both have linear genomes and share high degrees of similarity at both the nucleotide and amino acid levels [2,6]. In contrast, OP2 has a circularly permuted and terminally redundant genome, which differs in sequence from those of Xp10 and OP1 [2,6,8]. Xp15 is a phage of X. campestris pv. pelargonii; its genomic sequence (55,770 bp) is available in the NCBI database (AY986977).
We recently isolated a Xoo bacteriophage, Xop411, from rice plants from a rice paddy near National Chung Hsing University that showed serious symptoms of bacterial leaf blight [9]. During our sequencing of the Xop411 phage genome, the genomic sequences of Xp10 and OP1 were published [2,6]. Since comparative analysis of several bacteriophages from a single species offers a unique opportunity to study the mechanisms that drive prokaryotic genetic diversity [10], we compared the sequence of Xop411 with those of Xp10, isolated in Taiwan in 1967, and OP1, isolated in Japan in 1954 [2,4,6,7].
Results and discussion
Assignments of Xop411 genes
Assembly of over 450 overlapped sequences (over 6× coverage) of the Xop411 genome showed that it was linear and consisted of 44,520 bp. The terminal sequence, 5'-GGACAGTCT-3', is identical to the 9-bp 3'-protruding sequence of Xp10 but was not observed in OP1 [2,6]. The G+C contents of the three Xoo phages were similar, 52% each for Xop411 and Xp10 and 51% for OP1 [6], but deviated from the 65% of Xoo [11]. The three phages showed highly similar genomic organization and highly similar protein products (Figure 1, Table 1). The Xp10 gene numbers were used for the corresponding genes (58) in Xop411, with a two-part number assigned when an additional gene was present, for example 31.1 for the gene between 31 and 32 (Additional file 1). Some genes, encoding HNH endonucleases (underlined) or hypothetical proteins, were only found in Xop411 (p42.1, p55.1, p57.1), Xp10 (p03, p04, p40, and p59) or OP1 (ORF15, 16, and 32), or were missing only from Xop411 (p05/ORF3, p47/ORF47, and p50/ORF50), Xp10 (p27.1/ORF26.1, p31.1/ORF31) or OP1 (p17/p17) (Figure 1, Table 1). These findings indicate that numerous insertions/deletions have occurred in the Xoo phages. More deduced Xop411 proteins shared higher degrees of similarity with Xp10 than with OP1 proteins. Only 15 Xop411 proteins, most located between p23 and p31, shared higher identities with OP1 than with Xp10 proteins (Table 1). These findings suggest that Xop411 is more closely related to Xp10 than to OP1. Although sequence information from more phages is required, discrepancies in similarity indicate that geographical separation may have limited lateral gene transfer between phages and other sources.
Figure 1.
Genomic organization of phages Xop411, Xp10 and OP1. Colored arrows indicate the directions and categories (denoted below) of the genes. The bars between the genomic maps indicate the identities at the nucleotide level between Xop411 and Xp10 (upper) and between Xop411 and OP1 (lower); black denotes regions of > 80% identity; grey denotes regions of 65–80% identity; and white denotes regions of < 65% identity. Insertions are indicated with filled arrowheads and deletions with empty arrowheads. Knobs indicate the positions of predicted terminators.
Table 1.
Comparison of proteins deduced from the genes of X. oryzae phages Xop411, Xp10, and OP1.
| Xop411 | Xp10 | OP1 | Xop411 with Xp10 | Xop411 with OP1 | Xp10 with OP1 | |||
| Gene | Length (aa) | Gene | Length (aa) | Gene | Length (aa) | id (%)/aligned aa | id (%)/aligned aa | id (%)/aligned aa |
| p01 | 72 | p01 | 72 | ORF1 | 72 | 98/72 | 88/72 | 88/72 |
| p02 | 101 | p02 | 101 | ORF2 | 101 | 90/101 | 81/80 | 75/79 |
| p03 | 154 | 61/134 (p17)a | 65/134 (ORF31)b | |||||
| 64/130 (p31.1)a | 55/134 (ORF58)b | |||||||
| 61/134 (p42.1)a | ||||||||
| 63/142 (p55.1)a | ||||||||
| p04 | 64 | 100/64 (p06)a | 69/56 (ORF4)b | |||||
| p05 | 67 | ORF3 | 65 | - | - | 65/49 (ORF3)b | ||
| 51/35 (ORF58)b | ||||||||
| p06 | 569 | p06 | 561 | ORF4 | 561 | 93/561 | 84/561 | 83/561 |
| p07 | 432 | p07 | 432 | ORF5 | 432 | 99/432 | 98/432 | 98/432 |
| p08 | 245 | p08 | 245 | ORF6 | 245 | 76/245 | 97/245 | 77/245 |
| p09 | 390 | p09 | 390 | ORF7 | 390 | 98/390 | 97/390 | 98/390 |
| p10 | 115 | p10 | 115 | ORF8 | 115 | 95/115 | 92/115 | 90/115 |
| p11 | 124 | p11 | 124 | ORF9 | 125 | 96/124 | 96/123 | 98/123 |
| p12 | 119 | p12 | 119 | ORF10 | 157 | 98/119 | 90/118 | 88/118 |
| p13 | 118 | p13 | 118 | ORF11 | 118 | 96/118 | 91/118 | 94/118 |
| p14 | 210 | p14 | 210 | ORF12 | 210 | 94/210 | 96/210 | 97/210 |
| p15 | 100 | p15 | 100 | ORF13 | 100 | 95/100 | 88/100 | 89/100 |
| p16 | 100 | p16 | 100 | ORF14 | 68 | 97/100 | 83/68 | 85/68 |
| ORF15 | 76 | - | - | |||||
| ORF16 | 510 | - | - | |||||
| p17 | 169 | p17 | 172 | 66/171 (p17)c | 62/163 (ORF31)b | 64/164 (ORF31)b | ||
| 65/164 (p50)c | 51/168 (ORF58)b | 57/169 (ORF58)b | ||||||
| 61/166 (p58)c | ||||||||
| 61/134 (p03)c | ||||||||
| p18 | 998 | p18 | 998 | ORF17 | 999 | 94/998 | 90/999 | 91/999 |
| p19 | 118 | p19 | 118 | ORF18 | 117 | 98/118 | 96/117 | 94/117 |
| p20 | 152 | p20 | 152 | ORF19 | 152 | 98/152 | 97/152 | 99/152 |
| p21 | 130 | p21 | 146 | ORF20 | 146 | 99/130 | 95/130 | 95/146 |
| p22 | 1573 | p22 | 1574 | ORF21 | 1571 | 97/1574 | 96/1573 | 96/1574 |
| p23 | 96 | p23 | 96 | ORF22 | 100 | 66/96 | 97/96 | 67/96 |
| p24 | 231 | p24 | 230 | ORF23 | 231 | 61/231 | 93/231 | 63/231 |
| p25 | 123 | p25 | 122 | ORF24 | 123 | 97/109 | 96/109 | 95/109 |
| p26 | 453 | p26 | 498 | ORF25 | 431 | 79/499 | 88/453 | 72/499 |
| p27 | 69 | p27 | 74 | ORF26 | 69 | 100/62 | 97/69 | 96/62 |
| p27.1 | 98 | p28 | 223 | ORF26.1d | 98 | 93/43 | 96/98 | 96/31 |
| p28 | 178 | p28 | 223 | ORF27 | 166 | 92/177 | 94/167 | 93/166 |
| p29 | 111 | p29 | 99 | ORF28 | 111 | 81/77 | 90/111 | 81/79 |
| p30 | 58 | p30 | 63 | ORF29 | 58 | 67/53 | 81/58 | 59/42 |
| p31 | 293 | p31 | 332 | ORF30 | 288 | 71/289 | 77/289 | 80/289 |
| p31.1 | 160 | ORF31 | 167 | 71/157 (p50)c | 68/158 (ORF31)b | 69/163 (p50)c | ||
| 67/157 (p58)c | 56/157 (ORF58)b | 69/166 (p58)c | ||||||
| 65/155 (p17)c | 64/164 (p17)c | |||||||
| 63/124 (p03)c | 65/134 (p03)c | |||||||
| ORF32 | 91 | - | - | |||||
| p32 | 746 | p32 | 746 | ORF33 | 774 | 91/746 | 89/746 | 87/746 |
| p33 | 255 | p33 | 255 | ORF34 | 188 | 94/255 | 74/189 | 75/189 |
| p34 | 86 | p34 | 172 | ORF35 | 86 | 98/72 | 97/72 | 96/82 |
| p35 | 245 | p35 | 245 | ORF36 | 279 | 100/245 | 97/270 | 97/245 |
| p36 | 134 | p36 | 135 | ORF37 | 135 | 99/116 | 97/116 | 96/116 |
| p37 | 309 | p37 | 309 | ORF38 | 309 | 97/309 | 94/309 | 94/309 |
| p38 | 266 | p38 | 201 | ORF39 | 288 | 98/188 | 93/265 | 91/188 |
| p39 | 793 | p39 | 794 | ORF40 | 793 | 98/794 | 95/792 | 95/792 |
| p40 | 139 | 100/112 (p41)a | 97/113 (ORF41)b | |||||
| p41 | 374 | p41 | 261 | ORF41 | 438 | 98/260 | 94/372 | 92/260 |
| 100/112 (p40)c | 97/113 (p40)c | |||||||
| p42 | 280 | p42 | 280 | ORF42 | 282 | 82/280 | 85/280 | 85/280 |
| p42.1 | 167 | 68/165 (p17)c | 65/163 (ORF31)b | |||||
| 65/164 (p58)c | 58/164 (ORF58)b | |||||||
| 66/163 (p50)c | ||||||||
| 61/134 (p03)c | ||||||||
| p43 | 53 | p43 | 50 | ORF43 | 81 | 81/49 | - | - |
| p44 | 222 | p44 | 222 | ORF44 | 255 | 89/222 | 78/222 | 81/222 |
| p45 | 105 | p45 | 104 | ORF45 | 105 | 76/102 | 83/105 | 79/102 |
| p46 | 73 | p46 | 73 | ORF46 | 73 | 71/73 | 72/73 | 84/73 |
| p47 | 67 | ORF47 | 90 | - | - | 67/67 | ||
| p48 | 134 | p48 | 138 | ORF48 | 108 | 71/134 | 53/107 | 70/108 |
| p49 | 179 | p49 | 151 | ORF49 | 161 | 80/150 | 59/159 | 63/151 |
| p50 | 174 | ORF50 | 52 | 65/164 (p17)a | - | - | ||
| 70/162 (p31.1)a | 69/163 (ORF31)b | |||||||
| 66/163 (p42.1)a | 53/163 (ORF58)b | |||||||
| 66/168 (p55.1)a | ||||||||
| p51 | 141 | p51 | 144 | ORF51 | 142 | 80/140 | 86/141 | 74/140 |
| p52 | 50 | p52 | 50 | ORF52 | 72 | 86/50 | - | - |
| p53 | 63 | p53 | 78 | ORF53 | 62 | 67/64 (p53)c | 74/58 (ORF53)b | 75/44 (p53)c |
| 43/55 (p17)c | 42/42 (ORF31)b | 50/36 (p58)c | ||||||
| 53/26 (p58)c | 50/22 (ORF50)b | 52/36 (p50)c | ||||||
| 44/43 (p50)c | 45/37 (p17)c | |||||||
| p54 | 111 | p54 | 111 | ORF54 | 120 | 87/111 | 80/110 | 70/110 |
| p55 | 56 | p55 | 55 | ORF55 | 35 | 67/55 | 62/37 | 88/35 |
| p55.1 | 189 | 66/168 (p50)c | 64/162 (ORF31)b | |||||
| 66/163 (p58)c | 55/163 (ORF58)b | |||||||
| 65/169 (p17)c | ||||||||
| 63/142 (p03)c | ||||||||
| p56 | 65 | p56 | 75 | ORF56 | 79 | 67/64 | 51/64 | 56/73 |
| p57 | 114 | p57 | 64 | ORF57 | 132 | - | 65/102 | - |
| p57.1 | 277 | 38/104 (p17)c | 35/138 (ORF31)b | |||||
| 39/104 (p03)c | 32/103 (ORF58)b | |||||||
| 31/173 (p50)c | ||||||||
| 37/103 (p58)c | ||||||||
| p58 | 174 | p58 | 167 | ORF58 | 172 | 37/114 (p17)c | 34/116 (ORF31)b | 57/169 (p17)c |
| 35/122 (p50)c | 35/110 (ORF58)b | 56/167 (p58)c | ||||||
| 36/98 (p58)c | 53/163 (p50)c | |||||||
| 35/98 (p03)c | 55/134 (p03)c | |||||||
| p59 | 124 | - | - | - | ||||
| p60 | 119 | p60 | 119 | ORF59 | 131 | 71/114 | 69/119 | 73/119 |
a, b, and c indicate sequence comparison with a protein from Xop411, OP1, and Xp10 itself, respectively. d, predicted ORF in this study. Symbol -, no similarity was found.
Holin genes required for host lysis were not assigned for Xp10 and OP1 [2,6]. These genes are usually small and adjacent to the cognate lysozyme genes, with their protein products usually containing at least one transmembrane domain (TMD) and a hydrophilic C-terminal domain [12]. In Xop411, p27.1 (98 aa, with one TMD at aa 25–47), located upstream of the previously characterized lysozyme gene (p28) [9], was assigned as the putative holin gene. However, since p27.1 overlaps with p28 by 104 bp and lacks a hydrophilic C-terminal domain, it is unclear whether it encodes holin function. A corresponding ORF was identified in OP1, but the corresponding region in Xp10 was assigned to the N-terminus of the lysozyme gene (Table 1).
The next best matched ORFs other than those from Xp10 and OP1
The deduced Xop411 proteins also share similarities with proteins other than those of Xp10 and OP1, and proteins encoded in five Xop411 regions are worth noting (see Additional file 2): 1) The tail-related proteins p19 to p22, encoded in a 5.9-kb region, share 33–44% identity (55–63% similarity) with ORFs of the X. campestris pv. pelargonii phage Xp15. 2) Proteins p26 to p28, encoded in a 2.3-kb region and including tail fiber and phage lysozyme, show 33–48% identity to proteins from Chromobacterium violaceum. 3) Proteins p35 to p37, encoded in a 2.1-kb region, share 30–47% identity with proteins from Pseudomonas aeruginosa. 4) Proteins p38 to p41, encoded in a 4.3-kb region, show 38–45% identity to proteins from Burkholderia pseudomallei. 5) Protein p33 shares 60% identity with a protein from Bradyrhizobium sp. In addition, Xop411 p08 (ClpP protease), p28 (lysozyme) and p39 (DNA polymerase I) are similar to proteins from Xylella fastidiosa (25–38% identity) and X. axonopodis pv. citri (42% identity) (see Additional file 2). These data suggest that Xoo phages have actively participated in gene transfer with several organisms. In contrast, the Xoo genome did not contain homologues with significant similarity (i.e. with expected values less than e-4) to the proteins of the three phages. Since the Xoo phages are lytic, opportunities to exchange genetic material with the host may have been rare.
Gene products related to endonucleases of the HNH family
Members of the HNH endonuclease family are encoded by free-standing ORFs between genes or within introns or inteins in viruses, bacteriophages, and bacteria, as well as in eukaryotic nuclear and organellar genomes [13]. Most of these proteins are homing endonucleases involved in the mobility of their own genes or of the introns/inteins in which they are located [13-15]. These HNH proteins are characterized by the motif His-Asn-His at the N-terminus but share little overall sequence similarity and can be classified into 8 subsets [16]. Proteins of the second subset usually consist of an HNN domain and an adjacent DNA-binding domain, AP2 (Pfam:PF00847) or IENRI (Smart:SM00479), and are found primarily in phage genomes [17,18]. For example, multiple copies of HNH endonuclease genes are present in the sequenced genomes of coliphages RB16 (DQ023482-7), RB43 (NC_007023), T1 [19], Rtp [20] and T5 [21] as well as in the lactophage bIL170 [22].
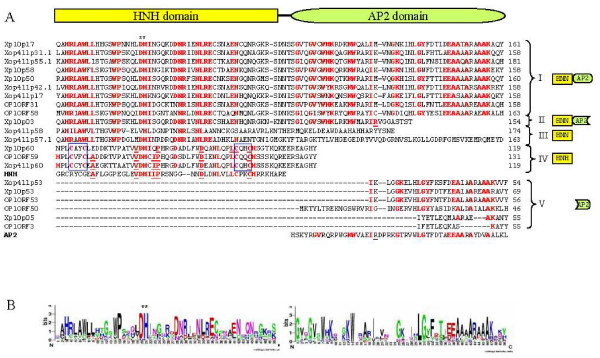
Xp10 and OP1 contain 7 (p03, p05, p17, p50, p53, p58, and p60) and 6 (ORF 3, 31, 50, 53, 58, and 59) genes encoding HNH endonucleases, respectively [2,6]. It was suggested that i) these proteins conserve many functionally important residues which may preserve their ability to bind DNA, ii) these genes may have populated the genomes through gene duplication and/or transposition, iii) their presence may account for the branched DNA structures observed by electron microscopy following denaturation and renaturation, and iv) one or more of these HNH family proteins may be involved in domain duplication of the tail fiber, which can alter the host range (see below) [2,6]. The Xop411 genome was found to contain 8 (p17, p31.1, p42.1, p53, p55.1, p57.1, p58, and p60) HNH endonuclease genes (Figures 1, 2). Using Weblogo to analyze these 21 Xoo phage proteins, consensus sequences were generated for the HNH and AP2 domains (Figure 2) [23]. They could be divided into 5 groups (Figure 2A). The HNN domain was found in proteins of groups I (9 proteins, each with intact HNN and AP2 domains), II (1 protein, with intact HNN and C-terminally truncated AP2 domains) and III (2 proteins, each containing only an HNN domain but no AP2 domain), whereas the HNH domain was detected only in the 3 proteins of group IV, which do not retain an AP2 domain. The 6 proteins in group V had degenerated, losing their HNH domains and over half of the N-terminus of their AP2 domains. A phylogenetic tree based on the alignment of 50 conserved amino acids of the HNH domain of the 15 proteins in groups I to IV suggests that the HNH type endonucleases may have arisen from an ancestor different from that of the HNN type endonucleases (see Additional file 3).
Figure 2.
Alignment of the 21 putative HNH endonucleases from the three Xoo phages. (A) Sequence alignment. The conserved residues are in bold-face and the boxes indicated the cysteine dyads (CX2C) flanking the conserved Asp/His residue (*). (B) Consensus sequences of HNH and AP2 domains, displayed using Weblogo.
We found that all the HNN domain-containing proteins of the Xoo phages have conserved Asp/His residues flanked by two quasi-conserved boxes (HRLAWLL and WP) at the N-terminus and three conserved boxes (DNR, NLRE and EN) at the C-terminus, but do not have either metal-binding cysteine-dyads (CX2C) or conserved GG motifs (Figure 2A). The lack of a metal-binding motif suggests that these HNN type endonucleases may not require zinc ion to function. Since most HNN-AP2/IENRI proteins are intron-encoded site-specific endonucleases [16], the presence of multiple HNN-AP2 endonuclease genes in all three Xoo phage genomes suggests that these genes, like the homing-endonuclease genes (HEGs), are able to self-duplicate in the genome. However, since no conserved sequences could be identified in the flanking regions of these endonuclease genes and their genomic locations varied among the three phages, it is likely that transmission of these HNN-AP2 endonuclease genes was sequence-independent.
The HNH domains of the group IV proteins, which share higher degrees of similarity with the consensus HNH domain, have two cysteine-dyads (CX2C) flanking the conserved Asp/His residues, suggesting that zinc ion is required for their function, as well as two boxes (DX2NL and CH) on the C-terminal side of each domain (Figure 2A). These group IV proteins are similar to the HNH-type protein (gp13) found in the lactophage bIL170, which has two cysteine-dyads (CX2CX36CX2C) and no DNA-binding motif [22]. As endonucleases of this type are present as unique copies at the analogous positions of the Xoo phage genomes (the right end), they may have specific functions other than transposition, similar to the HNN-AP2 type endonucleases.
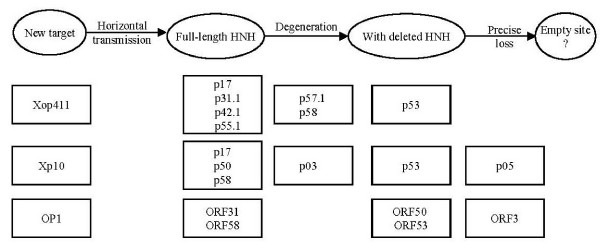
The HNN-AP2 type endonuclease genes may not only be able to transmit into multiple sites of the genome but may also degenerate. For example, although the genes hegG and hegJ are present in the genomes of three T5 phage strains, our sequence analyses showed that full-length genes are retained in the strains sequenced in France (GenBank accession numbers AY692264) and Moscow (AY543070), but that both genes had degenerated in the T5 strain ATCC11303-B5 (AY587007). Specifically, a short insertion disrupted hegG (AAX11946 and AAX11947) and two point deletions caused frame shifts in hegJ (AAX12048), suggesting that degeneration of HNH endonuclease genes may occur after a deleterious insertion/deletion. In addition, one T4 HNH type Mob endonuclease gene, mobA, was found to have degenerated into a pseudogene [24]. A cyclical model of gain and loss of HEGs [25,26] has been used to deduce the possible evolutionary path of the I-SceI endonucleases of a self-splicing group I intron in Saccharomyces cerevisiae and the intron/HEG of T-even-like phages [27,28]. For Xoo phages, however, the data may be better explained by a linear model of gain and loss, in which functional alien endonuclease genes would be fixed but start degenerating after successful incorporation (Figure 3). For example, in Xp10, proteins p17, p50 and p58, each with intact HNH and AP2 domains, may represent endonuclease family members that retain their functions; proteins p03 (with a complete HNN domain but lacking the C-terminus of AP2) and p53 (retaining only the C-terminus of AP2) may represent proteins after different degrees of progressive degeneration; and protein p05, which has only a small segment of a highly degenerated AP2 domain, may represent a gene product with the greatest extent of degeneration and the most ancient HNH-AP2 endonuclease in the genome (Figure 3). Similar clues to gene degeneration were observed in the HNH-AP2 endonuclease genes of the other two phages (Figure 3).
Figure 3.
Linear gains and losses of HNN-AP2 endonuclease genes in the three Xoo phages. The boxes contain the possible proteins at different stages of degeneration.
Promoters and terminators
We found that the nucleotide sequences between the end of p55 and the right end of the genome were highly variable in the three Xoo phages, with Xp10 and OP1 being more similar to each other than either were to Xop411, and segments with higher degrees of identity present at different positions (see Additional file 4). Mosaicism of the common segments suggests that these phages have undergone numerous recombination events, possibly during co-infections, resulting in gene rearrangements and insertion/deletion. In Xp10, the intergenic region between p57 and p58 separates the genes transcribed leftward and rightward and contains all six promoters [6,29,30]. Based on a similarity search, we located putative promoters resembling those of Xp10 in Xop411 and OP1. We found that the promoter sequences in Xp10 and OP1 were highly conserved, but shared lower degrees of identity with the Xop411 promoters (see Additional file 5). In addition, Xop411 had five sequences located between p56 and p57, and one sequence, P3, between p57.1 and p58, whereas OP1 had four sequences located between ORF57 and ORF58 and two, Pup and φP1, contained within ORF57 (see Additional file 4).
Similarity searches of the Xop411 and OP1 genomes for the five predicted terminators of Xp10 [29] revealed four corresponding sequences at analogous positions (Figure 1, Table 2). These predicted terminators in Xop411, TR2 to TR5, each shared high degrees of identity with the respective analogous terminators in the other phages. However, sequences similar to Xp10 TR1, which is thought to possess a low efficiency of termination [29], were not found in Xop411 and OP1, suggesting that such a low-efficiency terminator may not be essential.
Table 2.
Predicted terminators in Xp10, Xop411, and OP1 genome.
| Terminator name | Phage | Positions | Sequence | Energy score | Tail score | Identity/aligned base |
| TR1 | Xp10 | 1390~1411 | CTGCCCTACTTATGGGCAGTTT | 7.4 | 3.5 | 12/12 |
| TR2 | Xp10 | 6573~6603 | GGGAGGGGCTGGGAAACTGGCCCCTCTCTTT | 13.4 | 3.6 | 18/18 |
| Xop411 | 5704~5735 | GGGAGGGGCTGGGGGAACTGGCCCCTCTCTTT | 12.4 | 3.4 | 18/18 | |
| OP1 | 5842~5872 | GAGAGGGGCTAGGAAACTGGCCCCTCTCTTT | 17.8 | 3.6 | 18/18 | |
| TR3 | Xp10 | 19285~19308 | GGGGCAGGGTTTCCTGCCCCATTT | 15.0 | 3.3 | 16/16 |
| Xop411 | 18320~18343 | GGGGCAGGGTTTCCTGCCCCATTT | 15.0 | 3.3 | 16/16 | |
| OP1 | 19466~19489 | GGGGCAGGCTTTCCCTGCCCCTTT | 14.0 | 4.4 | 16/16 | |
| TR4 | Xp10 | 23718~23760 | GGGAGGGAGCTAAGCCTTAATGGCCTAGCCCCTCCCTTTTTTT | 15.5 | 5.9 | 28/28 |
| Xop411 | 22629~22670 | ATAGGGGACCTATTGCCTTTAATGGCAGGGTCCCCTTTTTTT | 24/28 | |||
| OP1 | 23709~23752 | GGGAGGGAGCTAAGCCTTTAATGGCCTAGCCCCTCCCTTTTTT | 14.5 | 5.7 | 28/28 | |
| TR5 | Xp10 | 42740~42759 | CTGAACGATCCGTTCAGTTT | 10.9 | 3.5 | 14/14 |
| Xop411 | 43861~43880 | CTGAACGGCTCGTTCAGTTT | 10.9 | 3.6 | 14/14 | |
| OP1 | 42375~42394 | CTGAACGATCCGTTCAGTTT | 10.9 | 3.8 | 14/14 |
Domain duplications in tail fiber and implications in host range
Japanese isolates of Xoo can be classified into four phagovars, based on their susceptibility to OP1, with host-range mutants of OP1 capable of infecting different phagovars [2]. Sequencing of the tail fiber genes from these phage strains revealed that changes in host range are due to duplications in at least one of three domains (domains 1, 2, and 3) in ca. 118 aa at the N-terminus (see Additional file 6). This is similar to findings in other phages; for example, the host range of T4 is expanded by duplications of a small region of the tail fiber adhesin [31]. Amino acid sequence alignments showed that OP1 possesses domains -1-2-3-, Xp10 has domains -1-2-2-2-3- [2] and Xop411 exhibits domains -1-2-3-3-3- (see Additional file 6). Interestingly, while OP1 and OPh1 have the same domain architecture (-1-2-3-) and no drastic changes in the surrounding amino acid residues, OP1 infects only phagovar A whereas OP1h infects only phagovar B (see Additional file 6) [2]. This finding suggests that these related Xoo phages might use a complex structure, also containing other component(s), to determine the host range, with mutations in the latter component(s) altering the host range. Further tests are needed to understand the host ranges of Xop411 and Xp10.
In mouse minisatellite Pc-1, tandem repeats of d(GGCAG)n, which can facilitate the formation of a telomere-like intra-molecular folded-back quadruplex structure, have been shown to be hotspots of recombination during meiosis [32-34]. The genes encoding the tail fibers of the Xoo phages contain many short repeats (see Additional File 7), including i) inverted repeats that are all located outside the domains, which may be important in the acquisition/loss of domain architectures, ii) direct G-rich pentanucleotide (GGCAG) repeats at both ends of domains 1 and 2, and iii) a direct G-rich octanucleotide (CAGGCCGC) repeat flanking domain 3. It is currently unclear whether the presence of these short direct repeats can facilitate the duplication/deletion of the tail fiber domains by recombination, as observed for mouse minisatellite Pc-1. Inoue et. al. proposed that the HNH-family proteins may be involved in domain duplication via recombination using Holliday junction structures as the intermediates [2], but it is not clear if this is the mechanism occurring here.
Identification of virion proteins
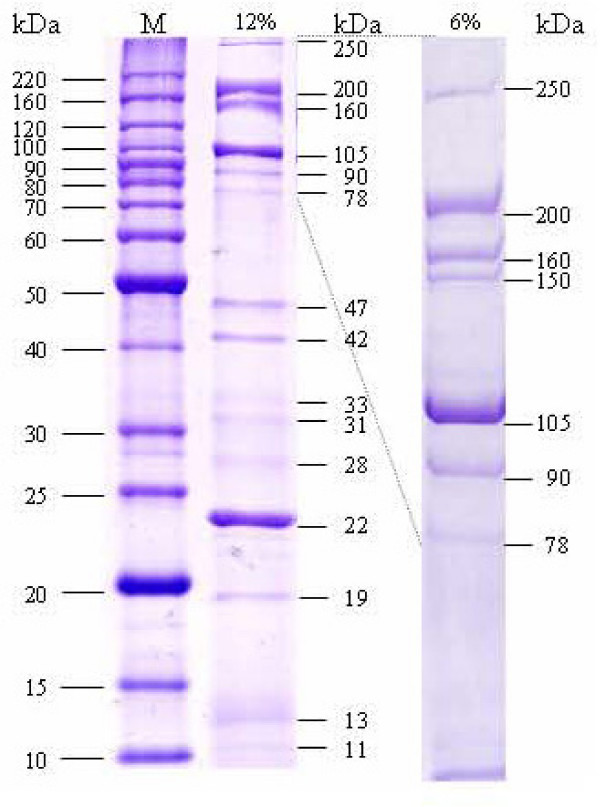
SDS-PAGE separation of the Xp10 virion proteins resulted in 6 major bands, three of which (p09, major head; p14, major tail; p26, tail fiber) were identified [6]. SDS-PAGE separation of the Xop411 virion proteins resulted in at least 16 discrete bands: 15 (of MW 250, 200, 160, 105, 90, 78, 47, 42, 33, 31, 28, 22, 19, 13, and 11 kDa) on 12% gels and 7 (of MW 250, 200, 160, 150, 105, 90, and 78 kDa) on 6% gels (Figure 4). LC MS/MS analysis (see Additional file 8) indicated that these bands contained 14 proteins, 9 from the virion and 5 from the host. The 250-, 200-, 150-, 78- and 42-kDa bands contain oligomers of p09, the 41.5-kDa major capsid subunit, of 2 to 6 subunits. Oligomerization of p09 was also observed in Xp10, but in the 140- and 165-kDa bands and in high MW materials in the gel wells. Xp10 p09 may be cleaved by a phage-encoded protease, p08, generating a mature major head protein of 283 aa, which is 170 aa less than the precursor protein [6]. In contrast, our N-terminal sequencing of the 42-kDa band gave a sequence, TDITSK, showing that only the N-terminal methionine was missing.
Figure 4.
SDS-polyacrylamide gel electrophoresis of Xop411 virion proteins. The purified Xop411 particles were separated in 12% (middle lane) or 6% (right lane) polyacrylamide gels and stained with Coomassie brilliant blue. The proteins are named by their apparent sizes. Lane M contained molecular weight markers.
The head portal protein, p07, with a calculated MW of 47 kDa, was found in the 47- and 31-kDa bands, suggesting that the unprocessed and processed forms co-exist in the virions. LC-MS/MS analysis showed that the 31-kDa band contained another protein, p26, which was identified as the tail fiber in Xp10 [6]. N-terminal sequencing showed that the 22-kDa band was p14, the major tail protein in Xp10. The 13-kDa band was also a doublet, containing p10 (phage conserved protein in Xp10) and p19 (tail protein). The 160-, 105-, and 11-kDa bands were identified as p22 (tail protein), p18 (tail length tape measure protein), and p13 (phage conserved protein in Xp10), respectively. In summary, six more proteins than those identified for Xp10 were found here, and the conserved proteins p10 and p13 in Xp10 were found to be phage coat proteins.
The 5 host proteins in the 4 bands were TonB-dependent receptor FyuA (90-kDa), outer membrane protein MopB and hypothetical protein XOO0584 (33-kDa), MopB and colicin receptor protein CirA (28-kDa), and hypothetical protein XOO4199 (19-kDa). Since the experiments were repeated four times using virions freshly purified by ultracentrifugation, the consistent presence of these proteins indicates that they were rather tightly associated with the phage particles.
Conclusion
Our results, showing that Xop411 and Xp10 have the same G+C content and that more of the deduced Xop411 proteins share higher degrees of identity with Xp10 than with OP1 proteins, indicate that the two phages isolated in Taiwan are more closely related to each other than they are to OP1. Thus, geographical separation may have limited lateral gene transfers between phages and other sources. However, our finding that more of the DNA sequences are conserved by Xp10 and OP1 in the region between p55 and the right end of the genome, a region containing the predicted promoters, suggests that Xop411 has undergone sequence rearrangements and insertions/deletions to a greater degree. The HNN-AP2 type endonucleases may have transferred their genes randomly and begun degenerating after successful horizontal transmission, whereas the HNH type endonucleases, each with one copy, were located within the same genome context. Comparison of the host range and the architecture of the duplicated domains in the N-terminus of the tail fiber proteins suggests that the Xoo phages may need additional components for adsorption. Some of the repeated sequences in and around the domains may be involved in duplication/loss of the domains. We identified 6 more proteins than those identified for Xp10, with p10 and p13 shown to be phage coat proteins.
Methods
Bacteria, bacteriophages, and growth conditions
X. oryzae pv. oryzae (Xoo) was cultivated in Tryptic Soy Broth or Agar (Bacto™) at 28°C and Escherichia coli was grown in LB medium at 37°C. Ampicillin (50 μg/ml) was added when necessary. The procedures described previously [9] were used for plaque assay, phage propagation (using Xoo strain 21 as the host), purification of phage particles, and isolation and restriction enzyme digestion of phage DNA.
Sequence analyses
The purified phage DNA was treated in a HydroShear (GeneMachines, San Carlos, CA). Fragments of 1.0 to 3.0 kb were isolated and ligated into the EcoRV site of pBluescript II SK. Clones were randomly picked and subjected to nucleotide sequencing (ABI 3700). To determine the 3'-protruding terminal sequences (gap closure), the Xop411 genomic DNA was treated with or without Klenow enzyme, using its 3'→5' exonuclease activity and ligated using T4 ligase, and the ligation products were PCR-amplified separately with a pair of primers annealed close to the ends, followed by sequencing of the amplicons. Thus the extra nucleotides, obtained from the PCR product amplified on the template that had not been treated with Klenow enzyme, represented the 3'-protruding sequence. A+T content was analyzed by using the program available online [35]. DNA sequences were assembled using the SeqMan program from the DNASTAR package (DNASTAR, Madison, WI) and analyzed with NCBI software [36]. ORF was predicted using GeneMark. The nucleotide sequence of phage Xop411 has been deposited in GenBank under accession no. DQ777876.
HNH endonucleases were identified by searching for conserved domains as well as similarities to the endonucleases identified in Xp10 [6]. The BLAST program was used to search for nucleotide and amino acid similarities, and phylogenetic analysis was performed using the parsimony method (Phylip package ver. 3.66). Bootstrap values were obtained for a consensus based on 1000 randomly generated trees using SEQBOOT and CONSENSE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and LC-MS/MS analysis
Phage particles purified by ultracentrifugation were mixed with sample buffer, heated in a boiling water bath for 3 min, and subjected to SDS-PAGE separation in 12% or 6% (w/v) polyacrylamide gel. Protein bands were visualized by staining the gels with Coomassie brilliant blue, excised from the gels and subjected to LC-MS/MS (ABI Qstar System) analysis at the Biotechnology Center, National Chung Hsing University.
N-terminal amino acid sequencing of proteins
The proteins from the Xop411 particles separated in SDS-PAGE were transferred to polyvinylidene difluoride membranes and stained with Coomassie brilliant blue. Membrane strips containing the isolated protein bands were excised and subjected to Edman degradation to determine their N-terminal sequences (477A sequencer, PE Applied Biosystems).
Authors' contributions
CNL, RMH, YHT, and SFW participated in genome analysis, data interpretation and manuscript preparation. CNL and HYC performed the SDS-PAGE and LC MS/MS analysis. RMH, TYC and JWL participated in the study design and data interpretation and helped to draft the manuscript. All the authors have read and approved the final manuscript.
Supplementary Material
Gene assignment of the Xoo phage Xop411.
Similarities shared between the Xop411 proteins and those of Xylella and Xanthomonas.
Phylogenetic tree based on alignments of 50 conserved amino acids from the HNH domains of proteins in groups I to IV.
The region between p55 and the right end of genome of the three Xoo phages. Thick arrows indicate the direction and length of the genes. Corresponding genes are in the same colors, except that yellow indicates an additional gene. The horizontal bars represent percent identity of the nucleotide sequence, with black denoting > 80%, grey 65–80%, and white < 65% identity. Blocks A, B, C, D, and E of Xop411 and Xp10 (total, 842 bp) showed 71–92% identity; blocks F, G, H, and I of Xop411 and OP1 (total, 669 bp) showed 77–88% identity; and blocks J, K, L, M, N, O, and P of Xp10 and OP1 (total, 1,636 bp) showed 81–94% identity. The horizontal red lines indicate AT-rich regions of Xp10 (262 bp, nt 42,929–43,190 with 68% A+T), Xop411 (240 bp, nt 43,921-44,160 with 72% A+T, including an 80-bp internal segment of 96% A+T from nt 43,974-44,033 with 4 perfect 15-bp tandem direct repeats ATTATTAATATTTAT), and OP1 (336 bp, nt 42,631-42,966 with 63% A+T). These AT-rich regions are worth testing for the possibility of containing replication origins of the Xoo phages. Bent arrows and knobs represent the predicted promoters and terminators, respectively.
Alignment of promoters found in Xp10 and predicted for OP1 and Xop411. Bases identical to those of the Xp10 are on black background. Ratios to the right are bases matched to the Xp10 sequences. Color of the bases: blue, -35; green, conserved to Xp10 RNA polymerase promoters; orange, extended -10 promoter elements; pink, -10; yellow, transcription start sites. The extended -10 elements were found to be resistant to p7, the inhibitor of transcription initiation, in Xp10.
Domain duplication at the N-terminus of the deduced tail fiber proteins of Xoo phages. (A) Alignment of the sequence of Xop411 p26 with its homologues in Xp10, OP1, and four OP1 host range mutants. A domain is indicated by a line above the sequences with an Arabic number in circle. Different amino acid residues within the duplicated domains are shaded. (B) Summary of domain duplications in the tail fiber proteins of Xop411, Xp10, OP1 and host range mutants of OP1. The relationships between domain duplication (number inside circle) and phagovars (letter in parenthesis) infected by OP1 phage strains are shown. Scheme representations are after Inoue et al [6].
Positions of the 2 direct and 4 inverted repeats in the duplicated domains of the tail fiber genes from Xop411 (A), Xp10 (B), OP1 (C), OP1hc (D), OP1h (E), OP1h2 (F), and OP1h2c (G). Shown are DNA regions containing the duplicated domains and the flanking sequences. Domains are in different colors: 1, red; 2, blue; 3, green. When the same domain runs consecutively, the alternate one(s) (i.e., 2nd and 4th if any) is underlined. Direction and position of the repeats are indicated by half arrows above the sequences. Direct repeats (DR) are mostly located inside the domains especially in the domain junctions, whereas all inverted repeats (IR) are outside the domains.
Identification of Xop411 virion proteins by mass spectrometry.
Acknowledgments
Acknowledgements
This study was supported by grant No. NSC93-2317-B-005-007- and NSC94-2317-B-005-009- from National Science Council of Republic of China and No. 93-B-FA05-1-4 from Program for Promoting University Academic Excellence, Ministry of Education, Republic of China.
Contributor Information
Chia-Ni Lee, Email: lml7796@hotmail.com.
Rouh-Mei Hu, Email: rmhu@asia.edu.tw.
Teh-Yuan Chow, Email: tychow@sinica.edu.tw.
Juey-Wen Lin, Email: jwlin@nchu.edu.tw.
Hui-Yi Chen, Email: yichen@dragon.nchu.edu.tw.
Yi-Hsiung Tseng, Email: yhtseng@ctust.edu.tw.
Shu-Fen Weng, Email: sfweng@dragon.nchu.edu.tw.
References
- Swings JG, Civerolo EL. Xanthomonas. London, Glasgow, New York, Tokyo, Melbourne, Madras , Chapman and Hall; 1993. pp. 30–40. [Google Scholar]
- Inoue Y, Matsuura T, Ohara T. Bacteriophage OP1, lytic for Xanthomonas oryzae pv. oryzae, changes its host range by duplication and deletion of the small in the deduced tail fiber gene. J Gen Plant Pathol. 2006;72:111–118. doi: 10.1007/s10327-005-0252-x. [DOI] [Google Scholar]
- Kuo TT, Huang TC, Wu RY, Chen CP. Phage Xp12 of Xanthomonas oryzae (Uyeda et Ishiyama) Dowson. Can J Microbiol. 1968;14:1139–1142. doi: 10.1139/m68-190. [DOI] [PubMed] [Google Scholar]
- Kuo TT, Huang TC, Wu RY, Yang CC. Characterization of three bacteriophages of Xanthomonas oryzae. Dowson Bot Bull Acad Sinica. 1967;8:246–257. [Google Scholar]
- Kuo TT, Huang TC, Chow TY. A filamentous bacteriophage from Xanthomonas oryzae. Virology. 1969;39:548–555. doi: 10.1016/0042-6822(69)90102-0. [DOI] [PubMed] [Google Scholar]
- Yuzenkova J, Nechaev S, Berlin J, Rogulja D, Kuznedelov K, Inman R, Mushegian A, Severinov K. Genome of Xanthomonas oryzae bacteriophage Xp10: an odd T-odd phage. J Mol Biol. 2003;330:735–748. doi: 10.1016/S0022-2836(03)00634-X. [DOI] [PubMed] [Google Scholar]
- Wakimoto SS. Classification of strains of X. oryzae on the basis of their susceptibility against bacteriophages. Ann Pytopathol pathol Soc Japan. 1960;25:193–198. [Google Scholar]
- Inoue Y, Matsuura T, Ohara T. Sequence analysis of the genome of OP2, a lytic bacteriophage of Xanthomonas oryzae pv. oryzae. J Gen Plant Pathol. 2006;72:104–110. doi: 10.1007/s10327-005-0259-3. [DOI] [Google Scholar]
- Lee CN, Lin JW, Chow TY, Tseng YH, Weng SF. A novel lysozyme from Xanthomonas oryzae phage varphiXo411 active against Xanthomonas and Stenotrophomonas. Protein Expr Purif. 2006;50:229–237. doi: 10.1016/j.pep.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci U S A. 2005;102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BM, Park YJ, Park DS, Kang HW, Kim JG, Song ES, Park IC, Yoon UH, Hahn JH, Koo BS, Lee GB, Kim H, Park HS, Yoon KO, Kim JH, Jung CH, Koh NH, Seo JS, Go SJ. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 2005;33:577–586. doi: 10.1093/nar/gki206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IN, Smith DL, Young R. Holins: the protein clocks of bacteriophage infections. Annu Rev Microbiol. 2000;54:799–825. doi: 10.1146/annurev.micro.54.1.799. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ, Moser MJ, Holley WR, Chatterjee A, Mian IS. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 1997;25:4626–4638. doi: 10.1093/nar/25.22.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J Mol Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- Mehta P, Katta K, Krishnaswamy S. HNH family subclassification leads to identification of commonality in the His-Me endonuclease superfamily. Protein Sci. 2004;13:295–300. doi: 10.1110/ps.03115604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani E, Sjolander K, Hake S. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell. 2004;16:2265–2277. doi: 10.1105/tpc.104.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigyo M, Hasebe M, Ito M. Molecular evolution of the AP2 subfamily. Gene. 2006;366:256–265. doi: 10.1016/j.gene.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Roberts MD, Martin NL, Kropinski AM. The genome and proteome of coliphage T1. Virology. 2004;318:245–266. doi: 10.1016/j.virol.2003.09.020. [DOI] [PubMed] [Google Scholar]
- Wietzorrek A, Schwarz H, Herrmann C, Braun V. The genome of the novel phage Rtp, with a rosette-like tail tip, is homologous to the genome of phage T1. J Bacteriol. 2006;188:1419–1436. doi: 10.1128/JB.188.4.1419-1436.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Jiang Y, Vincent M, Sun Y, Yu H, Wang J, Bao Q, Kong H, Hu S. Complete genome sequence of bacteriophage T5. Virology. 2005;332:45–65. doi: 10.1016/j.virol.2004.10.049. [DOI] [PubMed] [Google Scholar]
- Crutz-Le Coq AM, Cesselin B, Commissaire J, Anba J. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology. 2002;148:985–1001. doi: 10.1099/00221287-148-4-985. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov VM, Nolan JM, Bertrand C, Levy D, Desplats C, Krisch HM, Karam JD. Plasticity of the gene functions for DNA replication in the T4-like phages. J Mol Biol. 2006;361:46–68. doi: 10.1016/j.jmb.2006.05.071. [DOI] [PubMed] [Google Scholar]
- Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr Opin Genet Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Wessler SR. Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci. 2005;10:54–56. doi: 10.1016/j.tplants.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci U S A. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandegren L, Sjoberg BM. Distribution, sequence homology, and homing of group I introns among T-even-like bacteriophages: evidence for recent transfer of old introns. J Biol Chem. 2004;279:22218–22227. doi: 10.1074/jbc.M400929200. [DOI] [PubMed] [Google Scholar]
- Semenova E, Djordjevic M, Shraiman B, Severinov K. The tale of two RNA polymerases: transcription profiling and gene expression strategy of bacteriophage Xp10. Mol Microbiol. 2005;55:764–777. doi: 10.1111/j.1365-2958.2004.04442.x. [DOI] [PubMed] [Google Scholar]
- Djordjevic M, Semenova E, Shraiman B, Severinov K. Quantitative analysis of a virulent bacteriophage transcription strategy. Virology. 2006;354:240–251. doi: 10.1016/j.virol.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Tetart F, Repoila F, Monod C, Krisch HM. Bacteriophage T4 host range is expanded by duplications of a small domain of the tail fiber adhesin. J Mol Biol. 1996;258:726–731. doi: 10.1006/jmbi.1996.0281. [DOI] [PubMed] [Google Scholar]
- Mitani K, Takahashi Y, Kominami R. A GGCAGG motif in minisatellites affecting their germline instability. J Biol Chem. 1990;265:15203–15210. [PubMed] [Google Scholar]
- Tanaka E, Fukuda H, Nakashima K, Tsuchiya N, Seimiya H, Nakagama H. HnRNP A3 binds to and protects mammalian telomeric repeats in vitro. Biochem Biophys Res Commun. 2007;358:608–614. doi: 10.1016/j.bbrc.2007.04.177. [DOI] [PubMed] [Google Scholar]
- Nakagama H, Higuchi K, Tanaka E, Tsuchiya N, Nakashima K, Katahira M, Fukuda H. Molecular mechanisms for maintenance of G-rich short tandem repeats capable of adopting G4 DNA structures. Mutat Res. 2006;598:120–131. doi: 10.1016/j.mrfmmm.2006.01.014. [DOI] [PubMed] [Google Scholar]
- A. B. I. M. http://www.iut-arles.up.univ-mrs.fr/w3bb/d_abim/riche-adn.html
- NCBI http://www.ncbi.nlm.nih.gov/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene assignment of the Xoo phage Xop411.
Similarities shared between the Xop411 proteins and those of Xylella and Xanthomonas.
Phylogenetic tree based on alignments of 50 conserved amino acids from the HNH domains of proteins in groups I to IV.
The region between p55 and the right end of genome of the three Xoo phages. Thick arrows indicate the direction and length of the genes. Corresponding genes are in the same colors, except that yellow indicates an additional gene. The horizontal bars represent percent identity of the nucleotide sequence, with black denoting > 80%, grey 65–80%, and white < 65% identity. Blocks A, B, C, D, and E of Xop411 and Xp10 (total, 842 bp) showed 71–92% identity; blocks F, G, H, and I of Xop411 and OP1 (total, 669 bp) showed 77–88% identity; and blocks J, K, L, M, N, O, and P of Xp10 and OP1 (total, 1,636 bp) showed 81–94% identity. The horizontal red lines indicate AT-rich regions of Xp10 (262 bp, nt 42,929–43,190 with 68% A+T), Xop411 (240 bp, nt 43,921-44,160 with 72% A+T, including an 80-bp internal segment of 96% A+T from nt 43,974-44,033 with 4 perfect 15-bp tandem direct repeats ATTATTAATATTTAT), and OP1 (336 bp, nt 42,631-42,966 with 63% A+T). These AT-rich regions are worth testing for the possibility of containing replication origins of the Xoo phages. Bent arrows and knobs represent the predicted promoters and terminators, respectively.
Alignment of promoters found in Xp10 and predicted for OP1 and Xop411. Bases identical to those of the Xp10 are on black background. Ratios to the right are bases matched to the Xp10 sequences. Color of the bases: blue, -35; green, conserved to Xp10 RNA polymerase promoters; orange, extended -10 promoter elements; pink, -10; yellow, transcription start sites. The extended -10 elements were found to be resistant to p7, the inhibitor of transcription initiation, in Xp10.
Domain duplication at the N-terminus of the deduced tail fiber proteins of Xoo phages. (A) Alignment of the sequence of Xop411 p26 with its homologues in Xp10, OP1, and four OP1 host range mutants. A domain is indicated by a line above the sequences with an Arabic number in circle. Different amino acid residues within the duplicated domains are shaded. (B) Summary of domain duplications in the tail fiber proteins of Xop411, Xp10, OP1 and host range mutants of OP1. The relationships between domain duplication (number inside circle) and phagovars (letter in parenthesis) infected by OP1 phage strains are shown. Scheme representations are after Inoue et al [6].
Positions of the 2 direct and 4 inverted repeats in the duplicated domains of the tail fiber genes from Xop411 (A), Xp10 (B), OP1 (C), OP1hc (D), OP1h (E), OP1h2 (F), and OP1h2c (G). Shown are DNA regions containing the duplicated domains and the flanking sequences. Domains are in different colors: 1, red; 2, blue; 3, green. When the same domain runs consecutively, the alternate one(s) (i.e., 2nd and 4th if any) is underlined. Direction and position of the repeats are indicated by half arrows above the sequences. Direct repeats (DR) are mostly located inside the domains especially in the domain junctions, whereas all inverted repeats (IR) are outside the domains.
Identification of Xop411 virion proteins by mass spectrometry.