Abstract
Aphanomyces euteiches is an oomycete pathogen that causes seedling blight and root rot of legumes, such as alfalfa and pea. The genus Aphanomyces is phylogenically distinct from well-studied oomycetes such as Phytophthora sp., and contains species pathogenic on plants and aquatic animals. To provide the first foray into gene diversity of A. euteiches, two cDNA libraries were constructed using mRNA extracted from mycelium grown in an artificial liquid medium or in contact to plant roots. A unigene set of 7,977 sequences was obtained from 18,864 high-quality expressed sequenced tags (ESTs) and characterized for potential functions. Comparisons with oomycete proteomes revealed major differences between the gene content of A. euteiches and those of Phytophthora species, leading to the identification of biosynthetic pathways absent in Phytophthora, of new putative pathogenicity genes and of expansion of gene families encoding extracellular proteins, notably different classes of proteases. Among the genes specific of A. euteiches are members of a new family of extracellular proteins putatively involved in adhesion, containing up to four protein domains similar to fungal cellulose binding domains. Comparison of A. euteiches sequences with proteomes of fully sequenced eukaryotic pathogens, including fungi, apicomplexa and trypanosomatids, allowed the identification of A. euteiches genes with close orthologs in these microorganisms but absent in other oomycetes sequenced so far, notably transporters and non-ribosomal peptide synthetases, and suggests the presence of a defense mechanism against oxidative stress which was initially characterized in the pathogenic trypanosomatids.
Introduction
The legume root rot disease caused by the oomycete pathogen Aphanomyces euteiches is one major yield reducing factor in legume production including pea and alfalfa throughout the world [1]–[3]. A. euteiches can infect plants at any age. The first symptoms on pea may be discerned on roots which are softened and water-soaked. The pathogen spreads rapidly through the cortical tissue and the fine branches of feeding rootlets are destroyed. Then the epicotyls become dark and eventually collapse. Leaves progressively turn yellow, starting at the bottom of the shoot. In severe cases, the plants collapse and die before forming any pod.
Thick-walled oospores are resting stages and constitute the primary inoculum source in soil [4]. After germination, primary zoospores are released at the apex of sporangia. These bi-flagellate motile elements encyst at the root surface, germinate and infect the host root. After a few days, the pathogen can reproduce sexually in the host and form new oospores. These oospores can remain dormant in soil and organic debris for many years [5]. Effective chemical controls for Aphanomyces root rot of legumes are not available. Crop rotation and bioassay methods to assess the inoculum potential in the soil remain the only effective method to avoid disease. Using recurrent selection-based strategies, pea germplasms with partial resistance or tolerance were obtained [6].
Among Oomycetes, the genus Aphanomyces belongs to the Saprolegniales and comprises species that are destructive on plants, crustaceans and fishes such as A. astaci and A. invadans. Virulent genotypes of A. astaci were introduced in Europe giving rise to disease outbreaks among its crustacean host population, the freshwater crayfish [7]. This parasite is now considered as one of the 100 world's worst invasive species (Global Invasive Species Database, http//www.issg.org/database). For the last 30 years, the epizootic ulcerative syndrome (EUS) a disease caused by A. invadans, has affected wild and farmed fish in Asia and Indo-Pacific regions and has been recently detected in the eastern seaboard of the US [8]. Aphanomyces genus also includes significant plant pathogens such as A. euteiches and A. cochlioides causing the most serious seedling disease of sugar beet in terms of yield loss, persistence of the oospores in soil, and difficulty of control [9], [10]. By contrast to this diversity of hosts, some oomycete genera, such as the Peronosporale Phytophthora is composed of species which are only pathogenic to plants. Such diversity within the oomycetes could reflect different evolutionary histories and possibly different mechanisms of infection between Saprolegniales and Peronosporales [11].
Until recently, oomycetes genomics focussed only on Phytophthora spp. with the completion of genome sequencing projects and large scale identification of expressed sequences [12]–[17]. These data, combined to functional studies, led to the identification of new factors of pathogenicity such as the large family of RxLR effectors which are thought to manipulate the plant cell upon their translocation into the host cytoplasm [18]. However little is known about the biology and pathology of other oomycetes, notably those belonging to the Saprolegniales. To fill this gap, we have undertaken a large scale sequencing of A. euteiches cDNAs. This microorganism is of interest since it is able to interact with the model legume Medicago truncatula, offering the opportunity to engage genetic approaches for deciphering this interaction [9]. Here we report on the characterization of an EST collection comprising 18,684 EST assembled into 7,977 unique sequences from two different cDNA libraries. A systematic comparative strategy was engaged to identify specific A. euteiches gene sets absent in Phytophthora species. This work greatly expands our knowledge of this group of destructive pathogens and points to the large evolutionary divergence within the oomycete lineage.
Materials and Methods
Aphanomyces euteiches growth conditions
Aphanomyces euteiches (ATCC201684) was maintained on Corn Meal Agar medium (8.5 g/l) at 28°C in the dark. Synthetic medium [19] supplemented with pectin from apple (10 g/l) was used to perform growth assay on pectin.
cDNA libraries construction
Two unidirectional cDNA libraries from A. euteiches were prepared from mycelium grown on medium containing yeast extract/glucose (library MYC) and from mycelium in interaction with Medicago truncatula roots (library INT) [20]. A total of 18,684 expressed sequence tags (9,224 from library MYC and 9,460 from library INT) were submitted to the EBI databank for accession number assignation and were assembled into 7,977 unigenes [20].
Sequence analysis and database searches
Sequences describe here could be downloaded from the Aphanomyces database AphanoDB ([19], http://www.polebio.scsv.ups-tlse.fr/aphano/) and cDNA clones are available at the CNRGV (http://cnrgv.toulouse.inra.fr/ENG/). Data searches and analyses have been conducted on AphanoDB using local tools.
For detection of RxLR effectors, all open reading frames (ORFs) encoding at least 100 amino acids (delimited by two stops) were extracted. This led to 136,617 putative ORFs. Signal peptides were predicted with SignalP version 3.0 [21], with a hidden Markov model probability cutoff of 0.8 and 10,131 candidate secreted proteins were obtained.
Three independent strategies were used to look for the RxLR motif over all these sequences:
- Search for the regular expression RxLR-x(1,40)-[ED][ED][KR] (with the fuzzpro program from EMBOSS suite [22]) in the first 100 residues downstream of the signal peptide cleavage site.
This led to the identification of 5 sequences, among which only two were retained as being in the same frame as the BLASTX matches.
- Search with HMM profiles performed with the hmmsearch program from the HMMer 2.2 package [23] with two alternative profiles, RdEER [24] and cropped [18]. From the 2 sequences obtained with this method none was in the frame in agreement with BLASTX matches.
- Search for the regular expression RxLx[EDQ]-x(1,40)-[ED][ED][KR] (with the fuzzpro program from EMBOSS suite) in the first 100 residues downstream of the signal peptide cleavage site. The RxLx[EDQ] motif was the original signal required for secretion identified in Plasmodium falciparum [25]: 2 out of the 3 resulting sequences were discarded because of inconsistency of the strand relatively to similarity searches.
Multiple alignments were conducted using the program CLUSTALW [26] and visualized with BOXSHADE (http://www.ch.embnet.org/software/BOX_form.html). CLUSTALW results were submitted to the WebLogo server [27] (http://weblogo.berkeley.edu).
Results
cDNA libraries, cDNA sequencing and functional annotation
Two unidirectionnal cDNA libraries were constructed using mRNAs isolated from mycelium of A. euteiches cultivated either in vitro in a liquid medium (MYC library) or grown in contact to M. truncatula roots (INT library). A total of 18,684 high-quality sequences (9,224 for the MYC library and 9,460 for the INT library) corresponding to the 5′ end of cDNA inserts were acquired. A 7,977 unigene set was assembled from EST data. Among the unigenes, 2,843 consensus sequences were assembled from multiple sequence reads and 5,134 were singlets.
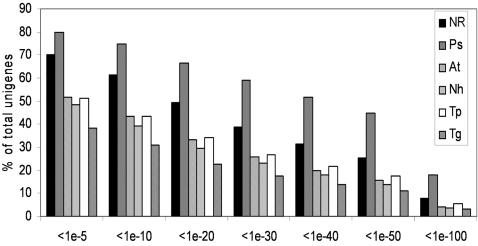
The 7,977 unigene set was annotated by comparison to the NCBI non-redundant (nr) protein database (5-17-2007 Version) using BLAST analyses [28]. Proteomes of seven fully sequenced organisms were added to the analysis. These include the proteomes from two oomycetes (P. sojae and P. ramorum), the diatom Thalassiosira pseudonana, the pathogenic fungus Nectria haematococca, the model plant Arabidopsis thaliana and the apicomplexa parasites Toxoplasma gondii and Plasmodium falciparum. Overall, about 70% of the sequences showed homology to previously described genes in the NCBI non-redundant protein database using a BLASTX E value cut-off of 10−5 (Figure 1). As expected, a large proportion of sequences (80%, E value cut-off of <10−5) had significant similarities to Phytophthora predicted proteins. Interestingly, about the same fraction (50%, E value cut-off of 10−5) of A. euteiches sequences showed homology to a plant, diatom, apicomplexa or fungal sequence whereas these organisms are distantly related (Figure 1). The unigene set was then annotated by comparisons with the Pfam database of protein domains [29]. 45% of the sequences showed homology with a protein domain with an E value<10−5. All the data were stored in a public database called AphanoDB (www.polebio.scsv.ups-tlse.fr/aphano/) which is described elsewhere [20]. Consensus and singlet sequences were named with a unique identifier with the prefix ‘Ae’ for the species, 3 digits for the number of EST included in the contig, the 2 digits ‘AL’ for the strain and 5 digits as unique number.
Figure 1. Comparison of A. euteiches sequences to the non redundant protein database at NCBI and proteomes of fully sequenced organisms.
Unigene sequences were blasted using the BLASTX software to the nr database and to the proteome of P. sojae (Ps), A. thaliana (At), N. haematococca (Nh), T. pseudonana (Tp) and T. gondii (Tg). For each class of e-value the percentage of unigenes showing homology is indicated.
Genes highly represented in the interaction library
Determination of the number of sequences specific of each library provided a good indication that the two growth conditions used for library construction allowed the expression of divergent transcriptomes. More than 40% of the sequences appeared to be specific for one library (Table 1). To identify unigenes with a statistical difference in transcript abundance, we used a method described in [30] based upon the frequencies which genes occur in each library and false discovery rate control for multiple test corrections [31]. Eight genes abundantly represented (p-value<B-H cutoff) in the interaction library were identified (Table 2). Interestingly, 4 genes encode putative proteins with significant similarity with transporters: 2 sucrose transporters (Ae_48AL7986 and Ae_29AL7339), a protein with an oligopeptide transporter domain (Ae_21AL7132; InterPro domain IPR004813), and a gene coding a probable mitochondrial substrate carrier (Ae_97AL5378). This latter gene is of interest since it has been shown that a gene encoding a mitochondrial carrier protein is required for pathogenicity of the soil-borne fungus Fusarium oxysporum [32].
Table 1. Number of expressed sequence tags and unigenes specific to a given library.
| Library | Singlets | Per contigs a | Total | Specific ESTs (%) | Specific unigenes b |
| INT | 2,932 | 1,652 | 4,584 | 48.40 | 3,576 |
| MYC | 2,202 | 1,772 | 3,974 | 43.08 | 2,755 |
Number of ESTs per contigs made of ESTs from a single library.
Total number of unigenes (singlets or contigs made of ESTs from a single library) found in a given library;
Table 2. Unigenes highly represented in the INT library.
| Unigene ID | ESTs number INT/MYC | Best BLASTX matcha | E value | Species | Pfam domain |
| Ae_97AL5378 | 93/4 | Solute carrier family | 5.10−41 | Danio rerio | Mitochondrial substrate carrier |
| Ae_48AL7986 | 45/3 | Probable sucrose transport protein | 7.10−12 | A. thaliana | No hit |
| Ae_21AL7132 | 20/1 | Metal-nicotianamine transporter | 1.10−70 | A. thaliana | Oligopeptide transporter |
| Ae_29AL7339 | 27/2 | Probable sucrose transport protein | 6.10−13 | A. thaliana | No hit |
| Ae_19AL5526 | 18/1 | No hit | Phosphatidylinositol kinase | ||
| Ae_57AL5693 | 44/13 | Elongation factor | 7.10−179 | Solanum lycopersicum | Elongation factor tu |
| Ae_44AL7441 | 39/5 | No hit | No hit | ||
| Ae_47AL5766 | 46/1 | No hit | No hit |
BLASTX was done against the SwissProt database
Genes encoding potential pathogenicity proteins
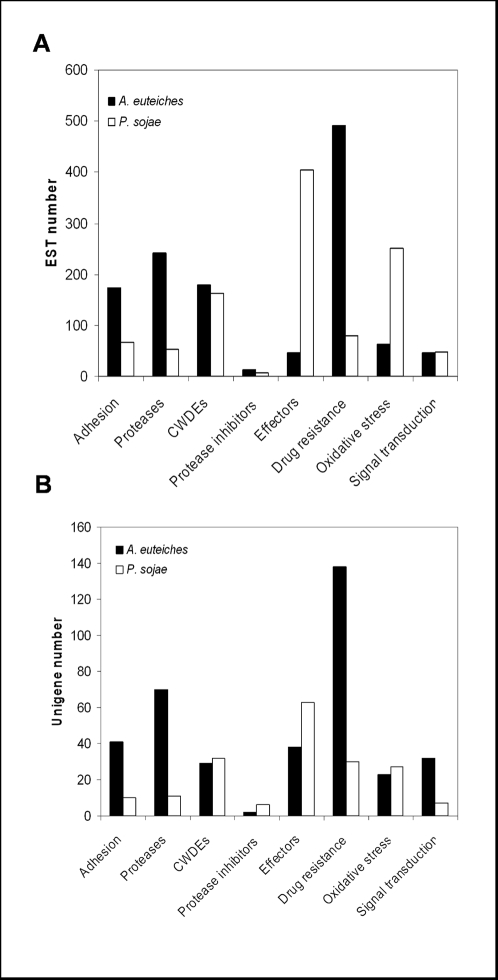
Plant pathogens express a large array of genes playing various roles in pathogenesis. These genes can be roughly classified into eight major categories according to a scheme established for P. sojae and P. parasitica [16], [17]. To facilitate comparative analyses, A. euteiches unigenes were classified following the same criteria. The results obtained with the A. euteiches ESTs were compared to those obtained from P. sojae ESTs described in [16] since in this later study about the same number of ESTs as in A. euteiches was analysed (26,943 ESTs assembled into 7,863 unigenes). This comparative approach revealed striking differences between A. euteiches and P. sojae (Figure 2). Gene families over-represented in A. euteiches compared to P. sojae include genes encoding proteins playing a role in adhesion (mucin-like proteins), in protein degradation and in drug resistance (ABC and PDR-like ABC transporter, cytochrome P450 enzymes).
Figure 2. Classification of unigenes based on eight functional categories involved in pathogenicity.
A. euteiches unigenes (black bars) for which a putative function was assigned were classified into defined categories according to [17] and [16]. For each category the number of ESTs (A) and the number of unigene (B) is shown. For comparison to Phytophthora, analysis of a EST collection from P. sojae was added to the figure [16] (white bars).
Adhesion
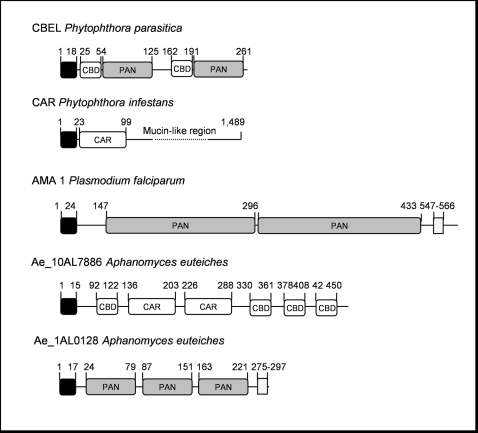
One of the most well-characterized protein playing a key role in oomycete adhesion to plant surface component is a cell wall protein isolated from P. parasitica named CBEL (for Cellulose Binding Elicitor Lectin) [33], [34]. CBEL contains two fungal type I Cellulose Binding Domains (CBD or CBM_1; InterPro domain IPR000254) which are involved in cellulose binding but which are also perceived by the host cells as pathogen associated molecular patterns [35]. CBEL also harbours two regions with similarities to the N/apple PAN domain (IPR000177), a conserved domain involved in protein-protein or protein-carbohydrate interactions [36]. cDNA sequences showing typical CBEL features, i.e CBDs associated to N/apple PAN domains, were not found in A. euteiches ESTs. Accordingly, antigens were not detected with antibodies directed against CBEL, when mycelium extracts were probed by western blot analysis (data not shown). However, CBDs were detected in a large gene family coding proteins showing similarities to a mucin-like protein identified in the soybean root nematode Heterodera glycines (GenBank accession number AAC62109). More than 150 ESTs were assembled into 32 unigenes, the largest contig containing 39 ESTs (Ae_39AL5321). Similarity between A. euteiches and H. glycines proteins is restricted to a domain originally found in cyst germination proteins of P. infestans (Figure 3; [37]). The full length sequence of Ae_10AL7886 was determined and domain organization of the predicted protein was deduced (Figure 3). SignalP analysis of the predicted protein identified a 15-amino acids putative signal peptide. Four domains showing typical features of fungal cellulose binding domains were found in the N-terminal and C-terminal ends of the protein. The central part showed similarities to the H. glycines mucin-like protein and the cyst germination proteins of P. infestans. The combination of these two modules was not detected so far and suggests a role of this protein in cell adhesion.
Figure 3. Protein domain organisation of cell surface proteins from oomycetes and apicomplexa.
Two proteins from Phytophthora, CBEL and CAR which are localized at the cell surface [33], [37] are represented. CBEL contains two closely associated domains, a cellulose binding domain (CBD) and a N/apple PAN domain (PAN). CAR proteins are composed of a mucin-like region and a N-terminal region (CAR) with conserved cysteine residues. AMA1 is a P. falciparum protein with two PAN domains [39].
Eight distinct cDNA sequences showed similarity to CBEL centered on the N/apple PAN domain. This domain is of interest since it has been found in surface proteins of apicomplexan parasites essential for attachment to host cells [38]. Whereas the role of this domain in oomycete proteins is still unclear, it can be speculated that it can interact with host targets to promote pathogenesis. cDNAs coding for proteins with N/Apple domains fell into two categories. The first category included predicted protein sequences for which no other protein domains were found except one or several N/Apple domains. A representative example is Ae_1AL0128 which was fully sequenced (Figure 3). It includes three N/Apple domains and a predicted transmembrane domain located at the C-terminus end. Interestingly, a transmembrane domain was also detected in the P. falciparum surface protein AMA1 which was shown to contain two N/Apple PAN domains (Figure 3, [39]). The second category included predicted protein sequences for which N/apple domains were associated with a catalytic domain. Two of these unigenes (Ae_7AL7989 and Ae_1AL5080) were predicted to encode proteins with a peptidase domain associated to one (Ae_1AL5080) or five (Ae_7AL7989) N/apple domains. Finally, three unigenes were identified showing several copies of a thrombospondin type I motif, a conserved 50 amino acid sequence found in surface proteins of Phytophthora and apicomplexan parasites [40], [41] and putatively involved in adhesion.
Cell wall degrading enzymes (CWDEs)
Sequences encoding enzymes potentially involved in degradation of plant cell walls were selected, by looking for the presence of glycoside hydrolase (GH) domains of enzymes hydrolysing substrates that can be found in plant cell walls, such as β-1,4- and β-1,3-glucanases, pectinases, xylosidases, arabinosidases and glucosidases of various specificities [42]. A collection of 53 potential CWDE unigenes out of 7977 unigenes, a proportion similar to what was recorded in P. sojae or ramorum [15] was obtained. All but one A. euteiches CWDE unigenes had a Phytophthora homolog with an E value lower than 10−40.
β-1,4-glucanases and β-1,3-glucanases mainly of GH families 5 and 81 potentially involved in degradation of plant cellulose and callose were represented by 29 unigenes. Among the putative β-1,3-glucanases from A. euteiches, an homolog (Ae_2AL6121) of a previously described GH family 17 endo-1,3-β-glucanase from Saprolegnia parasitica [43] and P. infestans (PiENDO1; [44]) was found, as well as two unigenes (Ae_2AL7302 and Ae_12AL7443) showing high similarity to the GH family 5 exo-1,3-β-glucanase PiEXO3 from P. infestans [44]. Interestingly, the latter unigenes are exclusively composed of ESTs from the interaction library, suggesting that they are involved in the interaction with the plant.
Other glycosidases from A. euteiches, potentially involved in degradation of plant cell wall polysaccharides such as xyloglucans, xylans or galacto(gluco)mannans, were represented by 24 unigenes encoding GH family 1, 3 and 30 enzymes. However, GH family 12 endoglucanases, potentially involved in xyloglucan degradation and represented by a family of 8 to 10 genes in P. ramorum and P. sojae [45], and GH family 10 xylanases, represented by more than 5 genes in the P. ramorum and P. sojae genomes, were missing in the database.
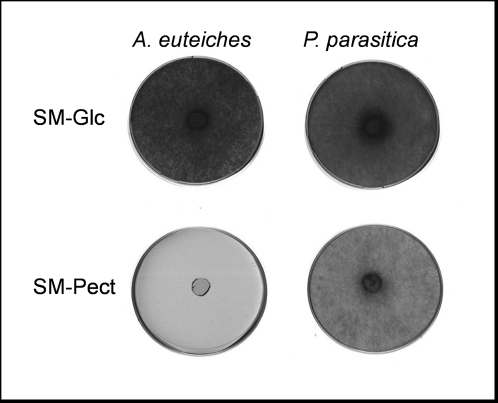
Surprisingly, no EST related to pectin metabolism was found, despite the fact that P. sojae and P. ramorum genomes contain numerous pectinase genes [15], and bona-fide pectinase genes were found in P. infestans, P. cinnamomi and P. parasitica [12], [46]–[48]. The lack of sequences related to pectin degradation is correlated with the lack of pectinase activity observed in yeast extract/glucose medium (data not shown) and the failure to grow A. euteiches on a synthetic medium containing pectin as sole carbon source, whereas P. parasitica, which produces an inducible polygalacturonase [47], grew equally well on glucose or pectin medium (Figure 4). These results showed that pectinase genes were poorly expressed in the growth conditions we used, and that pectinase genes are differently expressed in A. euteiches and Phytophthora species.
Figure 4. Growth of A. euteiches on synthetic media.
A. euteiches and Phytophthora parasitica mycelium were grown on synthetic medium containing Glucose (SM-Glc) or Pectin (SM-Pect) as carbon source. Photos were taken 15 days after inoculation.
Proteases and protease inhibitors
A large set of A. euteiches sequences were predicted to encode proteases (Figure 2). Serine, aspartyl and cysteine proteases were identified, the largest family being the cysteine protease family. Comparison with the P. sojae ESTs collection revealed that the number of protease genes is significantly higher in A. euteiches than in P. sojae. For example, 17 unigenes (67 ESTs) of A. euteiches were predicted to encode serine carboxypeptidase (IPR001563) whereas one unigene (1 EST) was found in P. sojae. Another example is represented by cysteine proteases (IPR000668) for which 25 unigenes (125 ESTs) were found whereas only 5 unigenes (41 ESTs) were detected in the P. sojae EST collection. Moreover, two types of proteases belonging to the cysteine protease family, peptidase c1b (IPR004134) and peptidase c14 (caspase, IPR011600) were found only in A. euteiches and not in other oomycetes. This analysis showed that A. euteiches is clearly distinct from Phytophthora species with respect to protease production, and this is in accordance with the detection of a high protease activity in culture medium (data not shown).
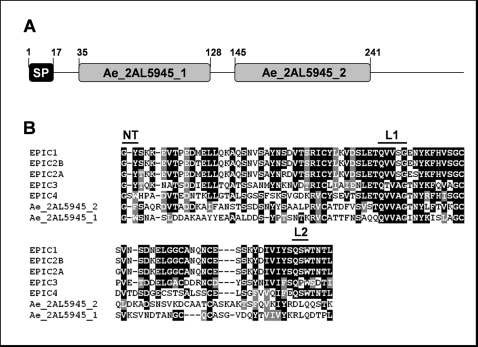
By contrast with the large number of proteases genes, only two genes (13 ESTs) encoding protease inhibitors were detected. These include a cystein protease inhibitor of the cystatin family (Ae_2AL5945; IPR000010) and a serine protease inhibitor (Ae_11AL6547) containing Kazal domains (IPR002350). These two types of protease inhibitors and their potential role in defense-counterdefense mechanisms were studied in P. infestans [49], [50]. Five domains containing typical features of Kazal motifs were found in Ae_11AL6547. However, more domains could be present since the sequence appeared to be incomplete. The predicted translation product of Ae_2AL5945 were analysed with SignalP leading to the identification of a 17 amino acid signal peptide with a significant mean S value of 0.90 and hidden Markov probability of 1.0. Similarity searches of the predicted protein against the NCBI non-redundant database using the BLASTP program revealed a weak similarity (E value = 6.10−3) to the P. infestans cystatin EPIC4, centered on the predicted active site. Multiple alignment using ClustalW [26] was done using cystatin domains from P. infestans (EPIC1, EPIC2A, EPIC2B, EPIC3 and EPIC4) and the two domains from Ae_2AL5945 (Ae_2AL5945_1 and Ae_2AL5945_2). Cystatin domains from A. euteiches are clearly distinct from P. infestans sequences except in the highly conserved sites of cystatins, i.e. the N terminal trunk and the L1 binding loop (Figure 5). However, the second binding loop is divergent in A. euteiches sequences.
Figure 5. A cystatin-like protease inhibitor in A. euteiches.
A, domain organisation of the deduced protein sequence of Ae_2AL5945. A putative signal peptide (SP) and two domains similar to cystatin domain are shown. B, alignment of cystatin domains from P. infestans cystatins (EPIC1, EPIC2A, EPIC2B , EPIC3 and EPIC4) and from Ae_2AL5945. The active sites of cystatins, including the N-terminal trunk (NT), first binding loop (L1), and second binding loop (L2), are shown.
Effectors
A class of oomycete effectors which could be delivered inside plant cells during infection was recently characterized. Effectors from Hyaloperonospora parasitica, P. infestans and P. sojae do not show extensive similarity except a conserved RxLR motif downstream of the signal peptide, followed by an EER motif. The A. euteiches unigene collection was mined for putative RxLR effectors looking for matches either to a sequence pattern or to HMM profiles. We end up with two unigenes containing the RxLR-dEER motif (Ae_1AL5059, Ae_1AL1390) and one with a RxLx[EDQ]-dEER motif (Ae_40AL5333), which is close to the Plasmodium motif. Ae_1AL5059 and Ae_1AL1390 showed homology to a putative P. sojae and P. ramorum genes but appeared to be incomplete. Ae_40AL5333 encodes a putative lysine rich protein with no homology to known protein sequence.
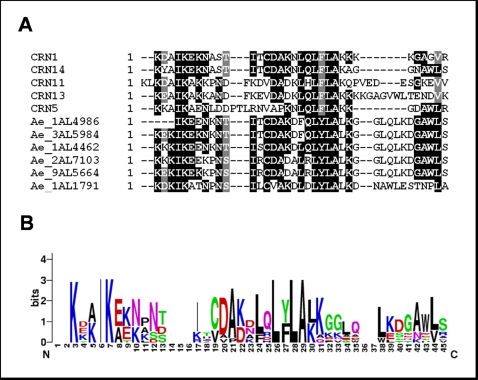
A distinct class of putative cytoplasmic effectors is the Crinkling and Necrosis (CRN) family first described in P. infestans [48]. While Phytophthora CRNs lack an RxLR motif, some H. parasitica CRNs show an RxRL sequence overlapping another conserved motif LxFLAK [24]. Several sequences showing high similarity with Phytophthora CRN were detected in A. euteiches. The largest groups of CRNs showed strong similarity to P. infestans CRN5 (14 unigenes) or CRN13 (9 unigenes). Interestingly, the conserved LxLFLAK sequence was not present in A. euteiches genes, but a closely related motif, F/LxLYLALK, was detected (Figure 6). This is the first time that CRN genes are found in an organism distinct from Peronosporales suggesting that this class of effectors plays an important role in oomycete pathogenicity.
Figure 6. Identification of conserved amino acid residues at the N-terminal extremities of P. infestans and A. euteiches CRN sequences.
A, multiple alignment of A. euteiches like CRNs and P. infestans CRNs (CRN1, CRN5, CRN11, CRN14) showing the highest homology to A. euteiches sequences. B, consensus sequence pattern calculated using Weblogo showing the most conserved amino acid residues.
Other oomycete effector genes include genes coding necrosis-inducing peptides (NPP family; [51], [52]) and elicitins [53], [54]. No ortholog of NPP was found and only one divergent sequence encoding a putative elicitin-like protein was detected (Ae_1AL4317).
Transporters
A large number of sequences were predicted to encode proteins involved in transport of molecules. The largest family encodes ABC transporters (84 unigenes and 342 ESTs) and PDR-like ABC transporters (28 unigenes and 86 ESTs). Interestingly, two oligopeptide transporter genes (Ae_21AL7132 and Ae_1AL0658) encoding putative proteins with an OPT domain (IPR004813) were found. Two distinct, proton-coupled peptide transport systems have been described in eukaryotic organisms: the PTR (peptide transport) system, specific for di- and tripeptides, and the OPT (oligopeptide transport) system, which transports tetra- and pentapeptides. Homolog of di-/tripeptide transporters are found in virtually all organisms, whereas the oligopeptide transporters are limited to fungi and plants (reviewed in [55], [56]). While the PTR system is well represented in Phytophthora proteomes (8 genes in P. sojae), the OPT proteins are absent, suggesting that OPT dependant oligopeptide transport is not widely found in oomycetes. However, Ae_21AL7132 and Ae_1AL0658 show strong similarity with an A. thaliana sequence (E value = 6.10−72 and 5.10−38 respectively) encoding a metal-nicotianamine transporter. This class of plant transporters is involved in iron–phytosiderophore uptake by roots but exhibits also significant sequence similarity to several functionally characterized yeast transporters of the OPT family [57], [58]. Thus it cannot be excluded that Ae_21AL7132 and Ae_1AL0658 encoded proteins are involved in uptake of phytosiderophore-Fe complex. Interestingly, ESTs assembled into Ae_21AL7132 were found almost exclusively in the interaction library (20 ESTs in the interaction library and 1 in the mycelium library; Table 2) suggesting a role for this class of transporters in pathogenesis.
Identification of A. euteiches genes without orthologs in Phytophthora
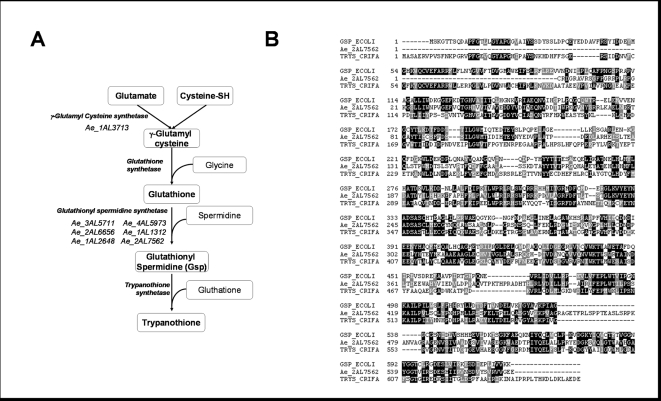
Identification of unigenes without any similarity to Phytophthora sequences but with similarity to sequences from trypanosamatid parasites led to the identification of a family of proteins putatively involved in the synthesis of trypanothione, a compound playing a key role in defense against oxidative stress in trypanosomatids [59]. Trypanothione is synthesized by conjugation of the polyamine spermidine and the tripeptide glutathione. The biosynthesis of trypanothione starts with the formation of glutathionylspermidine (Gsp) which then reacts with a second GSH molecule to form trypanothione (Figure 7A). These two biosynthetic steps are catalysed by two distinct enzymes, glutathionylspermidine synthetases and trypanothione synthetases. However, it has been shown that trypanothione syntethase can catalyse the entire synthesis of trypanothione in Trypanosoma cruzi and Crithidia fasciculata [60], [61]. Glutathionylspermidine has also been identified in E. coli where it could be involved in regulating the intracellular level of spermidine and glutathione. Six A. euteiches unigenes were found encoding proteins with a typical glutathionylspermidine syntethase domain (IPR005494) and similarity to trypanothione syntethase sequences from trypanosomatids (E value<10−10). Moreover, one sequence (Ae_2AL7562) contains a second domain found in trypanothione synthetases and bacterial bifunctional glutathionylspermidine synthetase/amidase, the CHAP (cysteine, histidine dependant amidohydrolase/peptidase) domain (IPR007921). Alignment of the protein sequence with an E. coli bifunctional glutathionylspermidine synthetase [62] and a trypanothione synthetase from the insect parasite C. fasciculata [63] is shown on Figure 7B. These results suggest the presence of a glutathionylspermidine based defense mechanism against oxidative stress in A. euteiches that is absent in other oomycetes such as P. sojae and P. ramorum.
Figure 7. Identification of A. euteiches genes putatively involved in trypanothione synthesis.
A, trypanothione biosynthesis starts with the formation of glutathione catalysed by g-glutamyl synthetase and glutathione synthetase. Trypanothione is synthesized from glutathione and spermidine either in two successive steps catalysed by glutathionyl spermidine synthase and trypanothione synthase. It has been shown in some cases that trypanothione synthase is able to catalysed the two reactions. Unigenes predicted to code enzymes belonging to this pathway are indicated. B, multiple alignment of gluthationyl spermidine synthetase from E. coli (GSP_ECOLI), from Crythidia fasciculata (TRYS CRIFA) and Ae_2AL7562.
To find A. euteiches genes which could be absent in other oomycetes but present in fungal plant pathogens, all the unigenes sequences were compared to the pea root pathogen N. haematococca. The sequences showing an E value greater than 0.1 to a Phytophthora sequence and lower than 10−10 to a sequence from the N. haematococca proteomes were selected. About half of the sequences (19 unigenes) matched to sequences of unknown function and without known protein domains. Interestingly, 3 of the remaining sequences (Ae_1AL0272, Ae_2AL7741 and Ae_1AL3374) showed a strong similarity to hydrolytic enzymes (glucan hydrolases and lipase) and 3 (Ae_1AL1763, Ae_1AL3400 and Ae_1AL0931) to non-ribosomal peptide synthetases (NRPS). These latter enzymes are of outstanding importance since their products have been shown to be involved in fungal pathogenesis [64], [65] (Table 3). Four putative NRPS genes were detected in P. sojae and P. ramorum genomes [15]. However these genes are not related to the A. euteiches sequences. These data suggests that A. euteiches could be able to produce secondary metabolites distinct from those synthesized by Phytophthora species.
Table 3. A. euteiches unigenes showing a BLASTX match to fungal, plant or diatom proteins (E value<1.10−10) and not to Phytophthora proteins (E value>0.1).
| Unigene ID | Best hit | Best BLASTX match nr databank | E value | Species | Pfam domain |
| Ae_1AL1763 | N. haematococca | nonribosomal peptide synthetase | 2.10−35 | Omphalotus olearius | Condensation domain |
| Ae_1AL0931 | non-ribosomal peptide synthetase | 8.10−46 | Granulibacter bethesdensis | No hit | |
| Ae_1AL3400 | unnamed protein product | 1.10−17 | Aspergillus oryzae | Condensation domain | |
| Ae_1AL0272 | β 1,3-glucanase | 3.10−19 | Strongylocentrotus purpuratus | No hit | |
| Ae_2AL7741 | hypothetical protein | 1.10−36 | Phaeosphaeria nodorum | GH 71 | |
| Ae_1AL3374 | hypothetical protein | 1.10−14 | Gibberella zeae | Lipolytic enzyme | |
| Ae_2AL7237 | A. thaliana | hypothetical protein | 9.10−23 | Oryza sativa | HMGCoA reductase |
| Ae_8AL7813 | HMG-CoA reductase | 6.10−98 | Gossypium hirsutum | No hit | |
| Ae_1AL1432 | HMG-CoA reductase | 5.10−82 | Gossypium hirsutum | HMGCoA reductase | |
| Ae_1AL0818 | cycloartenol synthase | 5.10−88 | Betula platyphylla | No hit | |
| Ae_1AL0450 | thiazole biosynthetic protein | 1.10−110 | Nicotiana tabacum | thiamine biosynthesis thi4 protein | |
| Ae_1AL1432 | T. pseudonana | HMG-CoA reductase | 5.10−82 | Gossypium hirsutum | HMGCoA reductase |
| Ae_2AL7237 | hypothetical protein | 9.10−23 | Oryza sativa | HMGCoA reductase | |
| Ae_3AL7274 | lanosterol synthase | 8.10−72 | Homo sapiens | Prenyltransferase/squalene oxidase | |
| Ae_2AL7738 | unnamed protein product | 8.10−63 | Tetraodon nigroviridis | Squalene/phytoene synthase |
Comparative analysis of A. euteiches sequences with a high BLASTX score to a A. thaliana or T. pseudonana sequences allowed the identification of two biosynthetic pathways which are absent in Phytophthora species but present in Aphanomyces (Table 3). One sequence (Ae_1AL0450) showed significant similarity to a thiamine biosynthetic enzyme from plants (top hit to protein AAP03875, E value = 1.10−110) and a thiamine biosynthesis Thi4 protein domain (IPR002922). This enzyme is involved in the synthesis of the thiamine precursor, thiazole. Members of the genus Phytophthora are thiamine auxotrophs and consequently lack this biosynthetic enzyme whereas A. euteiches and other Saprolegniales such as Saprolegnia parasitica are able to synthesize thiamine. Accordingly, the sequence from A. euteiches is highly similar to the S. parasitica gene (SPM2G2; 93% identity at the protein level) described in [43].
Phytophthora species being sterol auxotrophs, key genes coding for enzymes involved in sterol biosynthesis were not identified in Phytophthora genomes [15]. By contrast, Saprolegniales species such as A. euteiches are able to grow in minimal media without exogenous sterols. Accordingly, several genes coding for putative sterol biosynthesis enzymes were identified. These include two cycloartenol synthases (Ae_3AL7274; Ae_1AL0818) and a cytochrome P450 enzyme belonging to the CYP51 family (Ae_5AL7244) which strongly matched (E values<10−70) plant enzymes. Each gene has also closed orthologs in the diatom T. pseudonana suggesting that the sterol biosynthetic is ancient in the oomycete lineage and has been lost by Phytophthora species.
Discussion
In this study we describe an extensive characterization of an EST collection of A. euteiches, an economically important plant pathogen. The closest oomycete specie which was the subject of a genomic approach is the fish pathogen S. parasitica, with the sequencing of 1,510 cDNA clones (1,279 unigenes) obtained from one library constructed from mycelium grown axenically [43]. The current study is more expansive in terms of number of cDNA clones sequenced (18,684) and constructed libraries (2) and adds to the EST resources available for other oomycetes such as P. sojae (7,863 unigenes) [16], P. infestans (18,256 unigenes) [12] and P. parasitica (4734 unigenes from two libraries) [17], [66]. Comparison between the unigene contents of the two A. euteiches libraries showed that they reflected two divergent expression programs. The procedure used to cultivate A. euteiches mycelium in contact to M. truncatula roots avoided contamination of pathogen mRNAs with plant mRNAs [20]. Nevertheless, perception of plant root tissue by the pathogen occurred, leading to the induction of a specific subset of genes. These include 4 transporter genes which could be involved in nutrient uptake during infection.
Comparative analysis of A. euteiches unigene sequences to proteomes derived from fully sequenced organisms provided a global view of sequence similarities distribution among organisms which are distantly related. As expected, most of sequences showed similarity to a Phytophthora predicted protein sequence. About 20% of unigenes did not have any ortholog in P. sojae or P. ramorum. More surprisingly, we found that similar proportions of unigenes have an ortholog in the plant A. thaliana, the fungus N. haematococca, the diatom T. pseudonana and the apicomplexa T. brucei. These sequences were examined in more detail with a particular emphasis to those with no orthologs in Phytophthora genomes. This revealed the presence of genes involved in biosynthetic pathways which are known to be absent in Phytophthora. For example, it is known that Phytophthora species, but not Aphanomyces species, are thiamine auxotrophs. Accordingly, a gene coding a thiamine biosynthetic enzyme was identified, showing homology with a S. parasitica sequence [43]. Another highly significant example concerns sterol metabolism. Among oomycetes, members of the Saprolegniale order can synthesize sterols de novo whereas species belonging to Pythiales are sterol auxotrophs. We found several genes encoding proteins showing high similarity to enzymes involved in sterol biosynthesis. This pathway is a major target for chemical inhibitors and a large number of fungicides have been developed acting mainly on a demethylation step catalysed by a cytochrome p450 enzyme of the CYP51 family. The identification of this class of proteins in A. euteiches will allow the development of new compounds acting on Saprolegniale pathogens. The ability of A. euteiches to synthesize its own sterols could be correlated with the fact that one elicitin-like sequence has been detected and no antigen was detected with anti elicitin antibodies when culture filtrates were probed by western blot analysis (data not shown). Elicitins are proteins found only in oomycetes where they were proposed to act as sterol carriers [67]. In Phytophthora, elicitins represent a major class of secreted protein and are highly expressed. For example, 210 ESTs, corresponding to 16 elicitin genes were detected in the P. sojae EST collection [16]. Moreover, elicitins can be perceived by the host cell, leading to a strong induction of defense reactions, and are thought to play a role in plant-pathogen specificity [53]. Thus, the low expression of elicitins in A. euteiches could be linked to its ability to synthesize its own sterols.
Classification of putative pathogenicity factors into 8 categories revealed other striking differences between A. euteiches and Phytophthora species. These include qualitative and quantitative differences for gene encoding hydrolases, effectors or proteins involved in adhesion, drug resistance and oxidative stress. Adhesion of pathogens to the surface of host tissues is a critical step in the establishment of oomycete pathogenesis [68]. Although several adhesins produced by fungal pathogens have been characterized, molecular mechanisms involved in adhesion of oomycetes plant pathogens are still largely unknown. These include cyst germination proteins from P. infestans which share homology with mucins [37], Phytophthora surface proteins containing thrombospondin repeats [40] and P. parasitica CBEL [34]. A functional approach demonstrated the essential role of CBEL in adhesion. Inhibition of CBEL gene expression in transgenic P. parasitica strains led to a dramatic reduction in adhesion of the mycelium to cellulosic substrates [69]. Several sequences showing similarity with Phytophthora adhesins were found in A. euteiches but with specific features. Of particular interest is a highly expressed family of proteins composed of a combination of cellulose binding domains (CBM_1) similar to those present in CBEL, and acidic domains found in the N-terminal part of the P. infestans cyst germination proteins. These proteins can represent a novel class of oomycete adhesins.
Analysis of the EST collection revealed expansion of gene families in A. euteiches such as genes encoding proteases. Proteases and protease inhibitors play an important role in the molecular dialogue which establishes between the pathogen and its host. In Phytophthora, several protease inhibitors have been shown to target plant proteases during infection [49], [50]. Expansion of proteases gene families in A. euteiches indicates that these proteins are probably major players of A. euteiches pathogenicity. Thus, protein degradation and utilization is probably a major metabolic pathway related to pathogenesis in A. euteiches. Others genes showing expansion and diversification in A. euteiches comprise ABC transporters whose role in mediating resistance to plant toxins and fungicide has been suggested in Phytophthora [70]. By contrast, some gene families which are well represented in Phytophthora were not detected in A. euteiches. This is the case for pectinases for which their role as pathogenicity factors has been demonstrated for several plant pathogens. The lack of ESTs showing homology to pectinases is correlated to the weak production of extracellular pectinolytic activity in vitro and the absence of A. euteiches growth on a medium containing pectin as sole carbon source. Recently, sequencing of a 192 bp genomic DNA fragment of A. euteiches revealed similarity with a P. capsici polygalacturonase [71]. Thus it cannot be definitively concluded from our results that pectinase genes are not present in A. euteiches.
Another group of oomycete pathogenicity factors are proteins which are translocated into the cytoplasm of the host cell during infection. These gene products all contain an RxLR motif which has been shown to be essential for the effective transport of these effectors [18]. While no clear homolog of effectors belonging to the RxLR class were identified in A. euteiches, their presence cannot be fully excluded. It has been shown that these proteins are under a strong positive selection [24] and can thus escape in silico analyses. However, genes showing strong similarity with a second class of effectors belonging to the crinkler family were identified. Up to now, these proteins were found only in Phytophthora species where they represent a large superfamily with 40 genes in the genome of P. sojae [15]. CRNs were issued from a large screen aiming at identifying extracellular proteins from P. infestans able to induce symptoms when expressed in plant tissues [48]. While the exact function of CRNs is still unclear, conservation of these sequences in all pathogenic oomycetes examined so far indicates that they probably play a specific role in oomycete pathogenesis.
Finally, comparison of A. euteiches sequences to protein sequences from tryponosomatids led to the identification of protein sequences with strong similarity to enzymes involved in synthesis of antioxidant compounds, glutathionylspermidine and trypanothione. Trypanothione has been identified as an essential redox intermediate and its role in defense against oxidative stress has been demonstrated in the human pathogens Trypanosoma brucei and Leishmania donovani, [72], [73]. Since the oxidative burst is one of the earliest observable plant defense response against pathogen invasion (for review see [74]), the production of antioxidant compounds will constitute an efficient defense mechanism for the pathogen.
In conclusion this work greatly expands our knowledge of a genus which includes plant and animal pathogens and more generally illustrates the diversity which can exist among the oomycete. The repertoire of expressed genes described here will be used as source of candidate genes for functional studies aiming at identifying essential factors of A. euteiches pathogenicity.
Acknowledgments
We thank Didier Laborie, David Allouche, Jean-Marc Larre and all members of the Bioinformatic Platform of the Genopole (Toulouse, France) for computing technical support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by CNRS, University of Toulouse (Toulouse III) and Genoscope.
References
- 1.Delwich PA, Grau CR, Holub EB, Perry JB. Characterization of Aphanomyces euteiches isolates recovered from alfalfa in Wisconsin. Plant Dis. 1987;71:155–161. [Google Scholar]
- 2.Levenfors JP, Wikstrom M, Persson L, Gerhardson B. Pathogenicity of Aphanomyces spp. from different leguminous crops in Sweden. Eur J Plant Pathol. 2003;109:535–543. [Google Scholar]
- 3.Wicker E, Rouxel F. Specific behaviour of French Aphanomyces euteiches Drechs. populations for virulence and aggressiveness on pea, related to isolates from Europe, America and New Zealand. Eur J Plant Pathol. 2001;107:919–929. [Google Scholar]
- 4.Mitchell JE, Yang CY. Factors affecting growth and development of Aphanomyces euteiches. . Phytopathology. 1966;56:917–922. [PubMed] [Google Scholar]
- 5.Larsson M. Pathogenicity, morphology and isozyme variability among isolates of Aphanomyces spp. from weeds and various plants. Mycol Res. 1994;98:231–240. [Google Scholar]
- 6.Pilet-Nayel ML, Muehlbauer FJ, McGee RJ, Kraft JM, Baranger A, et al. Consistent quantitative trait loci in pea for partial resistance to Aphanomyces euteiches isolates from the United States and France. Phytopathology. 2005;95:1287–1293. doi: 10.1094/PHYTO-95-1287. [DOI] [PubMed] [Google Scholar]
- 7.Söderhäll K, Cerenius L. The crayfish plague fungus: history and recent advances. Freshwater Crayfish. 1999;12:11–35. [Google Scholar]
- 8.Blazer VS, Lilley JH, Schill WB, Kiryu Y, Densmore C, et al. Aphanomyces invadans in Atlantic menhaden along the east coast of the United States. J Aquat Anim Health. 2002;14:1–10. [Google Scholar]
- 9.Gaulin E, Jacquet C, Bottin A, Dumas B. Root rot disease of legumes caused by Aphanomyces euteiches. . Mol Plant Pathol. 2007;8:539–548. doi: 10.1111/j.1364-3703.2007.00413.x. [DOI] [PubMed] [Google Scholar]
- 10.Martin HL. Management of soilborne diseases of beetroot in Australia: a review. Aust J Exp Agri. 2003;43:1281–1292. [Google Scholar]
- 11.Kamoun S. Nonhost resistance to Phytophthora: novel prospects for a classical problem. Curr Opin Plant Biol. 2001;4:295–300. doi: 10.1016/s1369-5266(00)00176-x. [DOI] [PubMed] [Google Scholar]
- 12.Randall TA, Dwyer RA, Huitema E, Beyer K, Cvitanich C, et al. Large-scale gene discovery in the oomycete Phytophthora infestans reveals likely components of phytopathogenicity shared with true fungi. Mol Plant Microbe Interact. 2005;18:229–243. doi: 10.1094/MPMI-18-0229. [DOI] [PubMed] [Google Scholar]
- 13.Gajendran K, Gonzales MD, Farmer A, Archuleta E, Win J, et al. Phytophthora functional genomics database (PFGD): functional genomics of Phytophthora-plant interactions. Nucleic Acids Res. 2006;34:465–470. doi: 10.1093/nar/gkj119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waugh M, Hraber P, Weller J, Wu Y, Chen G, et al. The Phytophthora genome initiative database: informatics and analysis for distributed pathogenomic research. Nucleic Acids Res. 2000;28:87–90. doi: 10.1093/nar/28.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 16.Torto-Alalibo TA, Tripathy S, Smith BM, Arredondo FD, Zhou L, et al. Expressed sequence tags from Phytophthora sojae reveal genes specific to development and infection. Mol Plant Microbe Interact. 2007;20:781–793. doi: 10.1094/MPMI-20-7-0781. [DOI] [PubMed] [Google Scholar]
- 17.Panabières F, Amselem J, Galiana E, Le Berre J. Gene identification in the oomycete pathogen Phytophthora parasitica during in vitro vegetative growth through expressed sequence tags. Fungal Genet Biol. 2005;42:611–623. doi: 10.1016/j.fgb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 19.Papavizas G, Davey C. Some factors affecting growth of Aphanomyces euteiches in synthetic media. Amer J Bot. 1960;47:758–765. [Google Scholar]
- 20.Madoui MA, Gaulin E, Mathé C, San Clemente H, Couloux A, et al. AphanoDB: a genomic resource for Aphanomyces pathogens. BMC Genomics. 2007;8:471. doi: 10.1186/1471-2164-8-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 22.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 23.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 24.Win J, Morgan W, Bos J, Krasileva KV, Cano LM, et al. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell. 2007;19:2349–2369. doi: 10.1105/tpc.107.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 26.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Susko E, Roger AJ. Estimating and comparing the rates of gene discovery and expressed sequence tag (EST) frequencies in EST surveys. Bioinformatics. 2004;20:2279–2287. doi: 10.1093/bioinformatics/bth239. [DOI] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 32.Inoue I, Namiki F, Tsuge T. Plant colonization by the vascular wilt fungus Fusarium oxysporum requires FOW1, a gene encoding a mitochondrial protein. Plant Cell. 2002;14:1869–1883. doi: 10.1105/tpc.002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Séjalon-Delmas N, Villalba Mateos F, Bottin A, Rickauer M, Dargent R, et al. Purification, elicitor activity, and cell wall localization of a glycoprotein from Phytophthora parasitica var. nicotianae, a fungal pathogen of tobacco. Phytopathology. 1997;87:899–909. doi: 10.1094/PHYTO.1997.87.9.899. [DOI] [PubMed] [Google Scholar]
- 34.Villalba-Mateos F, Rickauer M, Esquerré-Tugayé MT. Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol Plant-Microbe Interact. 1997;10:1045–1053. doi: 10.1094/MPMI.1997.10.9.1045. [DOI] [PubMed] [Google Scholar]
- 35.Gaulin E, Dramé N, Lafitte C, Torto-Alalibo T, Martinez Y, et al. Cellulose binding domains of a Phytophthora cell wall protein are novel pathogen-associated molecular patterns. Plant Cell. 2006;18:1766–1777. doi: 10.1105/tpc.105.038687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepathocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999;461:63–67. doi: 10.1016/s0014-5793(99)01416-7. [DOI] [PubMed] [Google Scholar]
- 37.Gornhardt B, Rouhara I, Schmelzer E. Cyst germination proteins of the potato pathogen Phytophthora infestans share homology with human mucins. Mol Plant Microbe Interact. 2000;13:32–42. doi: 10.1094/MPMI.2000.13.1.32. [DOI] [PubMed] [Google Scholar]
- 38.Brecht S, Carruthers V, Ferguson D, Giddings O, Wang G, et al. The toxoplasma micronemal protein MIC4 is an adhesin composed of six conserved apple domains. J Biol Chem. 2001;276:4119–4127. doi: 10.1074/jbc.M008294200. [DOI] [PubMed] [Google Scholar]
- 39.Bai T, Becker M, Gupta A, Strike P, Murphy VJ, et al. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc Natl Acad Sci USA. 2005;102:12736–12741. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robold AV, Hardham AR. During attachment Phytophthora spores secrete proteins containing thrombospondin type 1 repeats. Curr Genet. 2005;47:307–315. doi: 10.1007/s00294-004-0559-8. [DOI] [PubMed] [Google Scholar]
- 41.Anantharaman V, Iyer LM, Balaji S, Aravind L. Adhesion molecules and other secreted host-interaction determinants in Apicomplexa: insights from comparative genomics. Int Rev Cytol. 2007;262:1–74. doi: 10.1016/S0074-7696(07)62001-4. [DOI] [PubMed] [Google Scholar]
- 42.Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 43.Torto-Alalibo T, Tian M, Gajendran K, Waugh M, van West P, et al. Expressed sequence tags from the oomycete fish pathogen Saprolegnia parasitica reveal putative virulence factors. BMC Microbiol. 2005;2:5–46. doi: 10.1186/1471-2180-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLeod A, Smart CD, Fry WE. Characterization of 1,3-β-glucanase and 1,3;1,4-β-glucanase genes from Phytophthora infestans. . Fungal Genet Biol. 2003;38:250–263. doi: 10.1016/s1087-1845(02)00523-6. [DOI] [PubMed] [Google Scholar]
- 45.Costanzo S, Ospina-Giraldo MD, Deahl KL, Baker CJ, Jones RW. Gene duplication event in family 12 glycosyl hydrolase from Phytophthora spp. Fungal Genet Biol. 2006;43:707–714. doi: 10.1016/j.fgb.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Gotesson A, Marshall JS, Jones DA, Hardham AR. Characterization and evolutionary analysis of a large polygalacturonase gene family in the oomycete plant pathogen Phytophthora cinnamomi. . Mol Plant Microbe Interact. 2002;15:907–921. doi: 10.1094/MPMI.2002.15.9.907. [DOI] [PubMed] [Google Scholar]
- 47.Yan HZ, Liou RF. Cloning and analysis of pppg1, an inducible endopolygalacturonase gene from the oomycete plant pathogen Phytophthora parasitica. . Fungal Genet Biol. 2005;42:339–350. doi: 10.1016/j.fgb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 48.Torto TA, Li S, Styer A, Huitema E, Testa A, et al. EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. . Genome Res. 2003;13:1675–1685. doi: 10.1101/gr.910003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian M, Huitema E, Da Cunha L, Torto-Alalibo T, Kamoun S. A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem. 2004;279:26370–26375. doi: 10.1074/jbc.M400941200. [DOI] [PubMed] [Google Scholar]
- 50.Tian M, Win J, Song J, van der Hoorn R, van der Knaap E, et al. A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol. 2007;143:364–377. doi: 10.1104/pp.106.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fellbrich G, Romanski A, Varet A, Blume B, Brunner F, et al. NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. . Plant J. 2002;32:375–390. doi: 10.1046/j.1365-313x.2002.01454.x. [DOI] [PubMed] [Google Scholar]
- 52.Qutob D, Kamoun S, Gijzen M. Expression of a Phytophthora sojae necrosis-inducing protein occurs during transition from biotrophy to necrotrophy. Plant J. 2002;32:361–373. doi: 10.1046/j.1365-313x.2002.01439.x. [DOI] [PubMed] [Google Scholar]
- 53.Ponchet M, Panabières F, Milat ML, Mikes V, Montillet JL, et al. Are elicitins cryptograms in plant-Oomycete communications? Cell Mol Life Sci. 1999;56:1020–1047. doi: 10.1007/s000180050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang RH, Tyler BM, Whisson SC, Hardham AR, Govers F. Ancient origin of elicitin gene clusters in Phytophthora genomes. Mol Biol Evol. 2006;23:338–351. doi: 10.1093/molbev/msj039. [DOI] [PubMed] [Google Scholar]
- 55.Stacey G, Koh S, Granger C, Becker JM. Peptide transport in plants. Trends Plant Sci. 2002;7:257–263. doi: 10.1016/s1360-1385(02)02249-5. [DOI] [PubMed] [Google Scholar]
- 56.Hauser M, Narita V, Donhardt AM, Naider F, Becker JM. Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. . Mol Membr Biol. 2001;18:105–112. [PubMed] [Google Scholar]
- 57.Le Jean M, Schikora A, Mari S, Briat JF, Curie C. A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J. 2005;44:769–782. doi: 10.1111/j.1365-313X.2005.02569.x. [DOI] [PubMed] [Google Scholar]
- 58.Yen MR, Tseng YH, Saier MH., Jr Maize Yellow Stripe1, an iron-phytosiderophore uptake transporter, is a member of the oligopeptide transporter (OPT) family. Microbiology. 2001;147:2881–2883. doi: 10.1099/00221287-147-11-2881. [DOI] [PubMed] [Google Scholar]
- 59.Fairlamb AH, Blackburn P, Ulrich P, Chait BT, Cerami A. Trypanothione: a novel bis(glutathionyl)spermidine cofactor for glutathione reductase in trypanosomatids. Science. 1985;227:1485–1487. doi: 10.1126/science.3883489. [DOI] [PubMed] [Google Scholar]
- 60.Oza SL, Tetaud E, Ariyanayagam MR, Warnon SS, Fairlamb AH. A single enzyme catalyses formation of trypanothione from glutathione and spermidine in Trypanosoma cruzi. . J Biol Chem. 2002;277:35853–35861. doi: 10.1074/jbc.M204403200. [DOI] [PubMed] [Google Scholar]
- 61.Comini M, Menge U, Wissing J, Flohe L. Trypanothione synthesis in Crithidia revisited. J Biol Chem. 2005;280:6850–6860. doi: 10.1074/jbc.M404486200. [DOI] [PubMed] [Google Scholar]
- 62.Bollinger JM, Jr, Kwon DS, Huisman GW, Kolter R, Walsh CT. Glutathionylspermidine metabolism in Escherichia coli. Purification, cloning, overproduction, and characterization of a bifunctional glutathionylspermidine synthetase/amidase. J Biol Chem. 1995;270:14031–14041. doi: 10.1074/jbc.270.23.14031. [DOI] [PubMed] [Google Scholar]
- 63.Alphey MS, Leonard GA, Gourley DG, Tetaud E, Fairlamb AH, et al. The high resolution crystal structure of recombinant Crithidia fasciculata tryparedoxin-I. J Biol Chem. 1999;274:25613–25622. doi: 10.1074/jbc.274.36.25613. [DOI] [PubMed] [Google Scholar]
- 64.Lee BN, Kroken S, Chou DY, Robbertse B, Yoder OC, et al. Functional analysis of all nonribosomal peptide synthetases in Cochliobolus heterostrophus reveals a factor, NPS6, involved in virulence and resistance to oxidative stress. Eukaryot Cell. 2005;4:545–555. doi: 10.1128/EC.4.3.545-555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson RD, Johnson L, Itoh Y, Kodama M, Otani H, et al. Cloning and characterization of a cyclic peptide synthetase gene from Alternaria alternata apple pathotype whose product is involved in AM-toxin synthesis and pathogenicity. Mol Plant Microbe Interact. 2000;13:742–753. doi: 10.1094/MPMI.2000.13.7.742. [DOI] [PubMed] [Google Scholar]
- 66.Le Berre JY, Engler G, Panabieres F. Exploration of the late stages of the tomato-Phytophthora parasitica interactions through histological analysis and generation of expressed sequence tags. New Phytol. 2007:1–11. doi: 10.1111/j.1469-8137.2007.02269.x. [DOI] [PubMed] [Google Scholar]
- 67.Mikes V, Milat ML, Ponchet M, Panabieres F, Ricci P, et al. Elicitins, proteinaceous elicitors of plant defense, are a new class of sterol carrier proteins. Biochem Biophys Res Commun. 1998;245:133–139. doi: 10.1006/bbrc.1998.8341. [DOI] [PubMed] [Google Scholar]
- 68.Hardham AR. Cell biology of plant-oomycete interactions. Cell Microbiol. 2007;9:31–39. doi: 10.1111/j.1462-5822.2006.00833.x. [DOI] [PubMed] [Google Scholar]
- 69.Gaulin E, Jauneau A, Villalba F, Rickauer M, Esquerré-Tugayé MT, et al. The CBEL glycoprotein of Phytophthora parasitica var-nicotianae is involved in cell wall deposition and adhesion to cellulosic substrates. J Cell Sci. 2002;115:4565–4575. doi: 10.1242/jcs.00138. [DOI] [PubMed] [Google Scholar]
- 70.Judelson HS, Senthil G. Investigating the role of ABC transporters in multifungicide insensitivity in Phytophthora infestans. . Mol Plant Pathol. 2006;7:17–29. doi: 10.1111/j.1364-3703.2004.00256.x-i1. [DOI] [PubMed] [Google Scholar]
- 71.Akamatsu HO, Grunwald NJ, Chilvers MI, Porter LD, Peever TL. Development of codominant simple sequence repeat, single nucleotide polymorphism and sequence characterized amplified region markers for the pea root rot pathogen, Aphanomyces euteiches. J Microbiol Methods. 2007;71:82–86. doi: 10.1016/j.mimet.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 72.Krieger S, Schwarz W, Ariyanayagam MR, Fairlamb AH, Krauth-Siegel RL, et al. Trypanosomes lacking trypanothione reductase are avirulent and show increased sensitivity to oxidative stress. Mol Microbiol. 2000;35:542–552. doi: 10.1046/j.1365-2958.2000.01721.x. [DOI] [PubMed] [Google Scholar]
- 73.Tovar J, Wilkinson S, Mottram JC, Fairlamb AH. Evidence that trypanothione reductase is an essential enzyme in Leishmania by targeted replacement of the tryA gene locus. Mol Microbiol. 1998;29:653–660. doi: 10.1046/j.1365-2958.1998.00968.x. [DOI] [PubMed] [Google Scholar]
- 74.Wojtaszek P. Oxidative burst: an early plant response to pathogen infection. Biochem J. 1997;322:681–692. doi: 10.1042/bj3220681. [DOI] [PMC free article] [PubMed] [Google Scholar]