Abstract
fl(2)d, the Drosophila homolog of Wilms'-tumor-1-associated protein (WTAP), regulates the alternative splicing of Sex-lethal (Sxl), transformer (tra), and Ultrabithorax (Ubx). Although WTAP has been found in functional human spliceosomes, exactly how it contributes to the splicing process remains unknown. Here we attempt to identify factors that interact genetically and physically with fl(2)d. We begin by analyzing the Sxl-Fl(2)d protein–protein interaction in detail and present evidence suggesting that the female-specific fl(2)d1 allele is antimorphic with respect to the process of sex determination. Next we show that fl(2)d interacts genetically with early acting general splicing regulators and that Fl(2)d is present in immunoprecipitable complexes with Snf, U2AF50, U2AF38, and U1-70K. By contrast, we could not detect Fl(2)d complexes containing the U5 snRNP protein U5-40K or with a protein that associates with the activated B spliceosomal complex SKIP. Significantly, the genetic and molecular interactions observed for Sxl are quite similar to those detected for fl(2)d. Taken together, our findings suggest that Sxl and fl(2)d function to alter splice-site selection at an early step in spliceosome assembly.
IN Drosophila melanogaster, sexual identity is initially determined by the X chromosome-to-autosome (A) ratio (Cline and Meyer 1996). The system that measures the X-to-A ratio turns on the Sex-lethal (Sxl) gene in female (XX) embryos by activating the Sxl establishment promoter, Sxl-Pe. In males (XY), this promoter is not activated and Sxl remains off. The establishment promoter is active for only a brief period in precellular blastoderm female embryos, and maintaining Sxl in the on state during the remainder of development in females depends upon an autoregulatory mechanism in which Sxl proteins direct their own synthesis by promoting the female-specific splicing of Sxl pre-mRNAs transcribed from the Sxl maintenance promoter (Sxl-Pm). In this autoregulatory loop, Sxl proteins bind to multiple sites in the large introns located upstream and downstream of the male-specific third exon and promote the joining of the 5′ splice site of exon 2 to the 3′ splice site of exon 4, skipping exon 3. The translation of the resulting Sxl mRNA ensures the maintenance of female identity by providing a continuous source of Sxl protein. In males, the third exon is incorporated into the Sxl mRNA by the default splicing machinery. The male-specific exon has several in-frame stop codons that prematurely truncate the Sxl open reading frame, which begins in exon 2, resulting in the production of a truncated, nonfunctional polypeptide. Sxl controls the majority of female development by regulating the female-specific splicing of transformer (tra) pre-mRNA. This female-specific Tra protein (TraF) then regulates the alternative splicing of the transcription factors doublesex and fruitless. In addition, Sxl expression is required for female viability because it turns off the gene msl-2. MSL-2 is a component of the male-specific dosage compensation complex that hypertranscribes genes on the X chromosome, a process that is necessary in males, but lethal to females. As a result of this essential function, mutations in genes that are necessary for regulating the female-specific splicing of Sxl are often female lethal. One allele in particular, fl(2)d1, is both female specific and temperature sensitive; females are inviable at 29° and sterile at 18° (Granadino et al. 1992). These female-specific effects have been shown to be due to the male-specific splicing of Sxl pre-mRNAs (Granadino et al. 1990, 1992).
Previous reports have indicated that fl(2)d is necessary for regulating the alternative splicing of multiple pre-mRNA targets. In addition to its role directing Sxl splicing in females, fl(2)d also functions at the next step in the sex determination hierarchy by regulating the female-specific splicing of tra (Granadino et al. 1996; Ortega et al. 2003). In addition, fl(2)d is required in both sexes, outside of the sex determination pathway, for regulating the alternative splicing of Ultrabithorax (Ubx) (Burnette et al. 1999). Remarkably, each of the three known fl(2)d targets are alternatively spliced via three distinct mechanisms. Female-specific Sxl splicing is accomplished by skipping an entire exon in Sxl pre-mRNAs by blocking that exon's 5′ and 3′ splice sites. By contrast, an alternative 3′ splice site is used to produce female-spliced tra mRNA, most likely by a process in which the default 3′ splice site is blocked and the female-specific 3′ splice site is activated (Sosnowski et al. 1989; Valcarcel et al. 1993; Deshpande et al. 1999). In yet a third alternative splicing paradigm, it is thought that fl(2)d promotes the inclusion of the small internal exons mI and mII of Ubx as fewer mature mRNAs contain these exons in animals compromised for fl(2)d. Since each of the three pre-mRNAs are processed in three different fashions, it is clear that fl(2)d's function is not limited to a specific alternative splicing event.
Much of the evidence demonstrating that Fl(2)d regulates splicing comes from experiments done in Drosophila. In support of an evolutionarily conserved role for fl(2)d in splicing regulation, one of its homologs, Wilms'-tumor-1-associated protein (WTAP) has been isolated from human spliceosomes (Zhou et al. 2002). Despite an abundance of evidence indicating that fl(2)d regulates pre-mRNA splicing, it has been difficult to elucidate a molecular mechanism that explains exactly how fl(2)d contributes to the alternative splicing process. Here we attempt to address this question by analyzing the ability of Fl(2)d protein to associate with general splicing regulators that have a clearly defined function in the splicing process. We have found that Fl(2)d interacts both genetically and physically with the early acting general splicing regulators Snf, U170K, U2AF50, and U2AF38, but does not interact with the later-acting general splicing regulators U540K or SKIP. We also performed a detailed analysis of the Sxl/Fl(2)d protein–protein interaction and provide evidence supporting a model whereby Sxl functions during the early stages of spliceosome assembly, rather than during the first and second catalytic steps, to regulate alternative splicing events.
MATERIALS AND METHODS
Fly stocks:
The following alleles were used for genetic analysis: fl(2)d1, fl(2)d2, Sxl7BO, Nβ-gal, snfJ210, snf148, U1-70k1, and U2af38ΔE18, and the following transgenic alleles were used: P{w+, snf5mer}, P{w+, otu∷Sxl}, and the deficiency Df 2RCX1 (Bloomington Stock Center).
Antibodies:
anti-Sxl m104 (1:10), anti-Sxl m114 (1:10), mouse anti-β-GAL (1:10) (Developmental Studies Hybridoma Bank, Iowa City, IA), anti-Snf 4G3 (1:10) (Flickinger and Salz 1994), anti-Scute5A10 (1:10) (Deshpande et al. 1995), anti-U1-70K (1:6000) (Nagengast et al. 2003), anti-U2AF50 (1:5000–50,000) (Rudner et al. 1998), anti-U2AF38 (1:10,000) (Rudner et al. 1996), anti-SKIPAb57 (1:2500) (Zhang et al. 2003), anti-U5-40k (1:50,000) (Achsel et al. 1998), anti-U5-116k (1:50,000) (Fabrizio et al. 1997). Monoclonal line anti-Fl(2)d 9G2 was generated similarly to that described by Deshpande et al. (1995) and used at a dilution of 1:10. His-tagged full-length Fl(2)d protein was produced using a baculovirus expression vector (a generous gift from Angeles Ortega) and purified on a Ni-NTA agarose column (QIAGEN, Valencia, CA).
Immunohistochemistry:
Embryos were fixed and stained as described previously (Deshpande et al. 1995). Alexa fluorophore-conjugated secondary antibodies were used at a 1:500 dilution (Invitrogen, San Diego). Hoechst was used to visualize DNA (1:1000, 5–10 min at room temperature).
Immunoprecipitations:
Nuclear embryonic extract was prepared similar to Samuels et al. (1994). Ovarian extract was prepared by dissecting ovaries from 20 females and following closely the protocol described by Tan et al. (2001). After two freeze/thaw cycles, the unfiltered ovarian extract was centrifuged twice at 3000 rpm for 5 min. For immunoprecipitations (Tan et al. 2001), extracts were incubated with 400 μl immunoprecipitation (IP) buffer, RNAsin, and 10–20 μl protein A/G beads crosslinked to the appropriate antibodies. Experiments with embryonic extract were carried out overnight at 4° or for 1–2 hr at room temperature and experiments with ovarian extract were carried out for 30 min at 29°. Beads were washed four to five times with 1 ml IP buffer, boiled in SDS/PAGE sample buffer for 5–10 min, and used in SDS/PAGE and Western blot analysis. (For all ovarian and embryonic extract preparations used in immunoprecipitation experiments, protein inhibitors were increased 10- to 40-fold to prevent Fl(2)d proteolysis.) GST pull-down experiments were performed as described previously (Chaouki and Salz 2006). In vitro immunoprecipitation experiments were performed as described by Deshpande et al. (1996). Expression and purification of recombinant full-length Sxl protein and GST-tagged Sxl fragments was described in Samuels et al. (1998). Expression and purification of Fl(2)d protein is described above.
RESULTS
fl(2)d suppresses female lethality caused by Nβ-gal transgene:
Sxl is an RNA recognition motif (RRM)-type RNA-binding protein and has two 90-aa RRM domains. While RRM domains are sufficient for mediating binding to the Sxl target RNA sequence (U8) (Samuels et al. 1998), they are not sufficient for directing the female-specific splicing of Sxl and tra pre-mRNAs. Splicing regulation also appears to depend upon the ∼100-aa N-terminal domain. Truncated Sxl proteins that lack 40 aa in the N terminus are defective in Sxl and tra splicing (Yanowitz et al. 1999). Conversely, a transgene that expresses a chimeric protein, Nβ-gal, consisting of the first 99 aa of Sxl fused to β-galactosidase, behaves like a dominant negative (Deshpande et al. 1999). In wild-type females, the Nβ-gal transgene interferes with Sxl autoregulation and causes partial female lethality. This lethality can be dominantly enhanced by reducing the dose of the Sxl gene or by mutations in known cofactors such as the U1/U2 snRNP protein Snf. To identify other genes involved in Sxl autoregulation, we screened the stock center deficiency kits for deletions that, when trans to the Nβ-gal transgene, either enhanced or suppressed its female lethal effects. One of deficiencies that suppressed the lethal effects of the Nβ-gal transgene was Df(2R)CX1. The interacting gene was identified as fl(2)d using smaller deficiencies and then by testing mutations in candidate genes that map to this region. Table 1 shows that female progeny from Nβ-gal/+ females and Nβ-gal/Nβ-gal males are only 87.9% as viable as their male siblings. fl(2)d1 suppressed this lethality as female progeny from fl(2)d1/+; Nβ-gal/+ females and Nβ-gal/Nβ-gal males are just as viable as their male counterparts. Similar results were obtained for the fl(2)d2 allele (data not shown).
TABLE 1.
fl(2)d1 suppresses partial female lethality of the dominant-negative Nβ-gal transgene
| Maternal genotype | % female viability | N |
|---|---|---|
| Nβ-gal/+ | 87.9 | 1565 |
| fl(2)d1/+; Nβ-gal/+ | 100* | 1692 |
Females of the genotype indicated were crossed to Nβ-gal/Nβ-gal males. Female progeny were scored relative to the number of male siblings. N, total progeny. *P < 0.0001, two-tailed Fisher's exact test.
fl(2)d1 appears to be an antimorph:
The fact that a reduction in fl(2)d activity suppressed rather than enhanced the lethal effects of the Nβ-gal transgene was unexpected. We had previously found that mutations in both Sxl and snf enhanced the female lethal effects of the Nβ-gal transgene (Deshpande et al. 1999) and we would have expected this to be true for fl(2)d as well since it is also an important cofactor for the regulatory activities of the Sxl protein. To investigate this anomaly further, we first tested for genetic interactions between Sxl and the two characterized fl(2)d alleles, fl(2)d1 and fl(2)d2. As described above, fl(2)d1 is a sex-specific, temperature-sensitive allele; homozygous females are sterile at 18° and inviable at 29°, while males are viable and fertile at both temperatures (Granadino et al. 1992). The phenotypic effects of fl(2)d1 are the result of a point mutation that causes an aspartic-acid-to-asparagine substitution at amino acid no. 180. fl(2)d2 has a stop codon close to the beginning of the open reading frame and should produce only a short (∼60 aa), and presumably nonfunctional, polypeptide (Penalva et al. 2000). We tested for genetic interactions by crossing females heterozygous for either fl(2)d1 or fl(2)d2 to Sxl− males. In these crosses, half of the progeny inherit the fl(2)d mutation and all of the progeny are maternally compromised for fl(2)d. Furthermore, since Sxl is on the X chromosome, all female progeny are heterozygous for Sxl. A reduced maternal dose of general splicing factors critical for the alternative splicing of Sxl pre-mRNAs, combined with half the normal zygotic dose of Sxl, can compromise the activation of the Sxl autoregulatory loop in early embryos, which results in female-specific lethality (Nagengast et al. 2003).
As shown in Table 2, we found that when mothers are heterozygous for the apparent null mutation, fl(2)d2, there is a small reduction (87%) in the viability of Sxl7B0/+ females compared to their male siblings. Since the protein null allele had only a mild effect on female viability, a strong maternal effect for the temperature-sensitive fl(2)d1 allele was not anticipated. However, contrary to this expectation, we found that fl(2)d1/+ females crossed to Sxl7BO/+ males gave rise to female progeny that were only 65% as viable as their male counterparts. Since fl(2)d1 is stronger than the null allele in this genetic test, this result suggests that fl(2)d1 is antimorphic.
TABLE 2.
Genetic interactions between fl(2)d and early acting general splicing regulators
| Maternal genotype | % female viability | N |
|---|---|---|
| fl(2)d1/+ | 65.3 | 329 |
| fl(2)d2/+ | 87.4 | 371 |
| snfJ210/+ | 83.6 | 465 |
| snfJ210/+; fl(2)d1/+ | 27.6**** | 680 |
| snfJ210/+; fl(2)d2/+ | 74.4*** | 863 |
| snf148/+ | 48.0 | 220 |
| snf148/+; fl(2)d1/+ | 11.1**** | 900 |
| snf148/+; fl(2)d2/+ | 45.6 | 1003 |
| snfJ210/+; +/+; P{w+, snf5mer/+} | 98.0 | 420 |
| snfJ210/+; fl(2)d1/+; P{w+, snf5mer/+} | 16.1**** | 419 |
| snfJ210/+; fl(2)d2/+; P{w+, snf5mer/+} | 79.3* | 945 |
| U2af38ΔE18/+ | 100 | 191 |
| fl(2)d1/U2af38ΔE18 | 25.8**** | 288 |
| fl(2)d2/U2af38ΔE18 | 90.1** | 346 |
Females of the genotype indicated were crossed to Sxl7BO males. Female progeny were scored relative to the number of male siblings. N, total progeny; two-tailed Fisher's exact test compared progeny from trans-heterozygous mothers to progeny from single-mutant heterozygous control mothers. ****P < 0.0001; ***P < 0.001; **P < 0.005; *P < 0.05.
Fl(2)d and Sxl form an RNA-independent complex and interact directly in vitro:
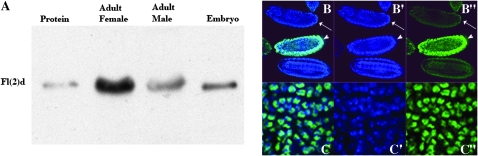
To be able to examine the relationship between Fl(2)d and Sxl in more detail, we generated monoclonal antibodies against the Fl(2)d protein. As shown in the Western blot in Figure 1A, our monoclonal antibody recognizes a protein of the expected size, ∼80 kDa (Penalva et al. 2000), in total extracts from adult males and females and from embryos. A protein of a similar size is also detected in nuclear extracts; however, Fl(2)d is quite sensitive to proteolysis and in some nuclear extract preparations a smaller ∼60-kDa protein is observed. To confirm the specificity of our monoclonal antibody, we used it to probe whole mounts of wild-type and fl(2)d2 embryos. The Fl(2)d protein localizes to the nucleus in wild-type embryos (Figure 1B, arrowhead), while it cannot be detected in homozygous fl(2)d2 embryos (Figure 1B, arrow). Figure 1C shows that Fl(2)d is not distributed uniformly throughout the nucleus but instead is concentrated in specific subregions as has been reported for other splicing factors.
Figure 1.—
Monoclonal antibody recognizes Fl(2)d protein on Western blots and whole-mount embryos. (A) Monoclonal anti-Fl(2)d 9G2 recognizes His-tagged purified Fl(2)d protein and 80-kDa protein in adult and embryonic extracts on Western blot. (B and C) Whole-mount embryos: Blue, DNA; Green, Fl(2)d. Wild-type embryos have Fl(2)d protein while the protein is absent from fl(2)d2/fl(2)d2 embryos. The higher magnification in C shows that Fl(2)d is not distributed evenly on the DNA in the nucleus.
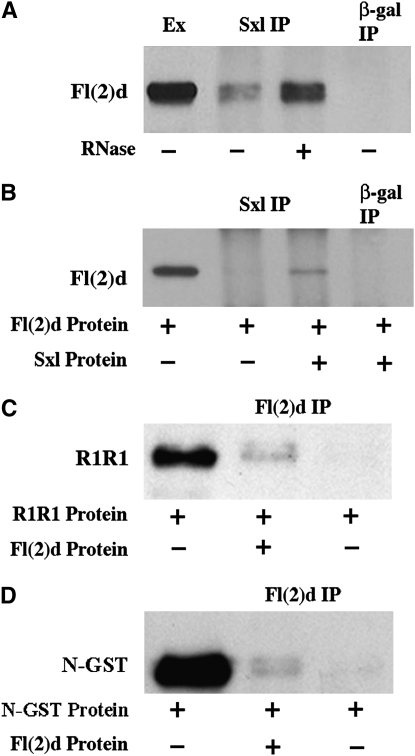
Previous studies with a polyclonal Fl(2)d antibody have suggested that Fl(2)d and Sxl are associated with each other in embryo nuclear extracts (Ortega et al. 2003). We have confirmed and extended these findings. Figure 2A shows that Sxl antibodies will co-immunoprecipitate Fl(2)d protein from embryonic extracts whereas Fl(2)d is not detected in the control immunoprecipitation with β-galactosidase antibody. Equivalent results were obtained in reciprocal immunoprecipitation using Fl(2)d antibody instead of Sxl antibody (Ortega et al. 2003). Sxl is also found in a complex with Snf, an RRM domain protein that is a component of both U1 and U2 snRNPs (Deshpande et al. 1996). Although recombinant Sxl and Snf can interact with each other directly (through the RRM domains) in vitro, the Sxl-Snf complex in nuclear extracts is sensitive to RNase A and disappears after RNase digestion (Deshpande et al. 1996; Samuels et al. 1998). To determine if this is also true for the Fl(2)d-Sxl complex, we performed Sxl co-immunoprecipitations with RNase-treated embryonic extracts. To our surprise, we found that Fl(2)d still co-immunoprecipitates with Sxl in the absence of RNA (Figure 2A). In fact, we consistently found that a greater amount of Fl(2)d associates with Sxl in RNase-treated extracts compared to the same extract that was instead protected from RNA degradation by an RNase inhibitor (compare lanes 2 and 3, Figure 2A).
Figure 2.—
Interactions between Sxl and Fl(2)d proteins in vivo and in vitro. (A) Fl(2)d-probed immunoprecipitations from wild-type nuclear embryonic extract. Lane 1, extract; lane 2, Sxl immunoprecipitation; lane 3, RNase-A-treated Sxl immunoprecipitation; lane 4, β-gal immunoprecipitation. (B) Fl(2)d-probed in vitro immunoprecipitations. Lane 1, Fl(2)d protein; lane 2, Sxl immunoprecipitation with Fl(2)d protein alone; lane 3, Sxl immunoprecipitation with Fl(2)d and Sxl proteins; lane 4, β-gal immunoprecipitation with Fl(2)d and Sxl proteins. (C) GST-probed Fl(2)d in vitro immunoprecipitations. Lane 1, R1R1-GST protein; lane 2, Fl(2)d immunoprecipitation with R1R1-GST and Fl(2)d proteins; lane 3, Fl(2)d immunoprecipitation with R1R1-GST protein alone. (D) GST-probed Fl(2)d in vitro immunoprecipitations. Lane 1, NSxl-GST protein; lane 2, Fl(2)d immunoprecipitation with NSxl-GST and Fl(2)d proteins; lane 3, Fl(2)d immunoprecipitation with NSxl-GST protein alone.
Since the interaction between Fl(2)d and Sxl does not require an RNA intermediate that could link the two proteins together, it seemed possible that the Fl(2)d-Sxl interaction might involve the direct binding of the two proteins to each other. To test this idea, we performed Sxl co-immunoprecipitations in vitro with purified Sxl and Fl(2)d proteins. Indeed, we found that Fl(2)d can be pulled down with Sxl antibody when full-length Sxl protein is included in the incubation mix. As expected, Fl(2)d does not immunoprecipitate with Sxl antibody in the absence of Sxl protein nor can it be immunoprecipitated with control beads containing β-galactosidase antibody (Figure 2B). We also performed Fl(2)d immunoprecipitation experiments with GST-tagged Sxl protein fragments to test whether Fl(2)d interacts with either the first (R1) or the second (R2) Sxl RRM domains. No interactions were detected with a GST fusion protein containing tandem RRM R2 domains (data not shown); however, as shown in Figure 2C, we did detect interactions between Fl(2)d and a GST fusion protein containing tandem Sxl RRM R1 domains. These findings suggest that the complexes containing Sxl and Fl(2)d in nuclear extracts could be generated by direct interactions between the two proteins and map one of the Sxl-Fl(2)d interaction domains to RRM R1. Interestingly, Sxl:Snf interactions are also mediated by the RRM domain R1 (Samuels et al. 1998).
Although these findings implicate the Sxl R1 RRM domain in protein–protein interactions with Fl(2)d, we wondered whether the Sxl N-terminal domain might also contribute to stabilizing the physical interactions between these two proteins. For one, the Fl(2)d protein interacted only poorly with a GST fusion protein containing a single R1 RRM domain (data not shown). In addition, one possible explanation for why a reduction in fl(2)d activity suppresses the female lethal effects of the Nβ-gal transgene is that the Fl(2)d protein binds to the Sxl Nβ-GAL fusion protein and recruits it to Sxl pre-mRNA splicing complexes. In this scenario, the female lethal effect of the transgene would be suppressed because less of the dominant-negative Nβ-GAL fusion protein would be incorporated into Sxl pre-mRNA splicing complexes when the activity of Fl(2)d is reduced. To explore this possibility, we tested whether the Fl(2)d can interact with the Sxl N terminus in vitro. In the experiment in Figure 2D, beads containing Fl(2)d antibody were used to pull down a GST-Sxl N-terminal fusion protein. When Fl(2)d was included in the incubation mix, a small amount of the GST-Sxl N-terminal protein could be pulled down by the Fl(2)d antibody beads, while the fusion protein was not pulled down when Fl(2)d was omitted from the incubation mix. While this finding would support the idea that the female lethal effects of the Nβ-GAL fusion protein might depend upon physical interactions with Fl(2)d, we were unable to demonstrate similar Nβ-GAL-Fl(2)d complexes in ovarian extracts from Nβ-gal transgenic females.
Sxl and Fl(2)d are associated in a protein complex in fl(2)d1 ovaries:
One model that could explain the apparent antimorphic activity of fl(2)d1 is that the aspartic-acid-to-asparagine substitution in the mutant protein disrupts Fl(2)d-Sxl protein interactions. If that is the case, Fl(2)d-Sxl complexes might not be formed or might be unstable in fl(2)d1 mutant females. Since fl(2)d1 females that survive at the permissive temperature are sterile, it is not possible to obtain a population of homozygous fl(2)d1 embryos. However, we thought that it would be possible to test for complexes with the mutant Fl(2)d1 protein in ovarian extracts. For this purpose, we raised fl(2)d1 flies at the permissive temperature. Following eclosion, we shifted the adults to 29° for 2–4 days. At the restrictive temperature, Sxl splicing in the mutant ovaries is in the male mode and little or no Sxl protein is expressed. To circumvent this problem, we rescued Sxl protein expression in the germline of the fl(2)d1 females by driving the expression of a female Sxl cDNA with the germline-specific otu promoter (P{w+, otu∷Sxl}) (Hager and Cline 1997).
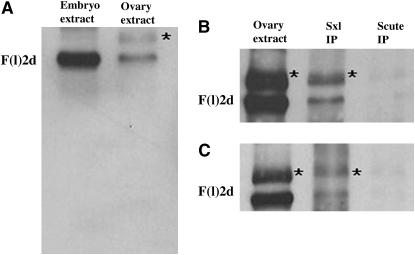
As shown in Figure 3A, wild-type ovarian extracts differ from extracts of embryos or adults in that two protein species are recognized by the Fl(2)d antibody. One is the same size, ∼80 kDa, as that observed in embryonic and adult extracts while the other is slightly larger. We suspect that this larger protein species is either a modified form of Fl(2)d or the product of an alternatively spliced fl(2)d transcript because we find that both proteins are present in Sxl immunoprecipitates from wild-type ovaries (Figure 3B). As shown in Figure 3C, both proteins are also present in Sxl immunoprecipitates from fl(2)d1 mutant ovaries. With the caveat that there may be small differences in the yield of the wild-type and Fl(2)d1 mutant complexes, this finding indicates that Fl(2)d1 protein is capable of assembling into a complex with Sxl. If there is no defect in the Fl(2)d-Sxl complex assembly, this would argue that Fl(2)d1 protein interferes with the splicing activity of the Sxl protein at a subsequent step in the splicing reaction.
Figure 3.—
Fl(2)d and Sxl form protein–protein complexes in wild-type and fl(2)d1 ovaries. An asterisk indicates an additional band picked up by Fl(2)d antibody in ovarian extract. (A) Fl(2)d-probed Western blot. Lane 1, embryonic extract; lane 2, ovarian extract. (B) Fl(2)d-probed immunoprecipitations from wild-type ovarian extract. Lane 1, ovarian extract; lane 2, Sxl immunoprecipitation; lane 3, Scute immunoprecipitation. (C) Fl(2)d-probed immunoprecipitations from fl(2)d1 ovarian extract. Lane 1, ovarian extract; lane 2, Sxl immunoprecipitation; lane 3, Scute immunoprecipitation.
Fl(2)d interacts with the U1 and the U2 snRNP protein Snf:
To better understand the role that Fl(2)d plays in Sxl-dependent alternative splicing, we looked for genetic interactions between fl(2)d and other components of the splicing machinery that specifically perturb the process of sex determination. We began by testing for interactions between fl(2)d and snf. Snf is the fly homolog of two mammalian snRNP proteins, U1A and U2B″, and it is found associated with both the U1 and the U2 snRNPs (Flickinger and Salz 1994; Polycarpou-Schwarz et al. 1996). Table 2 shows that females heterozygous for a null allele of snf crossed to Sxl− males gave rise to female progeny that were only 83% as viable as their male siblings. The comparable cross for fl(2)d1 generates females that are only 65% as viable as males (Table 2). To test for a genetic interaction between snf and fl(2)d, snfJ210/+; fl(2)d1/+ trans-heterozygous females were crossed to Sxl− males. Female progeny from this cross were only 27% as viable as their male siblings, indicating a synergistic interaction between snfJ210 and fl(2)d1. Genetic interactions were also observed between the null allele fl(2)d2 and snfJ210; however, they were not as strong as fl(2)d1 (see Table 2). This difference would provide additional support for the idea that fl(2)d1 is likely to be an antimorphic allele, at least with respect to sex determination.
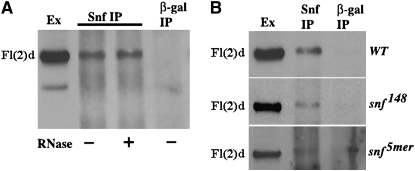
Since fl(2)d mutations enhance the female lethal effects of snf, we decided to test whether Fl(2)d is found in a complex with Snf as is observed for Sxl. We immunoprecipitated embryonic nuclear extracts with Snf antibody and then probed Western blots of the precipitated proteins with Fl(2)d antibody. Figure 4A shows that Fl(2)d is present in Snf immunoprecipitates. Given that the Sxl-Snf complex is unstable in the absence of RNA, while the Sxl-Fl(2)d complex is not, we tested whether RNase treatment disrupts the Fl(2)d-Snf complex in embryonic nuclear extracts. Like the Fl(2)d-Sxl complex, we found that the Fl(2)d-Snf complex is RNA independent. However, unlike the complex with Sxl, we did not see an enrichment of Fl(2)d in RNAse-treated extracts relative to extracts treated with an RNAse inhibitor (Figure 4A).
Figure 4.—
Fl(2)d and Snf form a protein–protein complex in embryos. (A) Fl(2)d-probed immunoprecipitations from wild-type nuclear embryonic extract. Lane 1, extract; lane 2, Snf immunoprecipitation; lane 3, RNase-A-treated Snf immunoprecipitation; lane 4, β-gal immunoprecipitation. (B) Fl(2)d-probed immunoprecipitations from wild-type (top), snf148 (middle), and snf5mer (bottom) nuclear embryonic extracts. Lane 1, extract; lane 2, Snf immunoprecipitation; lane 3, β-gal immunoprecipitation.
fl(2)d interacts genetically with Snf mutations that disrupt association with either U1 or U2 snRNPs:
As a component of both the U1 and the U2 snRNPs, the Snf protein participates in multiple steps of the splicing reaction, beginning with the initial assembly of U1 and U2 snRNPs on the pre-mRNA through the last step in splicing, the joining of the 5′ and 3′ splice junctions, and the release of lariat intron RNA. Consequently, the Fl(2)d-Snf interactions that are critical for the regulation of alternative splicing by Sxl are expected to be mediated by interactions involving either or both of these snRNPs and could potentially occur at a number of points in the splicing reaction. To determine which of the snRNPs might be the target for the Fl(2)d-Snf interactions, we took advantage of two snRNP-specific alleles of snf.
snf148 encodes a form of the Snf protein that can assemble into a U2 but not a U1 snRNP. Unlike wild-type Snf, the mutant Snf148 protein is also defective in its interaction with the Sxl protein, and little or no Sxl-Snf148 complexes are detected (Nagengast et al. 2003). Consistent with this defect in Sxl-Snf interactions, when snf148/+ females are crossed to Sxl− males, the female progeny are only 48% as viable as their male siblings (Table 2). The antimorphic fl(2)d1 allele enhances the female lethality of snf148 mutation and daughters arising from snf148/+; fl(2)d1/+ mothers and Sxl− fathers are only 11% as viable as their male counterparts (Table 2). By contrast, the null allele fl(2)d2 did not show any interaction with snf148.
snf5mer is the reciprocal allele to snf148 in that it encodes a form of Snf that assembles into U1 but not U2 snRNPs (Stitzinger et al. 1999). Moreover, unlike snf148, the Snf5mer protein can form a complex with Sxl and seems to have less of an effect on Sxl-dependent alternative splicing (Nagengast et al. 2003). As can be seen in Table 2, female progeny from snfJ210/+ ; +/+; P{w+, snf5mer}/+ mothers and Sxl− fathers are equally as viable as males. On the other hand, we detected a synergistic interaction between fl(2)d1 and snf5mer as snfJ210/+ ; fl(2)d1/+; P{w+, snf5mer}/+ females crossed to Sxl− males gave rise to females that were only 16% as viable as their male siblings. fl(2)d2 also interacted with the U2-defective snf5mer mutation; however, the effects were significantly weaker as the corresponding female progeny were 79% as viable (Table 2).
Since the mutant Snf148 and Snf5mer proteins exhibit differences in their ability to interact with the U1 and U2 snRNPs and with Sxl, we wondered whether they also differed in their ability to interact with Fl(2)d. In the experiments in Figure 4B, extracts prepared from wild-type, snf148, and snf5mer mutant embryos were immunoprecipitated with Snf antibody. We then probed Western blots of the immunoprecipitated proteins with Fl(2)d antibodies. Since we were able to detect Fl(2)d in all three Snf immunoprecipitates, it would appear that both snf148 and snf5mer are able to assemble into complexes containing Fl(2)d protein. On the other hand, it would appear that the Snf5mer:Fl(2)d complex is partially destabilized relative to wild type as it was generally more difficult to detect Fl(2)d in the Snf5mer immunoprecipitates than in the wild-type or Snf148 immunoprecipitates.
fl(2)d shows genetic interactions with U2-associated factor U2AF:
The fact that the U2-defective snf5mer shows female lethal interactions with fl(2)d even though snf5mer/+ results in little or no female lethality on its own suggests that one of the critical Fl(2)d-Snf interactions in the regulation of alternative splicing by Sxl is likely to involve the U2 snRNP. That this might correspond to a function of the U2 snRNP at a step early in the splicing reaction is suggested by the finding that the antimorphic fl(2)d1 allele enhances female lethal effects of the U1-defective snf148. To test these ideas further, we assayed for interactions between fl(2)d and other proteins that are associated or that function with either the U1 or the U2 snRNPs.
The U2-associated factor U2AF functions to recruit the U2 snRNP to the 3′ splice junction and is composed of two different subunits, U2AF38 and U2AF50 (Reed 2000). To look for genetic interactions between fl(2)d and U2AF, we tested whether reducing U2AF activity has female lethal effects. For this purpose, females heterozygous for a mutation in the U2AF38 subunit, U2AF38ΔE18 (Rudner et al. 1996), were mated with Sxl− males. As shown in Table 2, a reduction in the maternal contribution of U2AF38 had no apparent effect on the viability of the Sxl−/+ female progeny from this cross. However, as found for the U2-defective snf5mer mutation, female lethal interactions were observed when the mothers were simultaneously heterozygous for U2AF38ΔE18 and fl(2)d1. Female progeny from fl(2)d1/U2AF38ΔE18 mothers and Sxl− fathers were only 25% as viable as their sibling males (Table 2). As seen for snf5mer, U2AF38ΔE18 does not enhance the female lethal effects of the null mutation fl(2)d2 as strongly as it does the presumed antimorph fl(2)d1.
Fl(2)d associates with U1-70k and U2AF:
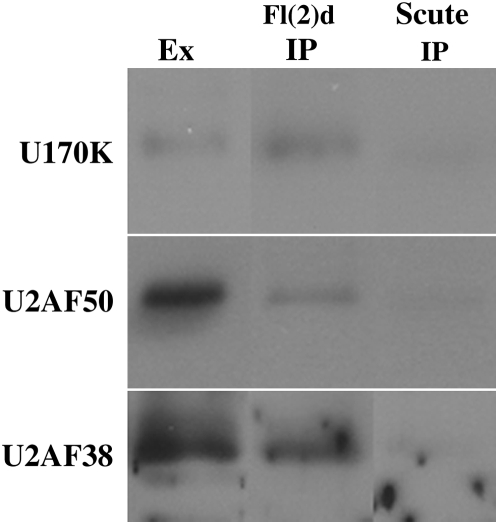
The genetic experiments described above suggest that Fl(2)d functions at a point early in the splicing reaction through interactions with proteins that function at the 5′ and perhaps also at the 3′ splice sites. The results of these genetic experiments are supported by the finding that Fl(2)d forms complexes with the U1 and the U2 snRNP protein Snf in nuclear extracts. To provide additional evidence for a physical association between Fl(2)d and factors important for the assembly or functioning of the U1 and U2 snRNPs, we immunoprecipitated wild-type nuclear extracts with Fl(2)d antibody and then assayed for the presence of U1- or U2-associated factors. Figure 5 shows that the U1-snRNP protein U1-70k is present in Fl(2)d immunoprecipitates, but absent from the negative control Scute immunoprecipitates. In addition, both subunits of U2AF, U2A50 and U2AF38, were present in Fl(2)d immunoprecipitates (Figure 5). These results provide additional evidence for a physical association between Fl(2)d and proteins that act at both the 5′ and the 3′ splice sites. Furthermore, these results support the idea that the function of Fl(2)d in Sxl-dependent regulation of alternative splicing occurs at an early step in the splicing reaction.
Figure 5.—
Fl(2)d forms protein–protein complexes with the early acting general splicing regulators U1-70K, U2AF50, and U2AF38. Immunoprecipitations of wild-type nuclear embryonic extract probed for U1-70K (top), U2AF50 (middle), and U2AF38 (bottom). Lane 1, extract; lane 2, Fl(2)d immunoprecipitation; lane 3, Scute immunoprecipitation.
Fl(2)d does not associate with U5-40K:
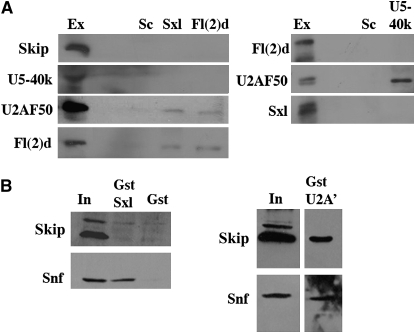
We wondered whether Fl(2)d remains associated with the spliceosome at later steps of the splicing reaction. To address this question, we assayed for complexes between Fl(2)d and proteins that function during middle and late, but not early, stages of splicing. We first tested for an association between Fl(2)d and U5-40k, a protein component of the U5 snRNP. The U5 snRNP, along with U4 and U6, is not recruited to the pre-mRNA splicing complex until after the A complex has formed via the binding of U1 and U2 snRNPs to the 5′ and 3′ splice sites, respectively (Jurica and Moore 2003; Stark and Luhrmann 2006). The association of U4/5/6 generates the B complex, which contains the full complement of snRNPs. This complex then undergoes a series of rearrangements beginning with the displacement of U1. This is followed with the formation of the U2/U6 hybrid and the displacement of U4 from the spliceosome. This generates the first activated complex, B*, which in turn catalyzes the phosphortransferase reaction that releases the 5′ exon and forms the intron lariat. Figure 6A shows that U5-40k is not present in Fl(2)d immunoprecipitates from embryonic nuclear extract. While it is difficult to conclude that two proteins do not interact on the basis of an inability to detect a complex, several lines of evidence suggest that the absence of U5-40k in Fl(2)d immunoprecipitates might be significant. Two different positive control proteins could be detected from the same Fl(2)d immunoprecipitations: Fl(2)d and U2AF50 (Figure 6A).
Figure 6.—
Fl(2)d and Sxl do not interact with the mid- and late-acting splicing regulators SKIP and U5-40k. (A) Immunoprecipitations from wild-type nuclear embryonic extract. (Left) Probed for SKIP (top), U5-40k (top middle), U2AF50 (bottom middle), and Fl(2)d (bottom). Lane 1, extract (lane 2, blank); lane 3, Scute immunoprecipitation; lane 4, Sxl immunoprecipitation; lane 5, Fl(2)d immunoprecipitation. (Right) Probed for Fl(2)d (top), U2AF50 (middle), and Sxl (bottom). Lane 1, extract (lane 2, blank); lane 3, Scute immunoprecipitation; lane 4, U5-40k immunoprecipitation. (B) GST pull downs from embryonic extract probed for SKIP (top) and Snf (bottom). (Left) Lane 1, embryonic extract input; lane 2, GST pull down with GST-Sxl; lane 3, GST pull down with GST protein alone. (Right) Lane 1, embryonic extract input; lane 2, GST pull down with GST-U2A′.
Furthermore, results from the reciprocal experiment are also consistent with the two proteins not being associated in an immunoprecipitable complex as Fl(2)d was not present in U5-40k immunoprecipitates while the U2 snRNP-associated factor, U2AF50, was (Figure 6A). Again, this result is validated by the presence of other general splicing regulators detected in the same U5-40k immunoprecipitations. We found that U5-116k, a component of the U5 snRNP, could be detected in the same U5-40k immunoprecipitates that did not contain Fl(2)d (data not shown). We also found that Snf protein was present in these same U5-40k immunoprecipitates (data not shown). Thus, even though Snf and Fl(2)d associate in a protein–protein complex, the absence of Fl(2)d in U5-40K immunoprecipitates suggests that the Snf-Fl(2)d complex is distinct from the Snf-U5-40K complex.
Fl(2)d does not associate with SKIP:
We also tested whether Fl(2)d is associated in an immunoprecipitable complex with the SKIP protein. SKIP is a late-acting splicing factor that has been found in B and C, but not in A, spliceosomal complexes (Hartmuth et al. 2002; Jurica et al. 2002; Makarov et al. 2002; Deckert et al. 2006). It then remains associated with the spliceosome through both the first and the second catalytic steps. As observed for U5-40k, we did not detect SKIP in the Fl(2)d immunoprecipitates from embryonic nuclear extracts, while U2AF50 was present (Figure 6A). Reciprocal SKIP immunoprecipitation experiments were consistent with these results (data not shown).
Sxl does not associate with either U5-40k or SKIP:
The results presented above indicate that Fl(2)d is associated with splicing complexes that are present during the early stages of spliceosome assembly, but does not seem to be in complexes that are formed during the actual catalytic splicing reaction. An important question is whether the Sxl protein is also primarily associated with complexes involved in the initial assembly of the U1 and U2 snRNPs on the pre-mRNA or remains present during the conversion of the A complex to the activated B complex. To address this question, we immunoprecipitated embryonic nuclear extracts with Sxl antibodies and then tested for the presence of U5-40k and SKIP proteins. As shown in Figure 6A, neither U5-40k nor SKIP could be detected in Sxl immunoprecipitates even though both U2AF50 and Fl(2)d could be detected in the same Sxl immunoprecipitations. Likewise, in the reciprocal IP with the U5-40k antibody, Sxl could not be detected, but U2AF50, Snf, and U5-116k were present (Figure 6A and data not shown).
Similar results were obtained in GST pull-down experiments from nuclear extracts using a GST-Sxl fusion protein. As shown in Figure 6B, GST-Sxl can pull down the Snf protein from nuclear extracts, while Snf is not pulled down by GST alone. In contrast, the SKIP protein is not pulled down by either GST-Sxl or GST. To test whether SKIP can be pulled down from nuclear extracts with a GST fusion protein, we generated a GST fusion to the U2 snRNP protein U2A′. As shown in Figure 6B, SKIP and also Snf can be pulled from nuclear extracts by the GST-U2A′ fusion protein. Taken together, these results suggest that, like Fl(2)d, Sxl is associated with splicing complexes that are formed during the early steps in the splicing reaction, while it does not remain associated with complexes that are present during the later stages of splicing.
DISCUSSION
Alternative splicing of pre-mRNAs requires the default splicing machinery to choose between different potential 5′ and 3′ splice-site combinations. Factors like Sxl that force the selection of alternative 5′ and 3′ splice-site combinations must exert their effects through interactions with components of the general splicing machinery. However, since the splicing of pre-mRNAs is a multi-step process that depends upon the assembly and remodeling of a large and highly dynamic RNA protein complex, these regulatory interactions could potentially occur at many different points in the processing reaction. Previous genetic and molecular studies have implicated several general splicing factors in Sxl-dependent alternative splicing. These include Snf, U1-70k, U2AF, and Spf45 (Deshpande et al. 1996; Salz and Flickinger 1996; Nagengast et al. 2003; Chaouki and Salz 2006). Of these proteins, only Snf is expected to be present in all of the intermediate steps in the splicing reaction. In contrast, studies on spliceosomal intermediates in humans indicate that the three other Sxl interactors are associated with the spliceosome only during the early stages of the splicing reaction (Jurica and Moore 2003; Deckert et al. 2006). Both Snf and U1-70k are components of the U1 snRNP and will be present when the U1 snRNP first associates with the 5′ splice site of the pre-mRNA to form the prespliceosome E complex. After U1 interacts with the 5′ splice site, U2AF is thought to bind to the polypyrimidine tract upstream of the 3′ splice site and recruit the U2 snRNP to the pre-mRNA to form spliceosome complex A (Jurica and Moore 2003; Stark and Luhrmann 2006). In addition to Snf, U1-70k, and U2AF, this complex in humans also includes the SPF45 protein (Hartmuth et al. 2002). In the next step, a complex containing three other snRNPs, the U4/U6,U5 tri-snRNP, associates with the spliceosome to form the B complex. This is followed by extensive structural rearrangements in which the U1 and subsequently U4 snRNPs are displaced. U170k together with the U1-associated Snf should be lost from the complex with disassociation of the U1 snRNP. The subsequent unwinding of the U4/U6 base pairs and dissociation of U4 permits base pairing between U6 and the 3′ splice site and the U2 snRNA (Stark and Luhrmann 2006). This generates the active complex B*, which catalyzes the first transesterification reaction to generate complex C. Both U2AF and SPF45 appear to be missing from complex B*, while the only Sxl cofactor that is expected to remain until the final splicing step should be the Snf protein associated with the U2 snRNP. Thus, with the exception of Snf, the proteins known to be important for Sxl-dependent alternative splicing appear to function prior to the formation of the activated B* complex and the first splicing reaction.
Several lines of evidence suggest that this is also likely to be true for Fl(2)d. First, we found that Fl(2)d is in an immunoprecipitable complex with Snf, U1-70K, and both of the U2AF subunits, U2AF50 and U2AF38. All of these proteins are expected to be present in one or more of the complexes (E, A, or B) that are formed early in the splicing reaction. Second, it has recently been found that U2AF not only is present in the early complexes E and A, but also can be detected in the inactive B complex (Deckert et al. 2006). Consistent with this expectation, we were able to detect complexes between U2AF50 and the U5 snRNP protein U5-40k. On the other hand, while we could detect complexes between U2AF50 and Fl(2)d, we could not detect complexes between U5-40k and Fl(2)d. This finding would suggest that Fl(2)d is stably associated with the E and/or A complex, but is not stably associated with the B complex. Third, the SKIP protein associates with the inactive and activated B complexes, but is absent from the A complex (Jurica and Moore 2003; Deckert et al. 2006). As was the case for U5-40k, we could not detect interactions between SKIP and Fl(2)d. With the caveat that these negative results must be interpreted with caution, these findings, taken together, would argue that Fl(2)d functions at an early step(s) in the splicing reaction prior to the formation of complex B.
Since Fl(2)d is expressed in both sexes and has functions in alternative splicing that are not connected to Sxl, it could be argued that the physical interactions detected between Fl(2)d and different components of the splicing apparatus do not reflect its functioning in Sxl-dependent alternative splicing. While this is a potential concern, there are a number of reasons why we believe this to be unlikely. For one, these complexes appear to be physiologically relevant to Sxl-dependent alternative splicing as we observe female-specific genetic interactions between fl(2)d and the genes encoding several of these proteins. In the case of snf, we tested not only a null allele, but also two mutations, snf148 and snf5mer, which differentially affect Snf protein interactions with the U1 or the U2 snRNPs, respectively. When mothers heterozygous for the U1-deficient snf148 are mated to Sxl− fathers, there is a marked reduction in the viability of female offspring. This female lethality is enhanced by the antimorphic allele fl(2)d1, but not by the null allele fl(2)d2. In contrast to snf148, there is little, if any, female-specific lethality in the progeny of snf5mer/+ females and Sxl− fathers; however, the snf5mer allele shows a very strong synergistic female lethal interaction with fl(2)d1. Likewise, a strong synergistic interaction was observed between fl(2)d1 and U2AF38ΔE18.
Another reason to believe that the association of Fl(2)d with early splicing regulators is relevant to how it promotes Sxl-dependent alternative splicing is the fact that Sxl is found in complexes with the same set of splicing factors in nuclear extracts as Fl(2)d. As noted above, these factors include Snf, U170k, and the two U2AF subunits U2AF38 and U2AF50. In the case of Snf, we have previously shown that Sxl is in an immunoprecipitable complex with Snf in nuclear extracts. Although this is also true for Fl(2)d, there are some differences in how Fl(2)d and Sxl interact with Snf. For one, Fl(2)d:Snf interactions in nuclear extracts are insensitive to RNase, while Sxl:Snf interactions are RNase sensitive. While we did not test whether Fl(2)d:Snf interactions involve direct protein contacts, both Fl(2)d and Snf can interact directly with Sxl in vitro. Fl(2)d and Sxl also differ in their interactions with the Snf mutant proteins 148 and 5mer. Sxl can associate with the Snf5mer protein, but does not form a complex with Snf148. By contrast, Fl(2)d complex formation with the Snf148 mutant protein appears to be equivalent to that observed for wild-type Snf, while complexes with Snf5mer appear to be destabilized and in our hands are present in reduced yield. In addition, we also found that Sxl resembles Fl(2)d in that it is not stably associated either with the U5 snRNP protein U5-40k or with SKIP.
It has been suggested—on the basis of in vitro splicing experiments using a chimeric pre-mRNA consisting of an adenovirus 5′ exon/intron fused to a short sequence spanning the 3′ splice site of the Sxl male exon—that Sxl autoregulation depends upon Sxl inhibition of the second catalytic step of splicing, i.e., the joining of the 5′ splice site of the Sxl second exon to the 3′ splice site of the male exon and the release of the second intron lariat intermediate (Lallena et al. 2002). In this model, Sxl was proposed to block this catalytic step by inhibiting the SPF45 factor bound to one of the two AG sequences in the male exon 3′ splice site. It was suggested that this would force the splicing machinery to bypass the male exon 3′ splice site and instead join the free 5′ splice site of exon 2 to the 3′ splice of exon 4 located slightly more than a kilobase downstream of the male exon. In addition to the fact that using a 3′ AG for the second catalytic step that is located ∼1 kb from the branch point would be highly unusual, it is difficult to reconcile this model for Sxl autoregulation with the results presented here, which argue that Sxl must act at a much earlier point during the initial assembly of the splicing apparatus on target pre-mRNAs. Other findings also seem to be inconsistent with this model. For one, the Sxl-binding sites located in the polypyrimidine tract of the male exon 3′ splice site that were used in the in vitro splicing experiments are completely dispensable for female splicing of the Sxl pre-mRNAs in vivo. In fact, the critical sites for Sxl binding are located in the upstream and downstream intron sequences flanking the male exon >200 bases from the male 3′ splice site. In addition, Sxl regulation in vivo seems to pivot on blocking the use of the male exon 5′ splice site, while controlling the use of the male exon 3′ splice site plays at most only a subordinate role in regulation (Horabin and Schedl 1993b). Finally, although SPF45 is present in purified B spliceosome complexes from humans, it is apparently absent from the catalytically active B* and C complexes (Jurica et al. 2002; Makarov et al. 2002; Deckert et al. 2006).
Another question of interest is the nature of the relationship between Sxl and Fl(2)d. Like Snf, Fl(2)d can interact directly with Sxl in vitro. For Snf, the first Sxl RRM domain R1 mediates this interaction while for Fl(2)d the interaction appears to depend upon a combination of the Sxl N terminus and the R1 RRM domain. Although the in vitro interactions between recombinant Sxl and Snf proteins are not dependent on (or stimulated by RNA), RNase treatment completely disrupts Sxl:Snf interactions in nuclear extracts. By contrast, RNase treatment appears to significantly enhance Sxl:Fl(2)d interactions in nuclear extracts. Since the two Sxl RRM domains undergo a substantial rearrangement when they bind to RNA (Hughson and Schedl 1999), it is possible that Sxl:Fl(2)d interactions occur prior to the binding of the Sxl protein to the pre-mRNA, while Sxl:Snf interactions occur after Sxl has associated with its target sequences. If this is the case, then one plausible idea would be that Fl(2)d helps recruit Sxl into the assembling spliceosome. This mechanism could potentially account for the finding that fl(2)d mutations dominantly suppress the female lethal effects of the antimorphic Nβ-gal trangene: there would be less of Nβ-GAL fusion protein incorporated into the Sxl splicing complex when fl(2)d activity is reduced. However, since we were unable to demonstrate an association between Fl(2)d and the Nβ-GAL fusion protein in vivo, we cannot exclude other mechanisms for suppression and further studies will be required to fully understand how Fl(2)d functions in Sxl-dependent alternative splicing.
Given that Sxl and key cofactors like Fl(2)d associate with early acting general splicing regulators that function to define the 5′ and the 3′ splice sites, it is possible that Sxl promotes female-specific splicing of its own pre-mRNAs by inhibiting the process of exon definition. Presumably it would do so by specifically targeting the U1 snRNP associated with the male exon 5′ splice site and SPF45 and the U2AF heterodimer associated with the male exon 3′ splice site. Exon definition is thought to be particularly important when a small exon is surrounded by large introns as is the case for the Sxl male exon. It is only ∼190 bp in length and is flanked by large introns. Interestingly, exon definition cannot occur for exons >500 nucleotides and, if a large exon is surrounded by large introns, such an exon is often skipped entirely (Sterner et al. 1996; Fox-Walsh et al. 2005). Consistent with these studies, several of the gain-of-function mutations in Sxl are transposon insertions that increase the size of the male exon. In these mutants, Sxl is not required for female-specific splicing and the male exon is skipped, even in the absence of Sxl protein (Bernstein et al. 1995). Also supporting the notion that male exon definition might be an especially sensitive step that would make it a good target for Sxl regulation is the fact that both the 3′ and 5′ male exon splice sites are known to be suboptimal. In fact, when the male exon (plus the associated splice sites) is placed into a heterologous intron, the male exon is not recognized by the splicing machinery unless the splice sites are optimized to more closely resemble the consensus sequence (Horabin and Schedl l993a). Even then, the male exon is not efficiently recognized by the default splicing machinery and is skipped most of the time. Further studies will be required to explore this possible mechanism for Sxl regulation.
Acknowledgments
We thank Tom Cline, Paul MacDonald, Steve Mount, Angeles Ortega, Don Rio, Lucas Sanchez, Juan Valcarcel, Cindy Will, and the Bloomington Stock Center for fly stocks and antibodies. We also thank Tony del Rio and Toh Hean Ch'ng for assistance with baculovirus protein expression, Joe Goodhouse for help with the confocal microscope, and Gordon Gray for providing fly food. This work was supported by National Institutes of Health grants to P.S., H.K.S. (RO1-GM61039), and A.S.C. (T32-HD07104 and T32-GM07250).
References
- Achsel, T., K. Ahrens, H. Brahams, S. Teigelkamp and R. Luhrmann, 1998. The human U5–220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 18 6756–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein, M., R. A. Lersch, L. Subrahmanyan and T. W. Cline, 1995. Transposon insertions causing constitutive Sex-lethal activity in Drosophila melanogaster affect Sxl sex-specific transcript splicing. Genetics 139 631–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette, J. M., A. R. Hatton and A. J. Lopez, 1999. Trans-acting factors required for inclusion of regulated exons in the Ultrabithorax mRNAs of Drosophila melanogaster. Genetics 151 1517–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouki, A. S., and H. K. Salz, 2006. Drosophila SPF45: a bifunctional protein with roles in both splicing and DNA repair. PLoS Genet. 2 1974–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, T., and B. Meyer, 1996. Viva la difference: males vs females in flies vs worms. Annu. Rev. Genet. 30 637–702. [DOI] [PubMed] [Google Scholar]
- Deckert, J., K. Hartmuth, D. Boehringer, N. Behzadnia, C. L. Will et al., 2006. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26 5528–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, G., J. Stukey and P. Schedl, 1995. scute (sis-b) function in Drosophila sex determination. Mol. Cell. Biol. 15 4430–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, G., M. E. Samuels and P. Schedl, 1996. Sex-lethal interacts with splicing factors in vitro and in vivo. Mol. Cell. Biol. 16 5036–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande, G., G. Calhoun and P. Schedl, 1999. The N-terminal domain of Sxl protein disrupts Sxl autoregulation in females and promotes female-specific splicing of tra in males. Development 126 2841–2853. [DOI] [PubMed] [Google Scholar]
- Fabrizio, P., B. Laggerbauer, J. Lauber, W. S. Lane and R. Luhrmann, 1997. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 16 4092–4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flickinger, T. W., and H. K. Salz, 1994. The Drosophila sex determination gene snf encodes a nuclear protein with sequence and functional similarity to the mammalian U1A snRNP protein. Genes Dev. 8 914–925. [DOI] [PubMed] [Google Scholar]
- Fox-Walsh, K. L., Y. Dou, B. J. Lam, S. P. Hung, P. F. Baldi et al., 2005. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc. Natl. Acad. Sci. USA 102 16176–16181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granadino, B., S. Campuzno and L. Sanchez, 1990. The Drosophila melanogaster fl(2)d gene is needed for the female-specific splicing of Sex-lethal RNA. EMBO J. 8 2597–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granadino, B., A. San Juan, P. Santamaria and L. Sanchez, 1992. Evidence of a dual function in fl(2)d, a gene needed for Sex-lethal expression in Drosophila melanogaster. Genetics 130 597–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granadino, B., L. O. Penalva and L. Sanchez, 1996. The gene fl(2)d is needed for the sex-specific splicing of transformer pre-mRNA but not for double-sex pre-mRNA in Drosophila melanogaster. Mol. Gen. Genet. 253 26–31. [DOI] [PubMed] [Google Scholar]
- Hager, J. H., and T. W. Cline, 1997. Induction of female Sex-lethal RNA splicing in male germ cells: implications for Drosophila germline sex determination. Development 124 5033–5048. [DOI] [PubMed] [Google Scholar]
- Hartmuth, K., H. Urlaub, H. P. Vornlocher, C. L. Will, M. Gentzel et al., 2002. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. USA 99 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horabin, J., and P. Schedl, 1993. a Regulated splicing of the Drosophila Sex-lethal male exon involves a blockage mechanism. Mol. Cell. Biol. 13 1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horabin, J., and P. Schedl, 1993. b Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol. Cell. Biol. 13 7734–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughson, F. M., and P. Schedl, 1999. Two domains and one RNA: a molecular threesome. Nat. Struct. Biol. 6 499–502. [DOI] [PubMed] [Google Scholar]
- Jurica, M. S., and M. J. Moore, 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12 5–14. [DOI] [PubMed] [Google Scholar]
- Jurica, M. S., L. J. Licklider, S. R. Gygi, N. Grigorieff and M. J. Moore, 2002. Purification and characterization of native spliceosomes suitable for three-dimensional structural analysis. RNA 8 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallena, M. J., K. J. Chalmers, S. Llamazares, A. I. Lamond and J. Valcarcel, 2002. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell 109 285–296. [DOI] [PubMed] [Google Scholar]
- Makarov, E. M., O. V. Markarova, H. Urlaub, M. Gentzel, C. L. Will et al., 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298 2205–2208. [DOI] [PubMed] [Google Scholar]
- Nagengast, A. A., S. M. Stitzinger, C. H. Tseng, S. M. Mount and H. K. Salz, 2003. Sex-lethal splicing autoregulation in vivo: interactions between SEX-LETHAL, the U1 snRNP and U2AF underlie male exon skipping. Development 130 463–471. [DOI] [PubMed] [Google Scholar]
- Ortega, A., M. Niksic, A. Bachi, M. Wilm, L. Sanchez et al., 2003. Biochemical function of female-lethal (2)D/Wilms' tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J. Biol. Chem. 278 3040–3047. [DOI] [PubMed] [Google Scholar]
- Penalva, L. O., M. F. Ruiz, A. Ortega, B. Granadino, L. Vicente et al., 2000. The Drosophila fl(2)d gene, required for female-specific splicing of Sxl and tra pre-mRNAs, encodes a novel nuclear protein with a HQ-rich domain. Genetics 155 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polycarpou-Schwarz, M., S. I. Gunderson, S. Kadels-Lewis, B. Seraphin and I. W. Mattaj, 1996. Drosophila SNF/D25 combines the functions of the two snRNP proteins U1A and U2B′ that are encoded separately in human, potato, and yeast. RNA 2 11–23. [PMC free article] [PubMed] [Google Scholar]
- Reed, R., 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12 340–345. [DOI] [PubMed] [Google Scholar]
- Rudner, D. Z., R. Kanaar, K. S. Breger and D. C. Rio, 1996. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc. Natl. Acad. Sci. USA 93 10333–10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner, D. Z., K. S. Breger, R. Kanaar, M. D. Adams and D. C. Rio, 1998. RNA binding activity of heterodimeric splicing factor U2AF: at least one RS domain is required for high-affinity binding. Mol. Cell. Biol. 18 4004–4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salz, H. K., and T. W. Flickinger, 1996. Both loss-of-function and gain-of-function mutations in snf define a role for snRNP proteins in regulating Sex-lethal pre-mRNA splicing in Drosophila development. Genetics 144 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels, M. E., D. Bopp, R. A. Colvin, R. F. Roscigno, M. A. Garcia-Blanco et al., 1994. RNA binding by Sxl proteins in vitro and in vivo. Mol. Cell. Biol. 14 4975–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels, M., G. Deshpande and P. Schedl, 1998. Activities of the Sex-lethal protein in RNA binding and protein:protein interactions. Nucleic Acids Res. 26 2625–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski, B. A., J. M. Belote and M. McKeown, 1989. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell 58 449–459. [DOI] [PubMed] [Google Scholar]
- Stark, H., and R. Luhrmann, 2006. Cryo-electron microscopy of spliceosomal components. Annu. Rev. Biophys. Biomol. Struct. 35 435–457. [DOI] [PubMed] [Google Scholar]
- Sterner, D. A., T. Carlo and S. M. Berget, 1996. Architectural limits on split genes. Proc. Natl. Acad. Sci. USA 93 15081–15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitzinger, S. M., T. R. Conrad, A. M. Zachlin and H. K. Salz, 1999. Functional analysis of SNF, the Drosophila U1A/U2B″ homolog: identification of dispensable and indispensable motifs for both snRNP assembly and function in vivo. RNA 5 1440–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, L., J. S. Chang, A. Costa and P. Schedl, 2001. An autoregulatory feedback loop directs the localized expression of the Drosophila CPEB protein Orb in the developing oocyte. Development 128 1159–1169. [DOI] [PubMed] [Google Scholar]
- Valcarcel, J., R. Singh, P. D. Zamore and M. R. Green, 1993. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362 171–175. [DOI] [PubMed] [Google Scholar]
- Yanowitz, J. L., G. Deshpande, G. Calhoun and P. D. Schedl, 1999. An N-terminal truncation uncouples the sex-transforming and dosage compensation functions of sex-lethal. Mol. Cell. Biol. 19 3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., D. R. Dowd, A. Staal, C. Gu, J. B. Lian et al., 2003. Nuclear coactivator-62 kDa/Ski-interacting protein is a nuclear matrix-associated coactivator that may couple vitamin D receptor-mediated transcription and RNA splicing. J. Biol. Chem. 278 35325–35336. [DOI] [PubMed] [Google Scholar]
- Zhou, Z., L. J. Licklider, S. P. Gygi and R. Reed, 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419 182–185. [DOI] [PubMed] [Google Scholar]