Abstract
To identify cell wall biosynthetic genes in filamentous fungi and thus potential targets for the discovery of new antifungals, we developed a novel screening method for cell wall mutants. It is based on our earlier observation that the Aspergillus niger agsA gene, which encodes a putative α-glucan synthase, is strongly induced in response to cell wall stress. By placing the agsA promoter region in front of a selectable marker, the acetamidase (amdS) gene of A. nidulans, we reasoned that cell wall mutants with a constitutively active cell wall stress response pathway could be identified by selecting mutants for growth on acetamide as the sole nitrogen source. For the genetic screen, a strain was constructed that contained two reporter genes controlled by the same promoter: the metabolic reporter gene PagsA-amdS and PagsA-H2B-GFP, which encodes a GFP-tagged nuclear protein. The primary screen yielded 161 mutants that were subjected to various cell wall-related secondary screens. Four calcofluor white-hypersensitive, osmotic-remediable thermosensitive mutants were selected for complementation analysis. Three mutants were complemented by the same gene, which encoded a protein with high sequence identity with eukaryotic UDP-galactopyranose mutases (UgmA). Our results indicate that galactofuranose formation is important for fungal cell wall biosynthesis and represents an attractive target for the development of antifungals.
THE efficiency of penicillin as an antibiotic that inhibits cell wall biosynthesis in Gram-positive bacteria, has inspired fungal researchers to search for drugs that block cell wall biosynthesis in fungi. Like the bacterial wall, the cell wall of fungi is essential, and by interfering with its synthesis or assembly, the cells will lyse and die. The cell wall of Aspergillus niger is similar to the cell wall of A. fumigatus and contains several classes of polysaccharides, including β-glucans, chitin, α-glucans, galactomannan, and cell wall mannoproteins (Bernard and Latge 2001; Klis et al. 2007). Both the proper synthesis and the crosslinking of the components to each other are essential to form a sturdy cell wall. The composition and architecture of the cell wall are highly dynamic in response to both internal signals and external conditions (Lesage and Bussey 2006; Klis et al. 2006). Cell wall remodeling in response to cell wall stress is of great importance to yeasts and filamentous fungi. Inability of the cell to respond and adapt to cell wall threatening conditions might result in cell lysis. Thus, compounds that interfere with the synthesis or the crosslinking of cell wall polymers are potentially interesting as antifungal agents.
Exposure of fungi to sublethal concentrations of cell wall-targeting antifungals, triggers the cell wall integrity (CWI) signaling pathway in both yeasts and filamentous fungi (Levin 2005; Gerik et al. 2005; Damveld et al. 2005b). Activation of the pathway results in the induced expression of several genes involved in cell wall synthesis and remodeling. Both the composition and the architecture of the cell wall are changed in response to cell wall stress and the response is suggested to be evolved as a survival strategy (Levin 2005; Lesage and Bussey 2006). The CWI pathway is best studied in the yeast Saccharomyces cerevisiae and consists of a signal transduction network that is able to sense cell wall weakening (see for review Levin 2005). This weakening of the cell wall activates the PKC-MAPK signaling cascade and results in the activation of the Rlm1p transcription factor, which regulates the transcription of at least 25 genes involved in cell wall biogenesis (Jung and Levin 1999). The involvement of the Rlm1-like transcription factor in the cell wall stress response pathway in A. niger suggests that the cell wall integrity pathway is conserved in yeasts and filamentous fungi (Damveld et al. 2005b).
Activation of the CWI-signaling pathway can be achieved either by compounds that interfere with cell wall biosynthesis or assembly (De Nobel et al. 2000; Garcia et al. 2004) or by using mutants with a defective cell wall, resulting in constitutive activation of this pathway (Lagorce et al. 2003). In S. cerevisiae, genomewide analyses have indicated that the cell wall remodeling response consists of a number of alterations in the cell wall:
Higher expression of certain GPI-dependent cell wall mannoproteins: These mannoproteins are thought to have a structural role in the cell wall and by their higher abundance, increase the strength of the cell (Terashima et al. 2000). Alternatively, they might protect the underlying glucan/chitin layer from being attacked by glucanases and/or chitinases since these proteins are localized mainly on the outside of the cell wall surface.
Increased chitin synthesis has been shown to be an important compensatory response to cell wall stress. Both in S. cerevisiae and in filamentous fungi, increased chitin levels have been reported after the addition of cell wall disturbing compounds and in cell wall mutants (Ram et al. 1994; Popolo et al. 1997; Dallies et al. 1998; Osmond et al. 1999; Lagorce et al. 2002; Ram et al. 2004).
Activation of genes encoding α-1,3-glucan synthases is a third response of fungi in response to cell wall stress. This response does not occur in S. cerevisiae and Candida albicans as these yeasts lack the genes encoding the α-1,3-glucan synthases. A. niger contains a family of five ags genes. Two of them, agsA and to a lesser extent agsE, are induced in response to calcofluor white (CFW)-induced cell wall stress (Damveld et al. 2005a). Induced expression of agsA is dependent on the RlmA transcription factor, a homolog of the Rlm1p transcription factor in S. cerevisiae (Damveld et al. 2005b).
In this study, we have used the agsA promoter region (PagsA) to construct reporters that allow the identification of mutants with a constitutively active cell wall stress response pathway. Among these mutants, we expected to find genes that encode enzymes involved in cell wall biosynthesis. On the basis of its combination of cell wall-related phenotypes (CFW hypersensitivity, osmo-remediable temperature sensitivity), four of the isolated mutants were selected for further analysis. Three of the selected mutants were complemented by the same gene, which showed high sequence identity toward eukaryotic UDP-galactopyranose mutases (UgmA), confirming that our new screening method can lead to the discovery of new antifungal targets related to cell wall biosynthesis in fungi.
MATERIALS AND METHODS
Strains, transformations, growth conditions, and molecular techniques:
The Aspergillus strains used in this study are listed in Table 1. Strains were grown on minimal medium (MM) (Bennett and Lasure 1991) containing 1% (w v−1) glucose and 0.1% (w v−1) casamino acids or on complete medium (CM), containing 0.5% (w v−1) yeast extract in addition to MM. When required, plates were supplemented with uridine (10 mm) or hygromycin (100 μg ml−1). MM agar plates containing acetamide as a sole nitrogen source were made as described by Kelly and Hynes (1985). Transformation of A. niger was performed as described by Punt et al. (1987), using 40 mg lysing enzymes (L-1412, Sigma, St. Louis) per gram wet weight of mycelium. For transformations using the hygromycin selection marker, pAN7-1 (accession no. Z32698) was used. Targeted integration of constructs at the pyrG locus using the pyrG* allele was done according to van Gorcom and van den Hondel (1988). Escherichia coli strain DH5α was transformed by electroporation for the propagation and amplification of cosmids. XL1-Blue was transformed using the heat-shock protocol as described by Inoue et al. (1990) and used for the amplification of plasmids. Fungal chromosomal DNA was isolated as described by Kolar et al. (1988). [α-32P]dCTP-labeled probes were synthesized using the Rediprime II DNA labeling system (Amersham Pharmacia Biotech, Piscataway, NJ) according to the instructions of the manufacturer. All molecular techniques were carried out as described by Sambrook et al. (1989). Sequencing was performed by ServiceXS (Leiden, The Netherlands).
TABLE 1.
Strains used in this study
| Strain | Description | Reference |
|---|---|---|
| N402 | cspA1 derivative of ATCC9029 | Bos et al. (1988) |
| AB4.1 | pyrG− derivative of N402 | Van Hartingsveldt et al. (1987) |
| MA70.15 | kusA∷amdS in AB4.1 pyrG− | Meyer et al. (2007a) |
| MA86.1 | kusA∷amdS in N402 | A. F. J. Ram (unpublished strain) |
| AF09.97 | ugmA∷pyrG in AB4.1 | This study |
| MA87.6 | ugmA∷pyrG in MA70.15 | This study |
| MA88.7 | ugmB∷hygB in MA86.1 | This study |
| MA89.1 | ugmA∷pyrG, ugmB∷hygB in MA70.15 | This study |
| RD6.47 | pPagsA-amdS-TamdS-pyrG* and pPagsA-H2B-GFP-TtrpC/pAN7.1 | This study |
| RD6.13 | pPagsA-amdS-TamdS-pyrG* and pPagsA-H2B-GFP-TtrpC/pAN7.1 | This study |
| RD15.4 | pPagsA-H2B-GFP-TtrpC-pyrG* and pPagsA-amdS-TamdS/pAN7.1 | This study |
| RD15.8 | pPagsA-H2B-GFP-TtrpC-pyrG* and pPagsA-amdS-TamdS/pAN7.1 | This study |
Construction of recombinant plasmids:
For the construction of plasmid PagsA-amdS-TamdS, a 2010-bp SalI–EcoRI fragment containing the agsA promoter region was isolated from pRD12 (Damveld et al. 2005a) and ligated into SalI–EcoRI digested pBluescipt II SK (Stratagene, La Jolla, CA). The resulting plasmid was linearized with EcoRI and XbaI yielding the first fragment for a three-way ligation. The second fragment, a 587-bp EcoRI–BglI fragment containing 30 bp of the agsA promoter and ∼0.56 kb of the 5′ sequence of the amdS gene, was created by fusing PCR, with primers AmdS-agsAP1 and AmdS-agsAP2 and plasmid p3SR2 (Corrick et al. 1987) as a template. The resulting PCR product was cloned in pGEM-T Easy (Promega, Madison, WI) and verified by sequence analysis. The 587-bp fragment was isolated after digestion with EcoRI and BglI. The third fragment was obtained by digestion of p3SR2 with BglI and XbaI. The 1544-bp fragment, containing the 3′ part of the amdS gene and the amdS terminator sequence was isolated and ligated with the other two fragments to give vector PagsA-amdS-TamdS. After ligation, this vector was linearized with XbaI to introduce the pyrG* gene, isolated as a 3.8-kb XbaI fragment from pAN52-7pyrG* (R. A. Damveld, unpublished vector) to give PagsA-amdS-TamdS-pyrG*.
The plasmid PagsA-H2B-GFP-TtrpC was constructed by a three-way ligation. First, a 0.6-kb EcoRI–NcoI fragment containing the 30-bp promoter fragment of agsA fused to H2B was generated by PCR with primers AgsAH2BP1 and AgsAH2BP2, using pH 2BG (Maruyama et al. 2001) as a template. The PCR product was cloned in pGEM-T Easy and verified by sequencing. The second fragment containing GFP-TtrpC and the pUC18 backbone sequence was isolated as an NcoI–NotI fragment from PgpdA-H2B-GFP-TtrpC (M. Arentshorst and A. F. J. Ram, unpublished vector). The third fragment containing a ∼2-kb fragment of the agsA promoter sequence was obtained after ligation of a ∼2-kb SalI–EcoRI fragment from pRD12 into a SalI–EcoRI opened pUC21 and subsequent reisolation after NotI–EcoRI digestion. The three fragments were ligated to give PagsA-H2B-GFP-TtrpC. The unique XbaI site was used to introduce the pyrG* gene (van Gorcom and van den Hondel 1988), isolated as a 3.8-kb XbaI fragment from pAN52-7pyrG* to give PagsA-H2B-GFP-TtrpC-pyrG*.
To construct the plasmid for deleting the ugmA and ugmB genes, 5′ and 3′ regions flanking the genes were amplified by PCR, using primers listed in supplemental Table 1 at http://www.genetics.org/supplemental/. 8660P7 and 8660P8 were used to amplify a 1.2-kb fragment containing the 5′ flank of ugmA. This fragment was digested with XbaI and NotI and cloned into XbaI- and NotI-digested pBluescript-KS to give p5-8660. Primers 8660P9 and 8660P10 were used to amplify the 1.0-kb 3′ flank of the ugmA locus. The fragment was digested with XbaI and EcoRI and cloned into XbaI- and EcoRI-digested pBluescript-KS fragment to give p3-8660. The A. oryzae pyrG gene was isolated as a 3.4-kb XbaI fragment from pAO4-13 (de Ruiter-Jacobs et al. 1989). p5-8660 was linearized with XbaI and EcoRI and the AopyrG XbaI fragment and XbaI–EcoRI fragment from p3-8660 were ligated to give pΔ8660.
2380P1 and 2380P2 were used to amplify a 1.1-kb fragment containing the 5′ flank of ugmB. This fragment was digested with KpnI and HindIII (internal HindIII site) and cloned into KpnI- and HindIII-digested pBluescript-KS to give p5-2380. Primers 2380P5 en 2380P6 were used to amplify the 1.2-kb 3′ flank of the ugmB locus. The fragment was digested with XhoI and NotI and cloned together with the XhoI–HindII 3.0-kb fragment from pAN7.1 (containing the hygromycin cassette) into HindIII/XhoI-digested p5-2380 to give pΔ2380. Deletion plasmids were linearized with NotI and transformed. Transformants were purified two times and selected transformants were subjected to Southern blot analysis.
The DNA sequence of the ugmA gene in the parental strain RD6.13 and the mutant strains RD6.13#44 RD15.4#17 and RD6.13#50 was determined by sequencing three independent PCR products for each strain. The PCR products obtained with primers 8660P2 and 8660P3, using genomic DNA of various strains as template DNA, were cloned into pGEMT-easy and sequenced using appropriate primers (supplemental Table 1 at http://www.genetics.org/supplemental/). The pGEMT-easy plasmid containing the wild type ugmA gene (pUgmA) was used for complementation of the ΔugmA strain.
Mutagenesis and the primary mutant screen:
The strains used for mutagenesis are listed in Table 1 and were constructed by transforming the AB4.1 (pyrG−) strain with either PagsA-amdS-TamdS-pyrG* or PagsA-H2B-GFP-TtrpC-pyrG*. Transformants with a single copy of one of the constructs integrated on the pyrG locus based on Southern analysis (data not shown) were selected, and cotransformed with pAN7.1 (hygromycin cassette) and PagsA-H2B-GFP-TtrpC to give RD6.13 and RD6.47 or transformed with pAN7.1 and PagsA-amdS-TamdS to give RD15.4 and RD15.8. For the UV mutagenesis, freshly harvested spores were diluted to 1 × 107 spores ml−1 and 15-ml spore solutions were mutagenized in a Bio-Rad cross linker (maximum energy output at λ = 254 nm, UV dose 60 J sec−1 m2) for 0–100 sec at 10-sec intervals. Survival rates at the different time points were determined and the spore suspensions with a ∼66% survival rate were used for the primary screen. For each of the four strains, 60 MM plates with acetamide as the sole nitrogen source were inoculated with ∼1 × 104 conidia and incubated at 30°. After 5 days, a single fast growing colony from each plate was transferred to CM plates and purified two times, yielding 240 primary independently obtained mutants.
Secondary screens:
Growth on acetamide: The purified mutants were retested for their ability to grow on acetamide plates at 30°. Equal amounts of conidia (∼500) were spotted on MM plates, with acetamide as the sole nitrogen source and images were taken after 3 days.
Nuclear GFP levels: For microscopic images, conidia were grown on cover slips in MM with casamino acids at 30° for 18 hr. The cover slips with adherent conidia were placed on microscope slides and microscopic GFP images were taken on an Axioplan 2 (Zeiss) equipped with a DKC-5000 (Sony) digital photo camera using a fixed exposure time of 1 sec. The fluorescence of the mutants was compared to their nonmutagenized parental strains. Images were analyzed using Qwin Pro (LEICA, v2.2). In brief, the green channels of the images were analyzed by selecting all green pixels with an intensity value >130. The average GFP values (Mean Grn) and the maximum GFP values (Max Grn) were determined for these selections and compared to the values of nonmutagenized parental strains. Mutants in which the average or maximum GFP values were higher when compared to nonmutagenized strains were scored as mutants with increased GFP expression from the agsA promoter.
Temperature sensitivity: Mutant strains were grown at both 30° and 42° on MM plates.
Osmotic remediability: The effect of the addition of the osmotic stabilizer (1.2 m sorbitol), was examined by growing strains on MM with or without 1.2 m sorbitol at 30° or 42°.
Sensitivity toward SDS and CFW: MM plates containing 50 μg ml−1 SDS, or CFW (as indicated) were inoculated with the mutant strains and grown at 30° for 3 days.
Complementation of the cell wall mutants:
Mutants selected for complementation analysis were made pyrG− by selecting 5-fluoroorotic acid (5-FOA) resistant mutants as described (Gouka et al. 1995). PyrG− mutants were transformed with a genomic cosmid library in an AMA1 containing self-replicating vector using the pyrG selection marker (P. Punt, unpublished data). Complementation of the mutant phenotype was analyzed by screening for strains that had obtained the parental SDS sensitivity at 42°. Cosmids from the putative complemented A. niger strains were isolated using the protocol for isolation of genomic DNA (Kolar et al. 1988). The cosmids were transformed to E. coli (DH5α) via electroporation and grown on LB plates with ampicillin. Subsequent cosmid isolations from 40 ml of overnight cultures were performed using the small scale DNA isolation method as described by Sambrook et al. (1989). Primers cosT7 and cosUL (supplemental Table 1 at http://www.genetics.org/supplemental/) were used for sequencing the ends of the inserts.
RESULTS
Concept and setup of the cell wall mutant screen:
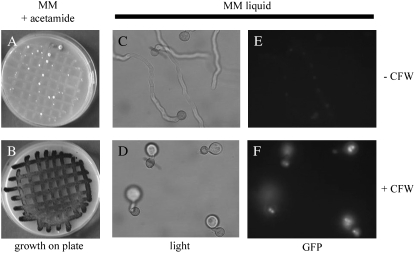
To identify genes involved in the synthesis of the fungal cell wall, we designed a positive screening procedure for the isolation of mutants disturbed in cell wall synthesis. For the genetic screen, reporter strains were constructed containing both the PagsA-amdS reporter as well as the PagsA-H2B-GFP reporter. When the parental strain was inoculated on MM agar plates containing acetamide as sole nitrogen source, only poor growth was observed, indicating that the basal activity of PagsA is not sufficient to allow efficient growth on acetamide (Figure 1A). As shown in Figure 1B, the addition of 0.5 mg ml−1 CFW to the plates resulted in growth and even sporulation of the reporter strain. The response of the second reporter to CFW-induced cell wall stress was also examined. As expected, the morphology of the germlings was normal and the fluorescence signal from the nuclear targeted H2B-GFP fusion protein was low in the control (Figure 1, C and E). The addition of CFW (0.2 mg ml−1) to the liquid minimal medium resulted in the formation of swollen hyphal tips (Figure 1D) and induction of agsA expression since the nuclear targeted GFP was clearly visible (Figure 1F). Using the PagsA-H2B-GFP reporter strain, we were able to show that the induction of agsA was not limited to CFW, but that the induction was also achieved by adding other cell wall disturbing compounds such as caspofungin and tunicamycin. In addition, the induction is specific for cell wall stress, since other forms of stress (high osmolarity stress, oxidative stress, or temperature stress) did not result in activation of the agsA promoter (Meyer et al. 2007b). Taken together, these results indicate that the reporter strain can identify mutants with increased levels of agsA expression, which probably result from a mutation that affects the integrity of the cell wall.
Figure 1.—
Phenotypic analysis of the cell wall stress reporter strain. Strain RD6.47, containing both the nuclear targeted GFP (H2B-GFP) and acetamidase (amdS) reporters under control of the agsA promoter, was grown for 3 days at 30° on plates containing acetamide as the sole nitrogen source under normal growth conditions (A) and with the addition of 0.5 mg ml−1 CFW giving an induction of the reporter (B). The reporter strain was also allowed to germinate on submerged cover slides. After 5 hr of growth at 37°, germlings were treated with 200 μg ml−1 CFW (D and F) or an equal amount on MilliQ water (C and E) and grown for another hour before microscopic observation. C and D show bright field; E and F, fluorescence microscopy.
Isolation and phenotypic characterization of the putative cell wall mutants:
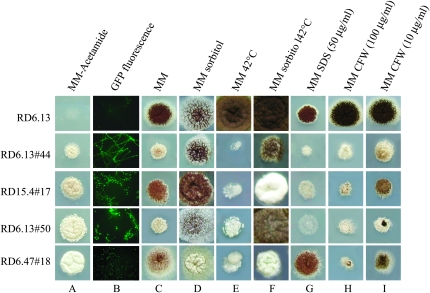
After mutagenesis, 240 mutants were purified and retested for the ability to grow efficiently on acetamide. From those mutants, 161 strains still grew well on acetamide. The 161 mutants were analyzed for the presence of increased nuclear GFP levels using fluorescence microscopy. Three mutants failed to show increased nuclear GFP levels compared to the parental strain and were not studied further. To investigate whether the remaining mutants had a defective cell wall, the mutants were tested for phenotypes that are indicative of a defective cell wall such as increased sensitivity toward elevated temperatures, CFW, and SDS (De Groot et al. 2001; Ram and Klis 2006). Of the 158 mutants, 27 mutants displayed a temperature-sensitive growth defect at 42°. Growth of 11 of the temperature-sensitive (ts) mutants was improved by the addition of sorbitol to the medium, indicating an osmotic-remediable, temperature-sensitive phenotype. Although we have also determined both CFW and SDS sensitivity for all 158 mutants (12 mutants showed a CFW-hypersensitive phenotype and 32 mutants an SDS-hypersensitive phenotype with an overlap of 6 mutants displaying hypersensitivity to both CFW and SDS), we have further focused on the temperature-sensitive, osmotic-remediable mutants. Four of these mutants displayed a higher sensitivity toward CFW and 6 were more sensitive toward SDS. The CFW-hypersensitive phenotype correlated with a higher sensitivity toward SDS in 3 of the mutants. We selected the 4 ts-osmotic-remediable, CFW-hypersensitive mutants (named RD6.13#44, RD15.4#17, RD6.13#50, and RD6.47#18) for further complementation analysis. As shown in Figure 2, all 4 mutants grow well on acetamide plates (Figure 2A), show induced nuclear GFP levels (Figure 2B), and are temperature-sensitive (compare Figure 2, C and E), a phenotype that could be (partially) remediated by the addition of an osmostabilizer (compare Figure 2, E and F). All 4 mutants show increased CFW sensitivity (compare Figure 2, C and H and I). RD6.13#44, RD15.4#17, and RD6.13#50 showed an increased sensitivity toward SDS (compare Figure 2, C and G). RD6.47#18 showed no SDS-hypersensitive phenotype.
Figure 2.—
Phenotypic analysis of the parental strain (RD6.13) and four selected mutants. Equal amounts of spores (5 × 105) were spotted on different types of minimal media (MM) under different conditions to determine the phenotype of the mutants. All growth experiments were performed at 30° and lasted for 3 days unless indicated differently.
Complementation analysis:
The available genomic cosmid library contained the pyrG gene as a selection marker and pyrG− derivatives of the mutants were obtained as described in materials and methods. The pyrG− strains from each mutant were transformed with the genomic cosmid library and grown at 30°, yielding between 400 and 7500 transformants per strain. Spores originating from one transformation plate (∼200 individual transformants) were pooled and subsequently analyzed for complementation of both the temperature-sensitive phenotype and the SDS-sensitive phenotype. Cosmids from putative complemented transformants were isolated and retransformed into their corresponding mutant strain and analyzed for complementation of the temperature-sensitive phenotype. The combined results from the restriction analysis and transformation experiments showed that mutants RD6.13#44, RD15.4#17, and RD6.13#50 were complemented by cosmids with overlapping inserts. Indeed, each cosmid that was isolated for each mutant also complemented the other two strains, suggesting that these three CFW-hypersensitive mutants might be affected in the same gene. The restriction pattern of the cosmid complementing RD6.47#18 was different from the other cosmids and this cosmid did not complement the other mutants, indicating that this mutant is altered in a different gene. End sequencing the cosmids complementing RD6.13#44, RD15.4#17, and RD6.13#50 and comparison of the cosmids to the A. niger genome sequence and to each other showed that the complementing cosmids shared a 35-kb region containing at least 9 predicted ORFs. Further subcloning and complementation analysis pointed to two candidate ORFs (An02g08650 and An02g08660) for complementation. These ORFs were PCR amplified from genomic DNA of the wild-type strain (N402) and transformed to RD6.13#44, RD15.4#17, and RD6.13#50. Only the PCR fragments with the An02g08660 ORF complemented all the phenotypes (temperature-sensitive growth defect at 42°, the SDS and CFW hypersensitivity) of the mutants. Thus, from the complementation analysis we conclude that RD6.13#44, RD15.4#17, and RD6.13#50 are each complemented by An02g08660. The cosmid complementing the RD6.47#18 mutant was also end sequenced. Comparison of the ends with the A. niger genome sequence indicated that the ends were 1.6 Mbp separated from each other, which did not match with the estimated insert size of ∼40 kb deduced from the digestion pattern. Complete characterization of the gene complementing this mutant is ongoing. In this article, we have further focused on An02g08660.
An02g08660 encodes an UDP-galactopyranose mutase:
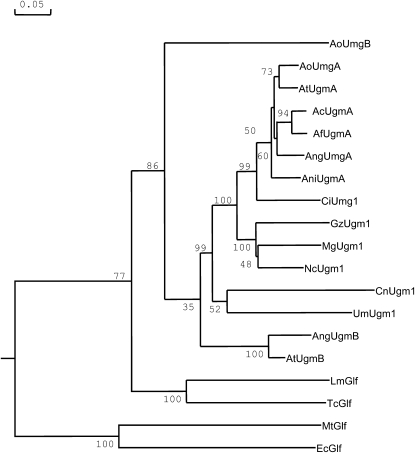
Sequence comparison of the protein sequence deduced from the An02g08660 gene revealed that the protein displayed strong sequence similarity to eukaryotic UDP-galactopyranose mutases and we will refer to this gene as ugmA. The gene encoding this protein has recently been identified and partially characterized from A. fumigatus, Cryptococcus neoformans, and Leishmania major (Bakker et al. 2005; Beverley et al. 2005). The enzyme catalyzes the conversion of UDP-galactopyranose into UDP-galactofuranose. UPD-galactofuranose is used as a sugar donor used by galactofuranose transferases for the synthesis of macromolecules containing galactofuranose. The A. niger ugmA gene contains five predicted introns that all contain consensus boundary sites for splicing (GATNGN … C/TAG). The predicted protein in A. niger is 510 amino acids in length and is 93% identical over the entire protein sequence compared to the A. fumigatus protein. BLAST searches against other fungal genomes indicate that UDP-galactopyranose mutases are found in many ascomycetes including other Aspergillus species, Neurospora crassa, Magnaporthe grisae, Gibberella zeae, Coccidioides immitis, as well as in the basidiomycetes C. neoformans and Ustilago maydis (Figure 3).
Figure 3.—
Bootstrapped phylogenetic tree of UDP-galactopyranose mutase enzymes from eukaryotic and bacterial origin. Accession numbers of the proteins used in the tree are listed in supplemental Table 2 at http://www.genetics.org/supplemental/. Bootstrap values are indicated on the node of each branch. The tree was created with DNAMAN 4.0 using gap and extension penalties of 10 and 0.5, respectively. The scale bar indicates 5% amino acid sequence difference.
As noted previously (Bakker et al. 2005), the eukaryotic enzymes show much less sequence similarity to the prokaryotic enzymes (Figure 3). Surprisingly, the A. niger genome contains a second gene encoding a putative galactopyranose mutase (An16g02380). The 492-amino-acid-long protein, designated UgmB, is 67% identical to the A. niger UgmA protein, but the level of identity between the A. niger UgmA and UgmB proteins is less than the identity between A. niger UgmA and the Ugm proteins from other Aspergilli (Figure 3 and data not shown). The only other Aspergillus species which contains two full-length Ugm proteins is A. terreus. The two Ugm proteins from A. terreus cluster in separate clusters in the tree and cluster either to the UgmA group or the UgmB protein of A. niger (Figure 3). The second ugm-encoding gene in A. oryzae (AoUgmB) does seem to encode a truncated protein of only 168 amino acids.
To confirm that ugmA was indeed the complementing gene and not a suppressor, the ugmA alleles of the mutants were sequenced. The ugmA locus in the RD6.13#44 strain contains a point mutation (T to C) at position 1756 (the A of the start codon ATG = 1), resulting in an amino acid change at position 462 (Phe to Ser). The ugmA locus in the RD6.13#50 strain contains two mutations at positions 725 and 727, (both T to C mutations), resulting in two amino acid changes at position 157 and 158 (Leu to Pro and Phe to Leu, respectively). The phenylalanine residue at position 462 is conserved in all fungi analyzed except in U. maydis, in which a serine residue is present at the aligned amino acid position. In all fungal UgmA proteins, the phenylalanine residue (position 158) is conserved. The leucine (position 157) can be replaced by an isoleucine. Surprisingly, no mutation was found in the ugmA gene of the RD15.4#17 mutant. The mutations found in the ugmA genes of RD6.13#44 and RD6.13#50 further confirm that the ugmA gene was not isolated as a suppressor. The situation concerning RD15.4#17 is not clear at this moment. The mutation might be located outside of the region that has been sequenced, although ∼1.2-kb promotor and 0–3-kb terminator sequence of the ugmA locus was determined. For the RD15.4#17 mutant we cannot exclude the possibility that the ugmA gene acts as a suppressor.
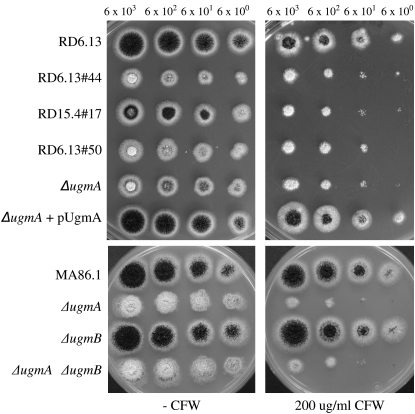
To examine whether the complete loss of function of the ugmA gene resulted in a more severe phenotype compared to the point mutants, an ugmA deletion strain (AF09#97) was constructed and compared to the growth phenotypes of the mutants (Figure 4A). Strain RD15.4#17 showed a slightly better growth and conidiation phenotype compared to the other three mutants (Figures 2 and 4A). The phenotype of RD6.13#44 and RD6.13#50 was identical to the ΔugmA strain (Figure 4A) indicating that the mutations in the ugmA gene result in an inactive protein. The CFW-hypersensitive and temperature-sensitive phenotypes of the ΔugmA strain were complemented by retransformation of the plasmid containing the ugmA gene (pUgmA) (Figure 4A and data not shown).
Figure 4.—
Susceptibility of the ugm mutants toward CFW. (Top) RD6.13#44, RD15.4#17, and RD6.13#50 and the ugmA knockout strain (ΔugmA) were assayed for CFW susceptibility compared to the parental strain (RD6.13) and the complemented knockout strain (ΔugmA+ pUgmA). (Bottom) CFW susceptibility of MA86.1 (parental strain), ΔugmA, ΔugmB, and ΔugmA/ΔugmB mutants. Tenfold serial dilutions of spores were spotted on complete medium plates containing 200 μg ml−1 CFW. Pictures were taken after 2 days of growth at 30°.
Finally, a possible redundant function of the A. niger ugmA and ugmB genes was examined by constructing a ugmB deletion strain (MA88.7) and a ugmA/ugmB double deletion strain (MA89.1). Proper deletion of the ugmA and ugmB genes in these mutants was confirmed by Southern blot analysis. Deletion of the ugmB gene did not result in any growth defect nor did it result in an altered sensitivity toward CFW (Figure 4B). The simultaneous deletion of ugmA and ugmB resulted in a growth phenotype and a CFW-sensitive phenotype that was indiscernible from the single ugmA deletion strain (Figure 4B). In addition, we examined ugmB expression during the exponential growth phase in both the wild type and the ugmA deletion strain using Northern blot analysis, but we were unable to detect ugmB expression in either of the two strains. These results indicate that the ugmB gene is not expressed, not even in the absence of ugmA, and that the two genes are not functionally redundant under the conditions tested.
DISCUSSION
To identify proteins involved in fungal cell wall assembly, we have designed a novel screening method for the identification of cell wall mutants. The screen is based on the observation that the agsA gene, which encodes a putative α-1,3-glucan synthase, is strongly induced in response to different forms of cell wall stress (Damveld et al. 2005a; Meyer et al. 2007b). Important factors for such a successful genetic screen are:
The induction of the reporter gene via the selected promoter should be specific for cell wall stress, because if additional forms of stress also induce the expression of the reporter, other mutants will be detected besides cell wall mutants.
The basal level of expression of the reporter gene driven by the promoter must be low under noninducing conditions and strongly induced upon stress.
The level of expression of the selection marker should result in a quantitative effect on growth. The AmdS marker is highly suitable for this purpose as increasing levels of the gene in A. niger result in an increasing ability to grow on acetamide as the sole carbon source (Kelly and Hynes 1985; Verdoes et al. 1993).
With respect to this latter point, the use of the pyrG gene was examined as a selection marker for the mutant screen instead of the amdS gene, but the basal activity of the agsA promoter already supplies the cell with a sufficient level of pyrG protein, since the transformant (pyrG−, PagsA-pyrG) could grow on plates without uridine (data not shown). As the agsA promoter fulfills the first two criteria (low basal level of expression and high induction specific for cell wall stress) and the AmdS marker meets the third criterion, the basis for a successful screening was provided.
To exclude cis-acting mutants mutated in the agsA promoter leading to high levels of transcription of the amdS gene, or to exclude mutants with additional copies of the amdS gene in the genome after mutagenesis, a second reporter construct was included in our reporter strain (PagsA-H2B-GFP). Of the 161 mutants with increased growth on acetamide, only three mutants did not show increased GFP levels, indicating that these might be cis mutants. The remaining 158 mutants were analyzed by several secondary screens to classify and group the mutants according to their phenotypes. The screens we have used consist of simple sensitivity assays that are indicative of cell wall defects. Although a significant number of mutants had cell wall-related phenotypes (45/158) in combination with increased agsA expression, the majority of the mutants (113/158) did not display additional phenotypes. However, the secondary screens were not exhaustive and additional secondary screens are currently considered.
To illustrate the potential of the screening method, we have focused on a subset of temperature-sensitive, osmotic-remediable mutants. These mutants were further hypersensitive to CFW and SDS and all three phenotypes are indicative of cell wall mutants (De Groot et al. 2001). Complementation analysis of four mutants showed that three mutants were complemented by the same gene, designated ugmA, encoding a UDP-galactopyranose mutase. Galactofuranose has been identified in the cell wall of A. niger both in the galactomannan fraction (Bardalaye and Nordin 1977; Barreto-Bergter and Travassos 1980) and as part of the N-glycan moiety of an extracellular enzyme, α-galactosidase (Wallis et al. 2001), or as part of O-linked glycans (Wallis et al. 1999). As all three processes (galactomannan biosynthesis, N- and O-linked glycosylation) are expected to be affected by the deletion of the ugmA gene, it is currently not known which of the three processes contributes most to the cell wall-related phenotypes.
A surprising finding was the occurrence of a second gene encoding a putative UDP-galactopyranose mutase, UgmB, in the A. niger genome. Other fungal genomes, except A. terreus, contain only a single copy of a gene encoding a full-length UDP-galactopyranose mutase. The presence of a second ugm gene in both A. terreus and A. niger might further illustrate that both Aspergilli are phylogenetically related as has been noted recently (Pel et al. 2007). The possible function of the A. niger UgmB protein was examined by deleting the ugmB gene. Since no phenotype was observed for the ugmB deletion strain and no additional phenotype was observed for the ugmA/ugmB deletion strain compared to the ugmA deletion strain, we have currently no indications for a role of UgmB in the formation of galactofuranose. The biochemical analysis of UgmB might further reveal its possible function. Expression analysis of the ugmB gene in our microarray data collection, which includes expression profiles in germinating spores, vegetatively grown mycelia using various carbon sources (glucose, maltose, xylose), and during conidiation, shows that the expression of the ugmB gene is below the detection level (absent call) in all arrays. UgmA expression was detected under all conditions (A. F. J. Ram, unpublished data). Interestingly, the ugmA gene was consistently higher expressed (approximately two-fold) on xylose compared to maltose and this might be indicative of a different cell wall composition between maltose- and xylose-grown cells.
Our results show that galactofuranose formation is important for fungal cell wall biosynthesis. Mutations in the UDP-galactopyranose mutase are likely to cause a cell wall integrity defect, which is counteracted by the fungal cell by activation of the cell wall stress response pathway, which includes the induced expression of agsA. Because growth of the ugmA deletion strain is strongly impaired, inhibitors of UDP-galactopyranose mutase or galactofuranose transferases might be effective against fungal pathogens.
Acknowledgments
We thank Stanley Brul, Suus Ooms, and Jaap Visser for sharing ideas and helpful advice. Gert Groot and Noël van Peij (DSM, The Netherlands) are acknowledged for providing early access to the A. niger genome sequence and K. Kitamoto for the pH2BG plasmid. This work was supported by a grant (LPB.5113) from Stichting Technische Wetenschappen (Dutch Foundation for Technical Research).
References
- Bakker, H., B. Kleczka, R. Gerardy-Schahn and F. H. Routier, 2005. Identification and partial characterization of two eukaryotic UDP-galactopyranose mutases. Biol. Chem. 386 657–661. [DOI] [PubMed] [Google Scholar]
- Bardalaye, P. C., and J. H. Nordin, 1977. Chemical structure of the galactomannan from the cell wall of Aspergillus niger. J. Biol. Chem. 252 2584–2591. [PubMed] [Google Scholar]
- Barreto-Bergter, E. M., and L. R. Travassos, 1980. Chemical structure of the D-galacto-D-mannan component from hyphae of Aspergillus niger and other Aspergillus spp. Carb. Res. 86 273–285. [Google Scholar]
- Bennett, J. W., and L. L. Lasure, 1991. More Gene Manipulations in Fungi, pp. 441–447, Academic Press, San Diego.
- Bernard, M., and J. P. Latge, 2001. Aspergillus fumigatus cell wall: composition and biosynthesis. Med. Mycol. 39(Suppl. 1): 9–17. [PubMed] [Google Scholar]
- Beverley, S. M., K. L. Owens, M. Showalter, C. L. Griffith, T. L. Doering et al., 2005. Eukaryotic UDP-galactopyranose mutase (GLF gene) in microbial and metazoal pathogens. Eukaryot. Cell 4 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos, C. J., A. J. Debets, K. Swart, A. Huybers, G. Kobus et al., 1988. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14 437–443. [DOI] [PubMed] [Google Scholar]
- Corrick, C. M., A. P. Twomey and M. J. Hynes, 1987. The nucleotide sequence of the amdS gene of Aspergillus nidulans and the molecular characterization of 5′ mutations. Gene 53 63–71. [DOI] [PubMed] [Google Scholar]
- Dallies, N., J. Francois and V. Paquet, 1998. A new method for quantitative determination of polysaccharides in the yeast cell wall. Application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast 14 1297–1306. [DOI] [PubMed] [Google Scholar]
- Damveld, R. A., P. A. VanKuyk, M. Arentshorst, F. M. Klis, C. A. van den Hondel et al., 2005. a Expression of agsA, one of five 1,3-alpha-D-glucan synthase-encoding genes in Aspergillus niger, is induced in response to cell wall stress. Fungal Genet. Biol. 42 165–177. [DOI] [PubMed] [Google Scholar]
- Damveld, R. A., M. Arentshorst, A. Franken, P. A. VanKuyk, F. M. Klis et al., 2005. b The Aspergillus niger MADS-box transcription factor RlmA is required for cell wall reinforcement in response to cell wall stress. Mol. Microbiol. 58 305–319. [DOI] [PubMed] [Google Scholar]
- de Groot, P. W. J., C. Ruiz, C. R. Vázquez de Aldana, E. Dueòas, V. J. Cid et al., 2001. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp. Funct. Genomics 2 124–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul et al., 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146 2121–2132. [DOI] [PubMed] [Google Scholar]
- de Ruiter-Jacobs, Y. M., M. Broekhuijsen, S. E. Unkles, E. I. Campbell, J. R. Kinghorn et al., 1989. A gene transfer system based on the homologous pyrG gene and efficient expression of bacterial genes in Aspergillus oryzae. Curr. Genet. 16 159–163. [DOI] [PubMed] [Google Scholar]
- Garcia, R., C. Bermejo, C. Grau, R. Perez, J. M. Rodriguez-Peña et al., 2004. The global transcriptional response to transient cell wall damage in Saccharomyces cerevisiae and its regulation by the cell integrity signaling pathway. J. Biol. Chem. 279 15183–15195. [DOI] [PubMed] [Google Scholar]
- Gerik, K. J., M. J. Donlin, C. E. Soto, A. M. Banks, I. R. Banks et al., 2005. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Mol. Microbiol. 58 393–408. [DOI] [PubMed] [Google Scholar]
- Gouka, R. J., J. G. Hessing, H. Stam, W. Musters and C. A. van den Hondel, 1995. A novel strategy for the isolation of defined pyrG mutants and the development of a site-specific integration system for Aspergillus awamori. Curr. Genet. 27 536–540. [DOI] [PubMed] [Google Scholar]
- Inoue, H., H. Nojima and H. Okayama, 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96 23–28. [DOI] [PubMed] [Google Scholar]
- Jung, U. S., and D. E. Levin, 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34 1049–1057. [DOI] [PubMed] [Google Scholar]
- Kelly, J. M., and M. J. Hynes, 1985. Transformation of Aspergillus niger by the amdS gene of Aspergillus nidulans. EMBO J. 4 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis, F. M., A. Boorsma and P. W. J. de Groot, 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast. 23 185–202. [DOI] [PubMed] [Google Scholar]
- Klis, F.M., A. F. J. Ram and P. W. J. de Groot, 2007. A molecular and genomic view of the fungal cell wall, pp 97–120 in The Mycota VIII Biology of the Fungal Cell, edited by R. J. Howard and N. A. R. Gow. Springer-Verlag, Berlin.
- Kolar, M., P. J. Punt, C. A. van den Hondel and H. Schwab, 1988. Transformation of Penicillium chrysogenum using dominant selection markers and expression of an Escherichia coli lacZ fusion gene. Gene 62 127–134. [DOI] [PubMed] [Google Scholar]
- Lagorce, A., V. Berre-Anton, B. Aguilar-Uscanga, H. Martin-Yken, A. Dagkessamanskaia et al., 2002. Involvement of GFA1, which encodes glutamine-fructose-6-phosphate amidotransferase, in the activation of the chitin synthesis pathway in response to cell-wall defects in Saccharomyces cerevisiae. Eur. J. Biochem. 269 1697–1707. [DOI] [PubMed] [Google Scholar]
- Lagorce, A., N. C. Hauser, D. Labourdette, C. Rodriguez, H. Martin-Yken et al., 2003. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 278 20345–20357. [DOI] [PubMed] [Google Scholar]
- Lesage, G, and H. Bussey, 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70 317–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin, D. E., 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69 262–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama, J., H. Nakajima and K. Kitamoto, 2001. Visualization of nuclei in Aspergillus oryzae with EGFP and analysis of the number of nuclei in each conidium by FACS. Biosci. Biotechnol. Biochem. 65 1504–1510. [DOI] [PubMed] [Google Scholar]
- Meyer V., M. Arentshorst, A. El-Ghezal, A. C. Drews, R. Kooistra et al., 2007. a Highly efficient gene targeting in the Aspergillus niger kusA mutant. J. Biotechnol. 10 770–775. [DOI] [PubMed] [Google Scholar]
- Meyer, V., R. A. Damveld, M. Arentshorst, U. Stahl, C. A. van den Hondel et al., 2007. b Survival in the presence of antifungals: genome-wide expression profiling of Aspergillus niger in response to sub-lethal concentrations of caspofungin and fenpropimorph. J. Biol. Chem. 282 32935–32948. [DOI] [PubMed] [Google Scholar]
- Osmond, B. C., C. A. Specht and P. W. Robbins, 1999. Chitin synthase III: synthetic lethal mutants and “stress related” chitin synthesis that bypasses the CSD3/CHS6 localization pathway. Proc. Natl. Acad. Sci. USA 96 11206–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pel, H. J., J. H. de Winde, D. B. Archer, P. S. Dyer, G. Hofmann et al., 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25 221–231. [DOI] [PubMed] [Google Scholar]
- Popolo, L., D. Gilardelli, P. Bonfante and M. Vai, 1997. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1delta mutant of Saccharomyces cerevisiae. J. Bacteriol. 179 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels and C. A. van den Hondel, 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 56 117–124. [DOI] [PubMed] [Google Scholar]
- Ram, A. F. J., and F. M. Klis, 2006. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor White and Congo Red. Nat. Protoc. 1 2253–2256. [DOI] [PubMed] [Google Scholar]
- Ram, A. F. J., A. Wolters, R. Ten Hoopen and F. M. Klis, 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 10 1019–1030. [DOI] [PubMed] [Google Scholar]
- Ram, A. F. J., M. Arentshorst, R. A. Damveld, P. A. vanKuyk, F. M. Klis et al., 2004. The cell wall stress response in Aspergillus niger involves increased expression of the glutamine: fructose-6-phosphate amidotransferase-encoding gene (gfaA) and increased deposition of chitin in the cell wall. Microbiology 150 3315–3326. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E. F. Fritsch and T. Maniatis, 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Plainview NY.
- Terashima, H., N. Yabuki, M. Arisawa, K. Hamada and K. Kitada, 2000. Up-regulation of genes encoding glycosylphosphatidylinositol (GPI)-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 264 64–74. [DOI] [PubMed] [Google Scholar]
- van Gorcom, R. F., and C. A. van den Hondel, 1988. Expression analysis vectors for Aspergillus niger. Nucleic Acids Res. 26 9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hartingsveldt, W., I. E. Mattern, C. M. van Zeijl, P. H. Pouwels and C. A. van den Hondel, 1987. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol. Gen. Genet. 206 71–75. [DOI] [PubMed] [Google Scholar]
- Verdoes, J. C., P. J. Punt, J. M. Schrickx, H. W. van Verseveld, A. H. Stouthamer et al., 1993. Glucoamylase overexpression in Aspergillus niger: molecular genetic analysis of strains containing multiple copies of the glaA gene. Transgenic Res. 2 84–92. [DOI] [PubMed] [Google Scholar]
- Wallis, G. L., R. J. Swift, F. W. Hemming, A. P. Trinci and J. F. Peberdy, 1999. Glucoamylase overexpression and secretion in Aspergillus niger: analysis of glycosylation. Biochim. Biophys. Acta 1472 576–586. [DOI] [PubMed] [Google Scholar]
- Wallis, G. L., R. L. Easton, K. Jolly, F. W. Hemming and J. F. Peberdy, 2001. Galactofuranoic-oligomannose N-linked glycans of alpha-galactosidase A from Aspergillus niger. Eur. J. Biochem. 268 4134–4143. [DOI] [PubMed] [Google Scholar]