Abstract
Hfq is an RNA binding protein that has been studied extensively for its role in the biology of small noncoding RNAs (ncRNAs) in bacteria, where it facilitates post-transcriptional gene regulation during stress responses. We show that Hfq also binds with high specificity and nanomolar affinity to tRNAs despite their lack of a canonical A/U rich single-stranded sequence. This affinity is comparable to that of Hfq for its validated ncRNA targets. Two sites on tRNAs are protected by Hfq binding, one on the D-stem and the other on the T-stem. Mutational analysis and competitive binding experiments indicate that Hfq uses its proximal surface (also called the L4 face) to bind tRNAs, the same surface that interacts with ncRNAs but a site distinct from where poly(A) oligonucleotides bind. hfq knockout strains are known to have broad pleiotropic phenotypes, but none of them are easily explained by or imply a role for tRNA binding. We show that hfq deletion strains have a previously unrecognized phenotype associated with mistranslation and significantly reduced translational fidelity. We infer that tRNA binding and reduced fidelity are linked by a role for Hfq in tRNA modification.
Keywords: Sm-like protein, tRNA processing, translation fidelity
INTRODUCTION
Hfq was first discovered in the mid-1960s in a screen for host proteins required for replication of Q-beta bacteriophage in Escherichia coli (Franze de Fernandez et al. 1968, 1972; Shapiro et al. 1968). The physiological role of this highly conserved RNA binding protein was unclear at the time. Since it seemed unlikely that bacteria would retain a protein whose sole function was to make it susceptible to bacteriophage infection, it was widely believed that Hfq had important physiological functions waiting to be uncovered. In the 1990s, it became clear that Hfq plays an important role in the biology of bacterial noncoding RNAs (ncRNAs) (Masse et al. 2003; Gottesman 2004; Storz et al. 2004; Valentin-Hansen et al. 2004). Hfq binds many small ncRNAs and facilitates post-transcriptional gene regulation by helping these ncRNAs identify their mRNA targets during stress responses (Majdalani et al. 1998; Lease and Belfort 2000; Masse and Gottesman 2002). The ensuing structural rearrangements can lead to up- or down-regulation of translation or can alter the stability of the target mRNAs. Since Hfq ternary complexes (Hfq:ncRNA:mRNA) are stable (Moller et al. 2002; Zhang et al. 2002; Lease and Woodson 2004), it is possible that Hfq serves additional functions, helping to direct the appropriate regulatory response after target identification.
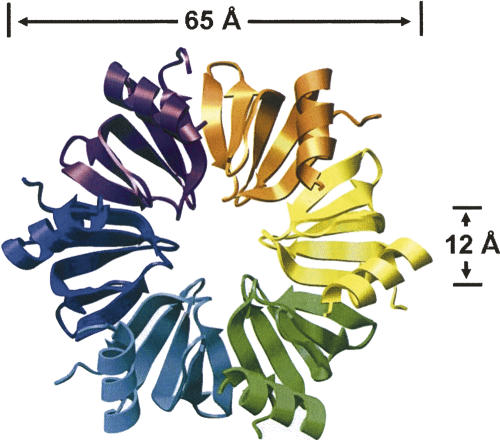
Structural and bioinformatic studies determined that Hfq is a prokaryotic homolog of Sm and Lsm proteins (Arluison et al. 2002; Moller et al. 2002; Schumacher et al. 2002; Sun et al. 2002; Zhang et al. 2002; Sauter et al. 2003; Wilusz and Wilusz 2005). Crystal structures of Hfq have been solved showing that it assembles into the characteristic doughnut-shaped structures of the Lsm proteins (Fig. 1). In the case of Hfq, they form a homohexameric ring rather than heteroheptamers found in eukaryotes or the homoheptamers of archael Lsms. These toroidal complexes bind RNAs on both faces although the binding specificity of the two surfaces differs from one another (Mikulecky et al. 2004; Sun and Wartell 2006). The ncRNAs typically bind to the proximal surface (also called the L4 face) whereas poly(A) sequences typically interact with the distal face. Mutational analysis has implied that mRNAs can interact with both proximal and distal surfaces simultaneously, although it seems unlikely that such mRNAs pass through the central cavity since the preassembled hexameric structure is exceedingly stable in vitro and retains binding activity.
FIGURE 1.
Structure of Hfq hexamers from Staphylococcus aureus (Schumacher et al. 2002). Image prepared with Chimera (Pettersen et al. 2004) based on PDB: 1KQ2.
In addition to binding RNAs, Hfq has been shown to interact with a variety of proteins (Sukhodolets and Garges 2003; Mohanty et al. 2004; Butland et al. 2005; Morita et al. 2005; T. Lee and A.L. Feig, unpubl.). In some cases, these interactions are direct contacts whereas in other cases, the binding appears to be indirect, as if Hfq were part of a larger ribonucleoprotein (RNP) particle. While investigating these protein–protein interactions of Hfq, we found that it associates with a variety of proteins that participate in tRNA maturation and modification, implying the potential involvement of Hfq in this process. Additional evidence supported this potential new role for Hfq in tRNA metabolism: (1) in microarray studies aimed at identifying all possible ncRNAs to which Hfq might bind, tRNAs were among the species identified (Zhang et al. 2003); (2) in yeast, depletion of Lsm complexes dramatically affects pre-tRNA processing (Kufel et al. 2002); (3) there is an unexplained genetic linkage between hfq and miaA, a gene encoding a tRNA isopentenyltransferase responsible for modification of A37 (Bjork 1996; Leung et al. 1997); and (4) defective tRNAs in both prokaryotic (Li et al. 2002) and eukaryotic organisms (Kadaba et al. 2004; Vanacova et al. 2005) are degraded through tRNA surveillance systems involving 3′-polyadenylation, and previous work has shown a strong connection between Hfq and poly(A) polymerase I (PAP I) (O'Hara et al. 1995; Mohanty and Kushner 1999; Mohanty et al. 2004). Due to this large body of circumstantial evidence, we began to explore the possibility that Hfq specifically interacts with tRNAs and that it plays a role in tRNA metabolism. In the present study we show that tRNAs bind to Hfq with high affinity and that it interacts with two independent sites on tRNA—located on the T-stem and D-stem, respectively. These Hfq binding sites on tRNA are different from the conventional Hfq interaction motifs observed on small regulatory RNAs involved in post-transcriptional gene regulation, but it uses part of the same RNA binding surface. In addition, evidence is presented suggesting that Hfq may be required for proper tRNA modification.
RESULTS
Hfq binds specifically to tRNAs
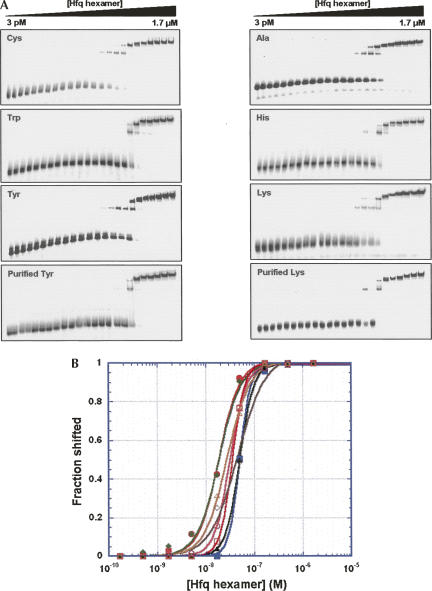
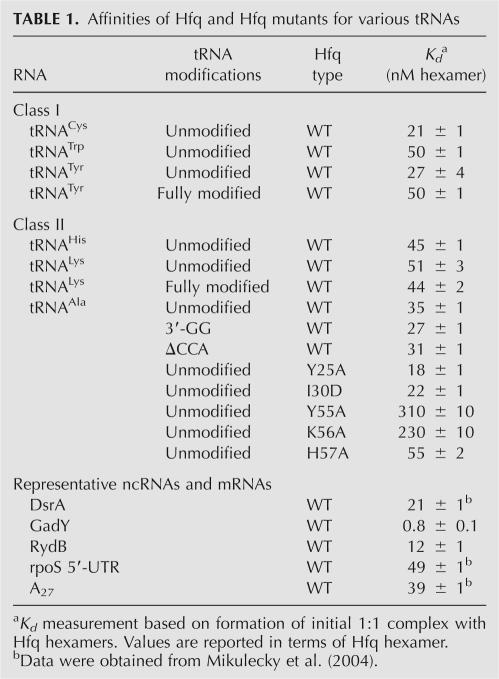
Electrophoretic mobility shift assays were used to probe the interaction between Hfq and tRNAs. As a preliminary test, a tRNA mixture extracted from E. coli was titrated with Hfq. Most tRNAs bound Hfq. The small fraction, which remained unbound, was presumed to be misfolded (data not shown). To measure the dissociation constant of each tRNA, a series of tRNAs was assayed, including substrates for both class I and class II aminoacyl synthetases (Fig. 2; Table 1). The results show that Hfq binds effectively to all of the tRNAs tested, with Kd values in the range of 20–50 nM (in hexamer). This affinity compares favorably with the Kd for known Hfq ligands like the ncRNA DsrA and the rpoS mRNA 5′-untranslated region (5′-UTR), which have affinities of 21 nM and 49 nM, respectively (Table 1). Two classes of complexes can be observed on these gels. The complexes that migrate relatively fast appear initially at low concentration of Hfq, and additional higher-order complexes form at higher Hfq concentrations. Such higher-order complexes are almost universally observed by experimental techniques when RNA concentrations are limiting (such as in the gel shift experiments utilizing trace 32P-labeled material) but are typically not observed when high concentrations of RNA are present, such as in the ITC experiments that have been previously reported (Mikulecky et al. 2004). All quantitative data reported herein reflect the initial binding event.
FIGURE 2.
(A) Electrophoretic mobility shift experiments used to measure the affinity of Hfq for various tRNA species. Gels on the left and the right represent tRNA substrates for Class I and Class II tRNA synthetases, respectively. In the case of tyrosine and lysine, binding is shown for both unmodified tRNAs prepared by T7 transcription as well as fully modified tRNAs obtained from E. coli. (B) Quantitative analysis of the gel shift data from panel A. Binding constants for the interactions were determined by nonlinear least-squares analysis as described in the Materials and Methods section and the values are summarized in Table 1. tRNA members of Class I and Class II were shown by closed and open marks, respectively. (Closed circles) tRNACys; (closed squares) tRNATrp; (closed diamonds) tRNATyr; (closed triangles) fully modified tRNATyr; (open circles) tRNAAla; (open squares) tRNAHis; (open diamonds) tRNALys; (open triangles) fully modified tRNALys.
TABLE 1.
Affinities of Hfq and Hfq mutants for various tRNAs
One possible physiological function of Hfq in tRNA metabolism might be related to a quality control by selectively binding either mature or pre-tRNAs. Toward that end, the in vitro binding of Hfq to several tRNA variants was tested. For lysyl and tyrosyl tRNAs, the affinities of Hfq for T7 transcripts were within twofold of the fully modified tRNAs isolated from E. coli. Thus it appears that Hfq cannot effectively distinguish modified tRNAs from their unmodified relatives. Other forms of defective tRNAs Hfq might encounter contain 3′ extensions resulting from incomplete processing by RNases or exonucleolytic degradation of the CCA end due to a 3′ → 5′ exonuclease activity. Using tRNAAla as a test case, constructs were prepared with altered 3′ termini to mimic these defects. In every case, Hfq showed no preferential binding to either form (Table 1), making it clear that Hfq alone is incapable of using these determinants to distinguish mature tRNAs from defective ones.
Mapping the RNA binding surface on Hfq that interacts with tRNAs
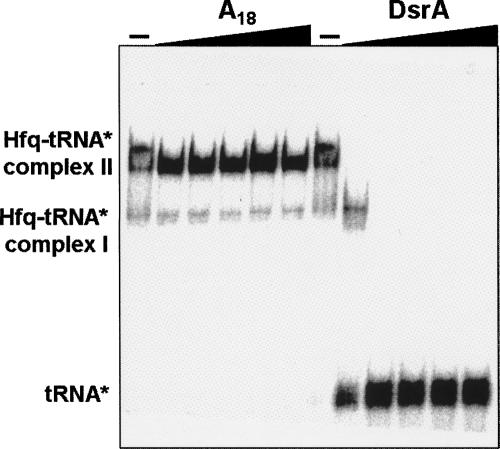
The overall affinity for these tRNAs is quite respectable relative to other cellular targets to which Hfq binds (Table 1). However, it is still important to establish whether the interaction is specific and which surface of Hfq is used to recognize tRNAs. Hfq was previously shown to have at least two independent RNA binding faces, and some RNAs prefer one binding site over the other (Mikulecky et al. 2004). Competitive binding assays were used to look at the issue of site specificity (Fig. 3). After preforming the 32P-tRNA:Hfq adduct, increasing concentrations of unlabeled competitor RNA were added to displace the tRNA. In one experiment, homopolymer A18 [18-nucleotide (nt) synthetic poly(A) oligomer] was used. The interaction between Hfq and A18 has been studied previously and shown to interact specifically via the distal face of the hexamer (Mikulecky et al. 2004). The addition of even very high concentrations of A18 was unable to displace tRNA from the surface of Hfq. Separate samples of the Hfq:tRNA adduct were treated with increasing concentrations of the ncRNA DsrA (Fig. 3). DsrA and many other ncRNAs that have been studied bind to Hfq via the proximal face (Sledjeski et al. 2001; Mikulecky et al. 2004). In the presence of excess DsrA, tRNA was readily displaced from Hfq. This behavior is indicative of tRNA binding to the same site (or at least an overlapping site) on Hfq as DsrA.
FIGURE 3.
Competition binding experiments to determine the Hfq surface that interacts with tRNAs. The complex between Hfq and 32P-tRNAAla was preformed and then incubated with increasing amounts of A18 or DsrA. DsrA and other ncRNAs have been previously shown to interact with the proximal face of the Hfq hexamer (Sledjeski et al. 2001; Mikulecky et al. 2004; T. Lee and A.L. Feig, unpubl.). Since only DsrA was able to compete with tRNA for binding to Hfq, this experiment establishes that tRNAs interact with some or all of the same binding surface as the ncRNAs.
A complementary way to map the site of tRNA's interaction with Hfq is through the use of site directed mutants known to alter the RNA binding affinity. Binding of tRNAAla to several mutant Hfq species was probed. Mutation of key residues from the Sm2 motif (Y55A and K56A) located around the central cavity on the proximal face significantly reduced tRNA binding affinity, whereas mutations elsewhere on the proximal surface (H57A) and on the distal face (Y25A, I30D) showed wild-type affinities (Table 1). These data are completely consistent with the competition studies. Of the five mutants tested, only Y55A and K56A disrupted binding of the ncRNAs. In the case of DsrA, these mutations led to comparable binding defects (∼10-fold) while the H57A, Y25A, and I30D mutations did not alter DsrA affinity (Mikulecky et al. 2004). From these experiments, we conclude that tRNAs bind along the proximal surface.
Mapping of the tRNA interaction surface
The classical Hfq binding motif has been shown to be an A/U-rich single-stranded sequence adjacent to a helical element that can lie 3′ or 5′ of the single-stranded region (Moller et al. 2002; Zhang et al. 2002; Brescia et al. 2003; Geissmann and Touati 2004). Such structures have been observed in a variety of small ncRNAs involved in Hfq-dependent gene regulation and are reminiscent of the Sm binding sites found in eukaryotic snRNAs (Wilusz and Wilusz 2005). This type of motif, however, is not evident in tRNA. Thus it is important to map the interaction surface to obtain a picture of how Hfq interacts with tRNAs.
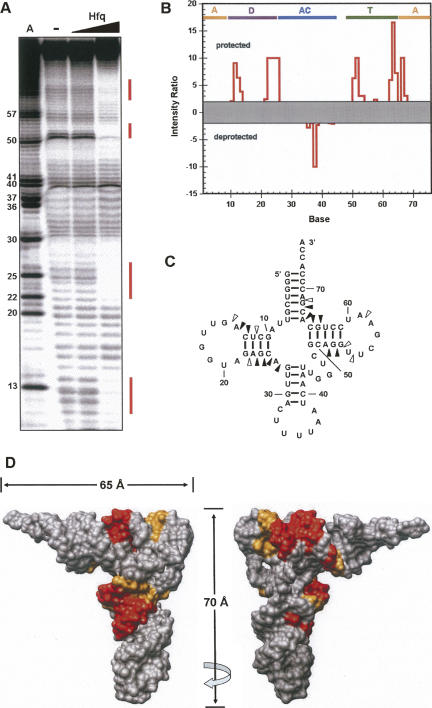
By using Tb(III) hydrolysis mapping techniques (Walter et al. 2000), the protection pattern for tRNAs in the presence and absence of Hfq was assessed. This technique is useful because the Tb(III) ion is quite small and can often provide higher resolution than nuclease mapping. A representative experiment using tRNALys is shown in Figure 4. The pattern does not vary significantly between the different tRNA species that were tested. Four significant regions of protection are observed, including nucleotides 11–14, 22–26, 51–54, and 63–68 (Fig. 4A–C). When mapped onto the three-dimensional structure, the protected nucleotides cluster into two discrete interaction sites (Fig. 4D). One region lies along the D-stem near the elbow region of the tRNA (D-site). The second set of protections cluster along the top surface of the T-arm (T-site). These sites point away from one another on the structure of the free tRNA, implying that they must be independent interaction surfaces unless the structure of the tRNA is dramatically distorted upon binding. Nuclease mapping studies using double- and single-stranded RNA specific nucleases did not provide any evidence for such large-scale distortions (data not shown). The deprotected position at base 38 was not reproducible.
FIGURE 4.
Footprinting of Hfq binding to tRNALys. (A) High resolution polyacrylamide gel showing the effect of Hfq binding on Tb(III) mediated to 5′-32P-tRNALys. Lane A is a marker lane prepared from 5′-32P-tRNALys transcribed with ATPγS and cleaved by treatment with I2/EtOH. Subsequent lanes show the Tb(III) mediated cleavage pattern in the absence and presence of Hfq. (B) Quantitative analysis of Hfq binding based on the gel in panel A. Data are represented as a ratio of the intensity of each band in the absence and presence of 1 μM Hfq hexamer. Values greater than 1 represent protection. For sites that showed more intense cleavage upon Hfq binding, the ratio is inverted (presence/absence) and the data are assigned a negative value. Data between +2 and −2 were considered to be no significant effect. Bars at the top indicate the secondary structure elements where A is the acceptor stem, D is the D stem, AC is the anticodon stem and TΨC is the TΨC stem. (C) Superposition of the footprinting data onto the tRNALys secondary structure. Open triangles represent modest protection (intensity ratio 2–5), and closed triangles represent sites of strong protection (intensity ratio >5). (D) Superposition of the footprinting data of tRNALys onto the crystal structure of tRNAPhe (PDB ID: 1EHZ), where orange and red indicate modest and strong footprinting, respectively, shows that there are two Hfq binding sites on the tRNA. Images are rotated by 180° as indicated by the arrow.
Tb(III) hydrolysis mapping under the different concentration of Hfq showed the appearance of the protected pattern on both D- and T-sites grew in concurrently rather than sequentially as Hfq concentration increased (data not shown). Thus, Hfq does not appear to bind preferentially to either the D- or T-site. Instead, it interacts with comparable affinity to both sites. In addition, minimal binding experiments showed that a tRNA fragment composed of the 5′ end through the anti-codon loop is sufficient to bind Hfq, suggesting that the binding sites act independently (data not shown).
Hfq appears to be involved in tRNA modification
A critical question regarding the interaction between Hfq and tRNA remains unanswered. What is its physiological role? Given that protein–protein interaction studies have shown that Hfq interacts with RNA modification enzymes (Butland et al. 2005; T. Lee and A.L. Feig, unpubl.), we hypothesize that Hfq is somehow involved in tRNA modification. Before attempting to specify the exact tRNA modification(s) mediated by Hfq, we tested whether Δhfq strains exhibit a more generic phenotype associated with incomplete tRNA modification—reduced translational fidelity. It has been shown previously that unmodified or hypomodified tRNAs exhibit defects in translational accuracy resulting from either mischarging of tRNAs or misreading of mRNAs (Hagervall et al. 1990; Cabello-Villegas et al. 2002). Defects in translational accuracy can be observed readily by isoelectric focusing (IEF) when the misincorporation alters the pI of the resulting gene product (Parker et al. 1978).
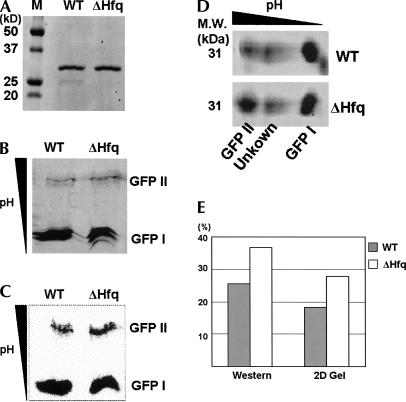
We used an N-terminally His6-tagged version of GFP as a reporter to study the translational fidelity in wild-type and Δhfq strains. GFPs from both strains were purified in two steps by using a Co2+ affinity column followed by size exclusion chromatography. The homogeneity of proteins was confirmed by denaturing SDS-PAGE (Fig. 5A). The purified GFPs from both wild-type and Δhfq strains were then analyzed on native IEF gels. Imaging based on the intrinsic GFP fluorescence revealed the presence of two bands (designated as GFP I and GFP II) with distinct pIs (Fig. 5B). GFP I with a pI ∼5 was highly fluorescent, but GFP II with a pI of ∼6.2 showed relatively low fluorescence. Based on previous reports of IEF analysis, it is clear that GFP I is the native material, and the observation of several closely spaced bands on IEF analysis is common (Richards et al. 1999; O'Neill et al. 2006). To ensure that GFP II was not an impurity resulting from incomplete purification, we performed a Western blot from the IEF gel using a probe specific for the His6-tag. As shown in Figure 5C, both GFP I and GFP II were recognized by the probe. Another possibility is that GFP II might be a degradation product of GFP I. Independent IEF analysis was performed using the same proteins, and each sample was further resolved in the second dimension by SDS-PAGE (Fig. 5D). Both GFP I and GFP II were found to have the same mass (∼31 kDa) based on protein standards, indicating that both bands in the IEF gel are full-length, mature GFP products (Fig. 5A). The 2-D gel shows two other features of the experiment. First, a small impurity was observed in the GFP isolated from wild-type E. coli. This material can be seen as a faint band (∼25 kDa) in Figure 5A and runs at lower molecular weight and slightly more acidic pI than GFP I in the 2-D analysis (Fig. 5D). Second, a nonfluorescent band is observed in the total protein staining of the 2-D gel at ∼31 kDa and intermediate pI between GFP I and GFP II. This band is highly variable between experiments and we have tentatively assigned it as the unoxidized GFP precursor lacking the proper mature chromophore.
FIGURE 5.
(A) SDS-PAGE analysis of GFPs purified from WT and Δhfq strains. (B) IEF analysis of GFPs from WT and Δhfq strains shows the two types of GFP. The location of proteins was detected by intrinsic fluorescence of GFP. The approximate pI values of GFP I and II are 5 and 6.2, respectively. (C) The N-terminal His6-tag was probed by Western blotting. Intensity of GFP II measured by this probing is much stronger than the one measured by fluorescence, indicating the majority of GFP II is defective. (D) 2-D gel analysis was performed by running the IEF followed by the SDS-PAGE. Both GFP I and II from WT and Δhfq were detected at the molecular weight of 31 kDa on the two-dimensional gel. Unidentified protein bands also appeared between GFP I and GFP II. A small impurity which is observed in panel A in the WT lane is shown at the right down corner of the WT panel. (E) Quantitation of the fraction of GFP II was carried out by applying different measurements on two independent experiments. Both measurements consistently show the increase of GFP II in Δhfq, indicating the involvement of Hfq in translational fidelity.
In wild-type E. coli about 20% of the total GFP was of the GFP II variety based on quantitation of the Western blots or total protein staining with Sypro Ruby. Considering the low fluorescence intensity of GFP II detected on the IEF gel, GFP II is likely a somewhat defective form of the GFP. When the Δhfq strain was assayed, the amount of GFP II increased by almost 50% relative to the wild-type strain. This result indicates a clear and consistent trend toward the translation of GFP II (Fig. 5E). These results strongly suggest that hfq knockout leads to a significant translational fidelity problem that may in turn derive from improper or incomplete tRNA modification.
Hfq may not be an essential factor for RNA quality control
Given the diverse participation of Lsm proteins in various aspects of RNA metabolism, it would not be surprising if Hfq played more than one role in tRNA biochemistry. Thus, we tested another hypothesis that Hfq might be part of a quality control checkpoint during tRNA maturation. In eukaryotes, when processing defects occur, Trf4p, a component of the TRAMP complex, polyadenylates the misfolded tRNA to initiate degradation (Kadaba et al. 2004; LaCava et al. 2005; Vanacova et al. 2005). Unlike cytoplasmic poly(A) tails, nuclear polyadenylation in eukaryotes targets the RNA for degradation in a process similar to bacterial RNA turnover, where mRNAs can be poly(A)-tailed to target them for degradeosome-mediated decay (Hajnsdorf et al. 1995; O'Hara et al. 1995; Ingle and Kushner 1996; Carpousis et al. 1999).
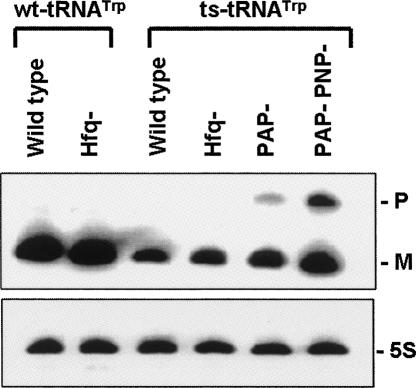
In a very nice series of studies, the Deutscher laboratory showed that defective tRNAs are degraded through the combined action of PAP I and polynucleotide phosphorylase (PNPase) (Li et al. 2002). They used a temperature sensitive mutant of tRNATrp (ts-tRNATrp) as a probe, since it is prone to be misfolded in vivo because of the disruption of GC base pair at the bottom of acceptor arm (Eisenberg et al. 1979; Eisenberg and Yarus 1980). In ΔPAPI, and even more dramatically in the case of the ΔPAPI/ΔPNPase, the accumulation of the misfolded ts-tRNATrp precursor was observed. Our logic was that Hfq might potentiate the interaction between the degradation machinery and the misfolded tRNA precursor because PAP I and PNPase have both been previously shown to bind Hfq and cooperate to degrade mRNAs (Mohanty et al. 2004). We used the same ts-tRNATrp strain to probe for the lifetime of the defective ts-tRNATrp precursor in the absence of Hfq, alone and in combination with PAP I and PNPase (Fig. 6; data not shown).
FIGURE 6.
Northern blot analysis of hfq − mutant in wild-type tRNATrp and ts-tRNATrp strains. Each strain was grown at 30°C, and total RNA was extracted when the OD600 value of the cell culture was 0.5 as described in Materials and Methods. Eight micrograms were resolved on the gel and probed by synthetic oligo complementary to residue 39–54 of tRNATrp. P and M indicate the positions of tRNATrp precursor and mature tRNATrp, respectively. The same membrane was probed for 5S rRNA as a loading control.
The results of these studies appear to disprove our hypothesis. While these strains readily reproduce the findings of the Deutscher laboratory and highlight the importance of PAP I and PNPase in degradation of defective ts-tRNATrp in E. coli, the presence or absence of Hfq does not appear to affect the lifetime of the defective ts-tRNATrp precursor. This result indicates that Hfq is not essential for the degradation of improperly processed tRNAs.
DISCUSSION
Our in vitro analysis clearly shows that tRNAs bind Hfq quite readily, with affinities rivaling those of the ncRNAs. We tested a variety of tRNA species. These include tRNA substrates for class I and class II aminoacyl synthetases, and defective versions of tRNAs derivatives prepared by removing the CCA end or adding two extra nucleotides to the 3′ end. In addition to these in vitro transcribed tRNAs that are not base-modified, we tested two fully modified tRNAs isolated from E. coli. All tRNAs tested showed strong binding affinities toward Hfq. The propensity of tRNA to bind Hfq makes it a poor choice for use as a nonspecific competitor in binding assays, as has been reported in a few instances (Moller et al. 2002; Geissmann and Touati 2004), because it may impact the measured affinity for Hfq.
Hfq uses its proximal face to bind either D- or T-sites on tRNAs. The nature of the interaction is still a little mysterious, however. Crystallographic and mutagenesis studies have shown the importance of the Sm2 domain and the central cavity in binding to canonical Sm-like binding sites containing of A/U-rich single-stranded sequences. Short model RNAs are trapped in the positively charged central cavity and exhibit base stacking with Y42 residues from the individual monomeric units as the RNA wraps around the cavity (Schumacher et al. 2002). This mechanism can be applied to interactions between Hfq and many ncRNAs that have conventional Hfq binding motifs. However, it seems unlikely that the same binding mechanism can be applied to the observed Hfq–tRNA interaction, as neither the D- nor T-site contains single-stranded regions. Furthermore, these sites exhibit little sequence conservation across the tRNA species that bind Hfq. Thus, it appears to be a structural effect that dictates this binding mode with little or no sequence specificity.
In previous studies using microarrays aimed at identifying all possible ncRNAs to which Hfq binds, tRNAs and a tRNA precursor were among the species identified (Zhang et al. 2003). It was speculated that Hfq might be involved in tRNA processing because the tRNA precursor lifetime depended on both Hfq and endonuclease RNase E (Zhang et al. 2003), which is an essential enzyme for pre-tRNA processing (Li and Deutscher 2002). In addition, it was reported that the precursor of tRNA but not the mature tRNA was enriched in the Hfq co-immunoprecipitated sample, especially in the case of proM tRNA. In contrast to this report, our in vitro tRNA binding assays imply that Hfq shows little preference for unmodified, modified, or even defective tRNAs. This discrepancy may be due to a difference between the cytosolic environment and the in vitro conditions used. Alternatively, there may be additional protein components that provide specificity in vivo that are missing in our in vitro assay.
Another factor might explain the preferential pull-down of pre-tRNAs. The majority of mature cellular tRNAs are charged in vivo, and charged tRNAs should be found as complexes with EF-Tu. The Hfq binding site on the T-stem overlaps with the site of EF-Tu binding (Nissen et al. 1995). Since EF-Tu binds charged tRNAs significantly more tightly than Hfq does (Louie et al. 1984), we expect it to readily displace Hfq upon charging. Thus, the pull-down studies may simply reflect the relative speciation of tRNA rather than inherent binding affinity. Mature tRNAs are fundamentally charged and bound to EF-Tu whereas immature and uncharged tRNAs might be found bound to Hfq.
Our studies presented evidence suggesting that hfq knockout alters translational fidelity in E. coli. It has been shown that base modifications of tRNA can play a crucial role in maintaining the high fidelity of protein synthesis. Some modified bases found in tRNAs are required for proper aminoacylation while others function to avoid recognition of mismatches between the codon and anticodon during decoding on the ribosome (Bjork 1996). The fidelity defect that we observed in the Δhfq strain, coupled with the observed binding of Hfq to tRNAs, provides strong evidence for a role of Hfq in tRNA modification. Ongoing studies are looking at whether there is a generalized defect in tRNA modification or if the effect is more localized on a small subset of the tRNA modifications found in E. coli. We also must determine if the mistranslation results from defects at the time of charging or on the ribosome during decoding.
The interaction of Hfq and tRNA and its potential physiological role in tRNA modification may also explain the functional relevance of the previously unexplained genetic linkage between miaA and hfq (Tsui et al. 1994). MiaA catalyzes the initial modification of A37 of those tRNAs that read codons starting with uridines. This modification is known to be important for increasing the translation fidelity by stabilizing the codon–anticodon interactions (Bjork 1996; Leung et al. 1997). The cooperative interaction between the two proteins, however, has remained a mystery. The tRNA binding we describe is the first evidence for a possible mechanism by which these proteins might join forces to modify appropriate tRNAs.
MATERIALS AND METHODS
Bacterial strains
Strain CA244 Trpts, which is E. coli K12 strain CA244(lacZ, trp, relA, spoT) carrying a G7 → A7 temperature-sensitive mutation in trpT encoding tRNATrp, and its derivatives CA244 PNP− Trpts, CA244 PAP− Trpts and CA244 PNP−PAP− Trpts were kindly provided by Dr. Murray P. Deutscher, University of Miami (Li et al. 2002). The Δhfq version of each strain was generated by replacing hfq with chloramphenicol cassette. Quick and Easy Conditional Knockout Kit (Gene Bridges) was used following the manufacturer's protocol. The hfq− phenotype was selected for based on chloramphenicol resistance and then verified by Western blot analysis using anti-Hfq polyclonal sera with the parental strains serving as positive controls. For the translational fidelity experiment, CP78 strain described by Sorensen (2001) was used as the wild-type. Δhfq strain of CP78 was prepared as described above, but kanamycin cassette was used instead of chloramphenicol.
Preparation of RNAs
Alanine-specific tRNAAla, its derivative tRNAAlaΔCCA, tRNAAla-GG, and tyrosine specific tRNATyr were independently cloned into pUC19 downstream of the T7 RNA polymerase promoter sequence. These tRNA constructs were verified by DNA sequencing. After cleaving the plasmid with the appropriate restriction enzyme (BglII), RNA was in vitro transcribed following the method of Milligan and Uhlenbeck (1989). tRNACys, tRNAHis, tRNALys, tRNATrp, and temperature-sensitive derivative ts-tRNATrp were prepared by in vitro transcription using template and top strand DNA oligonucleotides purchased from Integrated DNA Technologies. Fully modified tRNA species (tRNALys, tRNATyr, and various tRNA mixtures) were purchased from Sigma-Aldrich. DsrA RNA was prepared by in vitro transcription from a construct previously described (Mikulecky et al. 2004). The homopolymer RNA A18 was purchased from Dharmacon Research. In vitro transcribed RNAs and purchased tRNALys and tRNATyr quality were assessed by denaturing PAGE and gel purification.
Protein expression and purification
The expression and purification of C-terminal His6-tagged wild-type Hfq and its derivatives, Y25A, I30D, Y55A, K56A, and H57A, were described previously (Mikulecky et al. 2004). Concentration of protein and residual nucleic acid were assessed by the Warburg–Christian method (Stoscheck 1990). All Hfq samples used in these studies had <5% residual nucleic acid after purification and were free from nuclease contamination.
For the purification of GFP, the coding region of GFP that has N-terminal His6-tag was amplified from pRSET-EmGFP vector (Invtrogen) and subcloned into pBAD24. The DNA oligos used are 5′-CAGAACGGTACCTTCTCATCATCATCATCATC and 5′-GTCAACTCTAGATTACTTGTACAGCTCGTC. CP78 (wild type) and CP78 Δhfq stains transformed with this vector were grown in LB at 37°C overnight in the presence of 10 mM arabinose. Expressed proteins were purified by using Co2+ column. After washing the column with the buffer of 50 mM sodium phosphate (pH 8), 300 mM NaCl, and 50 mM imidazole, the proteins were eluted by applying the same buffer containing 200 mM imidazole. Gel filtration chromatography was done on an ÄKTA FPLC system. Proteins eluted from the Co2+ column were applied on a Superdex 200 HiLoad 16/60 column (Pharmacia) and separated in the same buffer lacking imidazole at 4°C with a flow rate of 0.5 mL/min. The concentration of the protein was measured as described above.
Gel shift experiments
5′ 32P-end-labeled RNAs that were heated at 90°C for 2 min and cooled down to room temperature were incubated with various concentrations of Hfq in binding buffer (50 mM Tris-HCl at pH 8.0, 250 mM NH4Cl, 10 mM MgCl2) for 30 min at room temperature. The sample was resolved on 5% (w/v) polyacrylamide native gel at 5 W at room temperature. Phosphorimagery was used to visualize the gels. Quantitation of gel bands was performed using ImageQuant 5.2 (Molecular Dynamics). Nonlinear least-squares analysis of the binding data using a cooperative binding model was done using Kaleidagraph 4 (Synergy Software).
Competitive binding analysis
5′ 32P-end-labeled tRNA was incubated with 1 μM Hfq in binding buffer (50 mM Tris-HCl at pH 8.0, 250 mM NH4Cl, 10 mM MgCl2). Competitors (A18 RNA and DsrA RNA) were added to a final concentration of 0.3, 1, 3, 10, and 30 μM. After incubation for 30 min at room temperature, these mixtures were resolved on 5% (w/v) polyacrylamide native gels at 5 W at room temperature. The visualization of gel was done as described above.
Tb(III) mediated RNA footprinting
The reaction was done following a published protocol (Walter et al. 2000). Briefly, 250K cpm of 5′ 32P-end-labeled tRNA was incubated in probing buffer (50 mM Tris-HCl at pH 8.0, 100 mM NaCl, 10 mM MgCl2) containing 0 nM, 20 nM, and 1 μM of Hfq hexamer at room temperature for 30 min. TbCl3 was added to a final concentration of 100 mM, and the reaction was incubated at room temperature for an additional 2 h. The reaction was quenched by addition of EDTA and SDS to final concentrations of 50 mM and 0.1%, respectively. Samples were then treated with proteinase K at 37°C for 30 min to destroy the Hfq in the samples, and the completion of the reaction was confirmed by gel analysis. The mixture without further treatment was directly resolved on an 8% (w/v) polyacrylamide denaturing sequencing gel at 60 W.
Isoelectric focusing and 2D gel analysis
We analyzed 3.5 μg of GFP in the buffer containing 8% glycerol on native IEF gel (BioRad, pH 5–8) following the manufacturer's instruction. In brief, 20 mM NaOH and 7% phosphoric acid were used as the running buffer for cathode and anode, respectively. Voltage was applied at 100 V for 1 h, 250 V for 1 h, and 500 V for 30 min using protean II cell (BioRad). After the IEF gel was equilibrated in a Tris-SDS buffer (0.123 M Tris at pH 6.8, 10% [v/v] glycerol, 2% [v/v] SDS, 7 mM DTT), the second dimensional gel was prepared by casting 12% SDS polyacrylamide gel with the IEF gel slice in place of the comb and resolved using standard protocols. The proteins on the SDS-PAGE gel were stained by Sypro Ruby protein gel stain (Molecular Probes).
Western blot analysis
Proteins on the IEF gel was blotted on the PVDF membrane, and the N-terminal His6-tag was probed by HisProbe-HRP (Pierce) following the manufacturer's instruction.
Northern blot analysis
Wild-type and temperature-sensitive cell strains were grown in LB media at 30°C to avoid the reversion of the slow-growing temperature-sensitive mutant to a fast-growing form (Li et al. 2002). Total RNA was extracted when the OD600 value of the cell culture was 0.5 and was further treated with DNase by using the DNA-free kit (Ambion). Total RNA (8 μg) was resolved on an 8% (w/v) polyacrylamide denaturing gel at 20 W. The gel was subsequently incubated in water for 15 min and then in 10× SSC (1× SSC = 0.15 M NaCl, 0.015 M sodium citrate) for 15 min with agitation. RNA was transferred to Hybond-XL membrane (Amersham Biosciences) by capillary blotting or electroblotting. Transferred RNA was fixed on the membrane, and the membrane was prehybridized at 37°C for 30 min in hybridization buffer (5× SSC, 0.1% [w/v] bovine serum albumin, 0.1% [w/v] ficoll, 0.1% [w/v] polyvinylpyrrolidone, 0.5% [w/v] SDS, 0.1 mg/mL salmon testes DNA). Hybridization with a 5′ 32P-end-labeled synthetic oligonucleotide complementary to residues 39–54 of tRNATrp was performed at 37°C overnight. After washing with 2×, 1×, and 0.1× SSC, each containing 0.1% (w/v) SDS solutions, the blot was dried and visualized by phosphorimaging.
ACKNOWLEDGMENTS
T.L. and A.L.F. thank Malcolm Winkler, Darren Sledjeski, and members of the Feig laboratory for helpful discussions and comments on the work. We also thank Murray P. Deutscher for supplying strains that were used as part of this work. We also thank Phil Cunningham and Tek Lamichhane for the constructive discussion on the work. This work was supported in part by NIH grant GM-075068 to A.L.F. A.L.F. is a Cottrell Scholar of Research Corporation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.531408.
REFERENCES
- Arluison, V., Derreumaux, P., Allemand, F., Folichon, M., Hajnsdorf, E., Regnier, P. Structural modeling of the Sm-like protein Hfq from Escherichia coli . J. Mol. Biol. 2002;320:705–712. doi: 10.1016/s0022-2836(02)00548-x. [DOI] [PubMed] [Google Scholar]
- Bjork, G.R. Stable RNA modification. In Escherichia coli and Salmonella: Cellular and molecular biology . In: Neidhardt F.C., et al., editors. 2d ed. Vol. 1. ASM Press; Washington, DC: 1996. pp. 861–886. [Google Scholar]
- Brescia, C.C., Mikulecky, P.J., Feig, A.L., Sledjeski, D.D. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butland, G., Peregrin-Alvarez, J.M., Li, J., Yang, W., Yang, X., Canadien, V., Starostine, A., Richards, D., Beattie, B., Krogan, N., et al. Interaction network containing conserved and essential protein complexes in Escherichia coli . Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Cabello-Villegas, J., Winkler, M.E., Nikonowicz, E.P. Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem–loops of Escherichia coli tRNAPhe . J. Mol. Biol. 2002;319:1015–1034. doi: 10.1016/S0022-2836(02)00382-0. [DOI] [PubMed] [Google Scholar]
- Carpousis, A.J., Vanzo, N.F., Raynal, L.C. mRNA degradation. A tale of poly(A) and multiprotein machines. Trends Genet. 1999;15:24–28. doi: 10.1016/s0168-9525(98)01627-8. [DOI] [PubMed] [Google Scholar]
- Eisenberg, S.P., Yarus, M. The structure and aminoacylation of a temperature-sensitive tRNATrp (Escherichia coli) J. Biol. Chem. 1980;255:1128–1137. [PubMed] [Google Scholar]
- Eisenberg, S.P., Soll, L., Yarus, M. The purification and sequence of a temperature-sensitive tryptophan tRNA. J. Biol. Chem. 1979;254:5562–5566. [PubMed] [Google Scholar]
- Franze de Fernandez, M.T., Eoyang, L., August, J.T. Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Franze de Fernandez, M.T., Hayward, W.S., August, J.T. Bacterial proteins required for replication of phage Q ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 1972;247:824–831. [PubMed] [Google Scholar]
- Geissmann, T.A., Touati, D. Hfq, a new chaperoning role: Binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman, S. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Hagervall, T.G., Ericson, J.U., Esberg, K.B., Li, J.N., Bjork, G.R. Role of tRNA modification in translational fidelity. Biochim. Biophys. Acta. 1990;1050:263–266. doi: 10.1016/0167-4781(90)90178-5. [DOI] [PubMed] [Google Scholar]
- Hajnsdorf, E., Braun, F., Haugel-Nielsen, J., Regnier, P. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli . Proc. Natl. Acad. Sci. 1995;92:3973–3977. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle, C.A., Kushner, S.R. Development of an in vitro mRNA decay system for Escherichia coli: Poly(A) polymerase I is necessary to trigger degradation. Proc. Natl. Acad. Sci. 1996;93:12926–12931. doi: 10.1073/pnas.93.23.12926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba, S., Krueger, A., Trice, T., Krecic, A.M., Hinnebusch, A.G., Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae . Genes & Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel, J., Allmang, C., Verdone, L., Beggs, J.D., Tollervey, D. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol. Cell. Biol. 2002;22:5248–5256. doi: 10.1128/MCB.22.14.5248-5256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava, J., Houseley, J., Saveanu, C., Petfalski, E., Thompson, E., Jacquier, A., Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Lease, R.A., Belfort, M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease, R.A., Woodson, S.A. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Leung, H.-C.E., Chen, Y., Winkler, M.E. Regulation of substrate recognition by the MiaA tRNA prenyltransferase modification enzyme of Escherichia coli K-12. J. Biol. Chem. 1997;272:13073–13083. doi: 10.1074/jbc.272.20.13073. [DOI] [PubMed] [Google Scholar]
- Li, Z., Deutscher, M.P. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Reimers, S., Pandit, S., Deutscher, M.P. RNA quality control: Degradation of defective transfer RNA. EMBO J. 2002;21:1132–1138. doi: 10.1093/emboj/21.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie, A., Ribeiro, N.S., Reid, B.R., Jurnak, F. Relative affinities of all Escherichia coli aminoacyl-tRNAs for elongation factor Tu-GTP. J. Biol. Chem. 1984;259:5010–5016. [PubMed] [Google Scholar]
- Majdalani, N., Cunning, C., Sledjeski, D., Elliott, T., Gottesman, S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse, E., Gottesman, S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli . Proc. Natl. Acad. Sci. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse, E., Majdalani, N., Gottesman, S. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 2003;6:120–124. doi: 10.1016/s1369-5274(03)00027-4. [DOI] [PubMed] [Google Scholar]
- Mikulecky, P.J., Kaw, M.K., Brescia, C.C., Takach, J.C., Sledjeski, D.D., Feig, A.L. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, J.F., Uhlenbeck, O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Mohanty, B.K., Kushner, S.R. Analysis of the function of Escherichia coli poly(A) polymerase I in RNA metabolism. Mol. Microbiol. 1999;34:1094–1108. doi: 10.1046/j.1365-2958.1999.01673.x. [DOI] [PubMed] [Google Scholar]
- Mohanty, B.K., Maples, V.F., Kushner, S.R. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli . Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- Moller, T., Franch, T., Hojrup, P., Keene, D.R., Bachinger, H.P., Brennan, R.G., Valentin-Hansen, P. Hfq: A bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- Morita, T., Maki, K., Aiba, H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes & Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B.F., Nyborg, J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- O'Hara, E.B., Chekanova, J.A., Ingle, C.A., Kushner, Z.R., Peters, E., Kushner, S.R. Polyadenylylation helps regulate mRNA decay in Escherichia coli . Proc. Natl. Acad. Sci. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, R.A., Bhamidipati, A., Bi, X., Deb-Basu, D., Cahill, L., Ferrante, J., Gentalen, E., Glazer, M., Gossett, J., Hacker, K., et al. Isoelectric focusing technology quantifies protein signaling in 25 cells. Proc. Natl. Acad. Sci. 2006;103:16153–16158. doi: 10.1073/pnas.0607973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J., Pollard, J.W., Friesen, J.D., Stanners, C.P. Stuttering: High-level mistranslation in animal and bacterial cells. Proc. Natl. Acad. Sci. 1978;75:1091–1095. doi: 10.1073/pnas.75.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., Ferrin, T.E. UCSF Chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Richards, D.P., Stathakis, C., Polakowski, R., Ahmadzadeh, H., Dovichi, N.J. Labeling effects on the isoelectric point of green fluorescent protein. J. Chromatogr. A. 1999;853:21–25. doi: 10.1016/s0021-9673(99)00687-1. [DOI] [PubMed] [Google Scholar]
- Sauter, C., Basquin, J., Suck, D. Sm-like proteins in Eubacteria: The crystal structure of the Hfq protein from Escherichia coli . Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, M.A., Pearson, R.F., Moller, T., Valentin-Hansen, P., Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq–RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro, L., Franze de Fernandez, M.T., August, J.T. Resolution of two factors required in the Q-β-RNA polymerase reaction. Nature. 1968;220:478–480. doi: 10.1038/220478a0. [DOI] [PubMed] [Google Scholar]
- Sledjeski, D.D., Whitman, C., Zhang, A. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen, M.A. Charging levels of four tRNA species in Escherichia coli Rel+ and Rel− strains during amino acid starvation: A simple model for the effect of ppGpp on translational accuracy. J. Mol. Biol. 2001;307:785–798. doi: 10.1006/jmbi.2001.4525. [DOI] [PubMed] [Google Scholar]
- Storz, G., Opdyke, J.A., Zhang, A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- Stoscheck, C.M. Quantitation of protein. Methods Enzymol. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- Sukhodolets, M.V., Garges, S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- Sun, X., Wartell, R.M. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–4887. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- Sun, X., Zhulin, I., Wartell, R.M. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, H.C., Leung, H.C., Winkler, M.E. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol. Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen, P., Eriksen, M., Udesen, C. The bacterial Sm-like protein Hfq: A key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- Vanacova, S., Wolf, J., Martin, G., Blank, D., Dettwiler, S., Friedlein, A., Langen, H., Keith, G., Keller, W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, N.G., Yang, N., Burke, J.M. Probing nonselective cation binding in the hairpin ribozyme with Tb(III) J. Mol. Biol. 2000;298:539–555. doi: 10.1006/jmbi.2000.3691. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., Wilusz, J. Eukaryotic Lsm proteins: Lessons from bacteria. Nat. Struct. Mol. Biol. 2005;12:1031–1036. doi: 10.1038/nsmb1037. [DOI] [PubMed] [Google Scholar]
- Zhang, A., Wassarman, K.M., Ortega, J., Steven, A.C., Storz, G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- Zhang, A., Wassarman, K.M., Rosenow, C., Tjaden, B.C., Storz, G., Gottesman, S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]