Abstract
The regions of single-stranded (ss) DNA that result from DNA damage are immediately coated by the ssDNA-binding protein (SSB). RecF pathway proteins facilitate the displacement of SSB from ssDNA, allowing the RecA protein to form protein filaments on the ssDNA region, which facilitates the process of recombinational DNA repair. In this study, we examined the mechanism of SSB displacement from ssDNA using purified Thermus thermophilus RecF pathway proteins. To date, RecO and RecR are thought to act as the RecOR complex. However, our results indicate that RecO and RecR have distinct functions. We found that RecR binds both RecF and RecO, and that RecO binds RecR, SSB and ssDNA. The electron microscopic studies indicated that SSB is displaced from ssDNA by RecO. In addition, pull-down assays indicated that the displaced SSB still remains indirectly attached to ssDNA through its interaction with RecO in the RecO-ssDNA complex. In the presence of both SSB and RecO, the ssDNA-dependent ATPase activity of RecA was inhibited, but was restored by the addition of RecR. Interestingly, the interaction of RecR with RecO affected the ssDNA-binding properties of RecO. These results suggest a model of SSB displacement from the ssDNA by RecF pathway proteins.
INTRODUCTION
DNA is constantly exposed to ultraviolet radiation, γ-radiation and chemical mutagens. As a result, DNA lesions such as nucleotide mismatches, the formation of pyrimidine dimers, occur. In all organisms, the repair of these DNA lesions is important for survival. DNA lesions are usually repaired using the complementary DNA strand as a template; however, lesions such as double strand breaks (DSBs) or single strand DNA gaps (SSGs), in which damage occurs on the ssDNA, cannot be repaired using the complementary strand. DSBs and SSGs often occur when the progression of the DNA replication fork is halted or collapsed due to the presence of base lesions or strand breaks. These types of DNA damage are repaired by homologous recombination, in which a homologous region of the genome is used as the template for repair, in a process called recombinational DNA repair (1).
The process of recombinational repair is initiated by the concerted action of a DNA helicase and a DNA nuclease, which process a DSB end into a 3′ ssDNA overhang. SSB (in prokaryotes) or RPA (in eukaryotes) then binds to the 3′ ssDNA overhang to prevent the formation of secondary structures and to protect it from nuclease digestion. Accessory proteins then facilitate the loading of the RecA family protein (RecA in prokaryotes or Rad51 in eukaryotes) onto SSB- or RPA-coated ssDNA. Upon displacement of SSB or RPA, the RecA family protein forms a filament on the ssDNA, which then catalyzes strand invasion at a homologous region in the genome (1,2).
The involvement of accessory proteins in the loading of the filamentous RecA family protein on the SSB- or RPA-coated ssDNA is conserved from prokaryotes to higher eukaryotes, although the proteins participating in the reaction are different. In higher eukaryotic cells, such as human cells, it is thought that the tumor suppressor protein BRCA2 is one of these accessory proteins. Brh2, the homologue of BRCA2 in fungus, has been implicated in Rad51-loading onto RPA-coated ssDNA (3). In yeast, Rad52 facilitates the formation of Rad51 filaments on RPA-coated ssDNA, as well as Brh2 (4,5). Rad52 also mediates the annealing of RPA-coated ssDNA, which is related to the annealing between the displaced strand after Rad51-mediated strand invasion and the second strand (6,7). Recently, it has been reported that the BRCA2 homologue of Caenorhabditis elegans, BRCA2 (CeBRC-2), and Brh2 can also anneal RPA-coated ssDNA like Rad52 (8,9). However, the mechanism of Rad51-loading onto RPA-coated ssDNA by Rad52 and/or BRCA2 homologues is still unknown. BRCA2 is a large protein, which complicates its functional analysis in vitro. In addition, eukaryotic proteins, including BRCA2 and Rad51, functionally interact with many other proteins, further complicating the analysis of their role in recombinational repair. To overcome some of these complications, we have studied the recombinational DNA repair pathway of prokaryotes as a model system. While it is a very simple system, it has many properties in common with the higher eukaryote.
The RecBCD and RecF pathways function in recombinational repair in bacteria. In Escherichia coli, the RecBCD pathway plays a central role in both DSB and SSG repair (10,11). However, many bacteria do not have the RecBCD pathway. In these bacteria, and in the recBC deficient E. coli mutants, DSBs can also be repaired by the RecF pathway. The RecF pathway is conserved in almost all bacteria and plays a central role in recombinational repair (12), suggesting that the RecF pathway constitutes a fundamental mechanism of DNA repair. Investigation of the proteins of the RecF pathway in bacteria that do not carry the components of the RecBCD pathway should provide fundamental information about a basic mechanism of DNA repair. Thermus thermophilus is one such model organism. In the current study, we analyzed the functions of the T. thermophilus HB8 RecF pathway proteins to clarify the mechanism of RecA-loading onto the SSB-coated ssDNA.
The proteins of the RecF pathway have been well studied in E. coli, which has led to a model of RecF-mediated DNA repair. At a DSB site, the DNA is unwound by the RecQ helicase and then degraded by the RecJ 5′ to 3′ exonuclease that generates a 3′ ssDNA overhang, which is bound and protected by SSB. SSGs are also protected by SSB. Although the SSB-coated ssDNA region is not a substrate of RecA, the concerted action of RecF, RecO and RecR enable RecA to form a filament on the SSB-coated ssDNA. The search for homologous sequences and strand invasion are then catalyzed by the RecA filament, and the region lost by the DSB or SSG is recovered by homologous recombination (13). In this process, it has been suggested that RecF or the RecFR complex recognizes the dsDNA–ssDNA junction, which prevents the RecA filaments from extending over the junction, and that the RecOR complex enhances the loading of RecA onto the SSB-coated ssDNA and stabilizes the RecA filaments at the dsDNA–ssDNA junction (13–16). RecO itself is also a functional homologue of Rad52 and mediates annealing of SSB-coated ssDNA (7). Recently, we examined the mechanism of recognition of the dsDNA–ssDNA junction by RecFOR proteins. Our results suggested that the RecR Toprim domain in the RecFR complex, which is located at the dsDNA–ssDNA junction, interacts with RecO on the ssDNA (17). Although the mechanism of recognition of the dsDNA–ssDNA junction by RecFOR is becoming clearer, the mechanism of SSB displacement remains uncertain. Since it appears that only RecO interacts with SSB, it may play an important role in the displacement of SSB from ssDNA (18–20).
In the current study, we purified T. thermophilus RecF pathway proteins and examined their role in the displacement of SSB from ssDNA. To our knowledge, this is the first report of the distinct functions of RecO and RecR in the RecF pathway of recombinational repair. We also show electron microscopy images of the RecO-ssDNA complex. Our results suggest that RecO displaces SSB on ssDNA through high-affinity binding to the DNA and that the displaced SSB remains on the ssDNA through its binding to RecO on the ssDNA. In addition, we show that the inhibitory effect of SSB on the activity of RecA is restored by the interaction of RecR with RecO in the SSB-RecO-ssDNA complex, and that the interaction of RecR with RecO simultaneously modulates the ssDNA-binding properties of RecO.
EXPERIMENTAL PROCEDURES
Preparation of T. thermophilus RecF pathway proteins
Thermus thermophilus (tt) RecA, RecR and SSB were prepared as previously described (21–23). The expression plasmids for ttRecO (pET11a-ttrecO) and ttRecF (pET11a-ttrecF) were a kind gift from Professor Seiki Kuramitsu (Osaka University).
To prepare T. thermophilus RecO protein, the E. coli strain Rosetta (DE3) pLysS (Novagen) was transformed with pET11a-ttrecO. Cells were grown in 1L of M9 medium (12% Na2HPO4, 6% KH2PO4, 1% NaCl, 1% NH4Cl, 2% glucose, 0.04% Thiamine sulfate, 2 mM MgSO4, 0.1 mM CaCl2, 2 μM FeCl3, 4 μM ZnSO4, 1 μM MnSO4, 4.7 μM H3BO3, 0.7 μM CuSO4, 100 μg/ml ampicillin) at 37°C to an OD600 of 0.6, then isopropyl β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM for 6 h. The cells were harvested (1 g) and re-suspended in 10 ml of buffer containing 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 2 mM EDTA, 2 mM β-mercaptoethanol, 100 μg/ml lysozyme, 2 μM amidinophenylmethylsulfonylfluoride (APMSF) and 0.1% (w/v) Brij58 on ice. The cells were disrupted using ultrasonic disruption (TOMY, UD201) and centrifuged at 72 000 × g for 1 h to remove insoluble debris. Subsequently, the supernatant was heat-treated at 80°C for 10 min and immediately placed on ice. The supernatant was applied to a DEAE-sepharose column (Amersham Biosciences, 8 ml) that was equilibrated with a buffer containing 20 mM Tris-HCl (pH 7.5), 2 mM EDTA, 2 mM β-mercaptoethanol and 500 mM NaCl. ttRecO was eluted in the same buffer. To dilute the concentration of salt, an equal volume of a buffer containing 20 mM Tris-HCl pH 7.5, 2 mM EDTA and 2 mM β-mercaptoethanol was added to the eluted fraction. The diluted fraction containing ttRecO was then applied to a SP-sepharose column (Amersham Biosciences, 8 ml) equilibrated with a buffer containing 20 mM Tris-HCl (pH 7.5), 2 mM EDTA, 2 mM β-mercaptoethanol and 200 mM NaCl. After washing the column with 32 ml of this buffer, ttRecO was eluted in a linear gradient of NaCl (0.2–1.5 M). Fractions containing ttRecO were collected and ammonium sulfate was added to a final concentration of 20% (w/v). This solution was applied to a Phenyl-toyopearl 650S column (TOSHO, 4 ml) equilibrated with a buffer containing 20 mM Tris-HCl pH 7.5, 2 mM EDTA, 2 mM β-mercaptoethanol and 20% (w/v) ammonium sulfate. After washing the column with 16 ml of this buffer, ttRecO was eluted in a linear gradient of ammonium sulfate (20–0%). Fractions containing ttRecO were collected, concentrated to a volume of 2 ml and then loaded onto a Superdex75 10/300GL column (Amersham Biosciences, 23 ml) equilibrated with a buffer containing 20 mM Tris-HCl (pH 7.5), 2 mM EDTA, 2 mM β-mercaptoethanol and 100 mM NaCl. Fractions containing ttRecO were eluted and then applied to a Resource S column (Amersham Biosciences, 6 ml) equilibrated with a buffer containing 20 mM Tris-HCl (pH 7.5), 2 mM EDTA, 2 mM β-mercaptoethanol and 100 mM NaCl. ttRecO was eluted in a linear gradient of NaCl (0.1–1 M). Fractions containing ttRecO were collected and concentrated. An equal volume of glycerol was added, and the samples were stored at −20°C until use. The concentration of ttRecO was determined using a molar absorption coefficient of 20 220 M−1 cm−1 at 280 nm, using the procedure described previously (24).
To prepare the T. thermophilus RecF protein, the E. coli strain BL21 (DE3) was transformed with pET11a-ttrecF. Cells were grown in 1.5 L of LB medium containing 100 μg/ml ampicillin at 37°C to an OD600 of 0.5, and then 1 mM IPTG was added for 6 h. The cells were harvested (1 g) and re-suspended in 5 ml of a buffer containing 50 mM Tris-HCl (pH 7.2), 1 mM EDTA, 25% (w/v) sucrose, 0.1 mM APMSF, 5 mM β-mercaptoethanol, 0.5 mg/ml lysozyme, 500 mM KCl and 0.5% (w/v) Brij58 on ice. After homogenization using a French press (OHTAKE), the cells were centrifuged at 70 000 × g for 1 h to remove insoluble debris. The supernatant was heat-treated at 80°C for 10 min, immediately placed on ice and then the supernatant was dialyzed against a buffer containing 20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA and 5 mM β-mercaptoethanol. The dialyzed solution was then applied to a DEAE-sepharose column (Amersham Biosciences, 8 ml) equilibrated with a buffer containing 20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 5 mM β-mercaptoethanol and 100 mM NaCl. ttRecF was eluted using the same buffer and collected as the flow-through fraction. This solution was then applied to an SP-sepharose column (Amersham Biosciences, 8 ml) equilibrated with a buffer containing 20 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 5 mM β-mercaptoethanol and 50 mM NaCl. After washing the column with 32 ml of this buffer, ttRecF was eluted in a linear gradient of NaCl (0.05–1.0 M). Fractions containing ttRecF were collected and stored at 4°C until use. The concentration of ttRecF was determined using a molar absorption coefficient of 31 380 M−1 cm−1 at 280 nm, using the procedure described previously (24).
Native PAGE analysis
ttRecA, ttRecR, ttSSB (10 μM) and ttRecO (1, 2, 5, 10, 20, 50 μM) or lysozyme (50 μM) were incubated in 100 μl of 50 mM Tris-HCl (pH 7.5) and 2 mM EDTA. After 10 min at 50°C, the samples were analyzed by polyacrylamide gel electrophoresis (PAGE) using a 10% gel under non-denaturing conditions. The proteins were visualized by Coomassie Brilliant Blue R-250 (CBB) staining. The interaction of ttRecR (10 μM) with ttRecF (1, 2, 5, 7 μM) or ttSSB (2, 10, 20 μM) was analyzed in a similar manner. The intensity of the protein bands was analyzed using a CS Analyzer version 2.0 (ATTO).
ssDNA-binding assay using fluorophotometry with etheno-modified ssDNA
The ssDNA-binding assay using etheno-modified ssDNA (εDNA) was performed as previously described (21,25). As a source of εDNA, calf thymus DNA was digested into ∼300 mer long fragments by using an ultrasonic disruptor and heat denatured. The DNA fragments were then etheno-modified as previously described (25). The concentration of εDNA (nucleotide concentration) was determined using an extinction coefficient of 9900 M−1 cm−1 at 260 nm, which reflects a low level of etheno-modified DNA. The fluorescence measurements were carried out using a Perkin Elmer LS 55 luminescence spectrometer equipped with a temperature-regulated holder. In the competition experiments, 4 μM of ttSSB or ttRecO was incubated in 200 μl of 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 100 mM KCl and 1 mM DTT in the presence of 10 μM εDNA. After 3 min at 25°C, the competing protein (either ttSSB or ttRecO at concentrations of 0.1, 0.2, 0.4, 0.8, 1, 1.5, 2 and 4 μM) was added to the reaction mixture. After 5 min at 25°C, the change in fluorescence that occurred because of the protein binding to the εDNA was measured and plotted against the concentration of the added protein.
Electron microscopy analysis
ttRecO or ttSSB (0.1 μM) was incubated with 2 ng of M13 ssDNA in 10 μl of 30 mM Tris–HCl (pH 7.5) and 5 mM MgCl2 for 10 min at 25°C. A 5 μl drop of the reaction mixture was placed onto a carbon-coated grid for 1 min. The excess drop was wiped off by filter paper and the grid was dried by air. Then the reaction mixture was negatively stained with 2% uranyl acetate and the excess stain was also wiped off by filter paper. After the grid was dried, it was rotary shadowed with platinum for 30 sec. The protein-ssDNA complexes were observed with a JEOL JEM 2000FX electron microscope operated at an acceleration voltage of 80 kV.
The ATP hydrolysis assay
The measurement of the ssDNA-dependent ATP hydrolysis of ttRecA in the presence of ttSSB, ttRecO and ttRecR was previously described (17). The reaction mixtures contained 30 μM ssDNA (nucleotide concentration) in 200 μl of a buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 50 mM KCl, 1 mM DTT, 1 mM ATP, 1.5 mM PEP, 0.3 mM NADH, 4.7 U/μl lactate dehydrogenase and 1.2 U/μl pyruvate kinase. ttSSB, ttRecO, ttRecR and ttRecA were added to the reaction mixture in different combinations and concentrations. After the addition of each protein, the mixture was incubated for 5 min at 37°C before adding the next protein. The combinations, the order of addition, and concentrations are indicated in the figure legends. The reactions were initiated by the addition of ttRecA. The kinetics of ATP hydrolysis were followed by measuring the absorption of NADH at 340 nm using an Ultrospec 4300 pro spectrometer (Amersham Pharmacia). The amount of hydrolyzed ATP was calculated by measuring the change in the absorbance at A340 per minute using as a conversion factor of ε340 value of 6.22 × 103 M−1 cm−1 for NADH. For the calculation of relative activity, the value of the decrease of the absorption of NADH during the stationary phase of RecA activity (last 5 min) was used. A 340-mer poly (dC) ssDNA fragment was used in all experiments.
The pull-down assay
Six picomoles of the 3′-biotinylated 60-mer ssDNA (GCC AAG CTT GCA TGC CTG CAG GTC GAC TCT AGA GGA TCC CCG GGT ACC GAG CTC GAA TTC) was incubated with 0.6 μl of Streptavidin sepharose (GE Healthcare) at room temperature for 5 min. To remove free ssDNA, the ssDNA-resin was suspended in 12.5 μl of a buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, 0.15% Tween20 and 0.2 mg/ml BSA and centrifuged at 2000 × g for 1 min. After centrifugation, the supernatant was discarded. This process was repeated three times. The ssDNA-resin was then suspended in 12.5 μl of the same buffer containing ttSSB and/or ttRecO and incubated at 50°C for 10 min. After three washes to remove unbound protein, the ssDNA resin was treated with SDS sample buffer and heated at 95°C for 5 min. The ssDNA-bound proteins were then analyzed using 12.5% SDS-PAGE. The same assays were also performed in 50 mM MES-NaOH (pH 6.5) and 50 mM Tris-HCl (pH 8.5) instead of 50 mM Tris-HCl (pH 7.5). Assays were also performed in the absence and presence of 1 mM MgCl2.
Agarose gel retardation assay
A radiolabeled 66-mer ssDNA fragment (CTT ATT AAA ACG GCA ACT CCT CCT CCG GCG GAA AGT CTT CCA AGC CTT CGT CAA TGT CCA CCC CAC) was prepared using [γ-33P] dATP (Amersham Pharmacia Biotech) and a commercial kit (MEGALABEL, Takara Bio). The 5′-33P-labeled 66-mer ssDNA (0.125 μM, nucleotide concentration) was incubated with ttRecO (1 μM) and ttRecR (1, 2, 4, 8 μM) for 5 min at 50°C in 10 μl of a buffer containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 100 mM KCl, 1 mM DTT and 2 mM ATP. The samples were then subjected to electrophoresis on a 2.5% agarose gel in TAE buffer (40 mM Tris-acetate (pH 8.0), 1 mM EDTA). The DNA and DNA-protein complexes were transferred from the gel onto Hybond-XL membranes (Amersham Biosciences). The membranes were dried and then exposed to an imaging plate over night. The complexes were analyzed using a BAS2000 image analyzer (Fuji Photo Film).
RESULTS
Purification of the T. thermophilus RecF pathway proteins
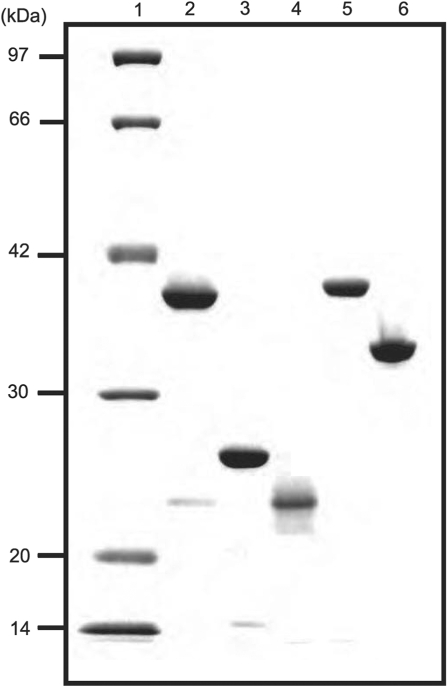
The proteins of the RecF pathway of T. thermophilus HB8, which included ttRecA, ttRecF, ttRecO, ttRecR and ttSSB, were overexpressed in E. coli and purified to homogeneity using heat treatment and column chromatography. The procedures for purifying ttRecF and ttRecO were established as part of this study and are described in the experimental procedures. Although none of the proteins contained a tag, we succeeded in purifying them to near homogeneity (Figure 1). Unless otherwise stated, the designations RecR, RecO, RecF, RecA and SSB refer to the proteins from T. thermophilus.
Figure 1.
Purification of ttRecF, ttRecO, ttRecR, ttRecA and ttSSB. The indicated samples (0.5 μg) were subjected to electrophoresis on 12.5% polyacrylamide gels containing 0.1% sodium dodecyl sulfate (SDS) under reducing conditions and visualized by staining with Coomassie Brilliant Blue R-250. The lanes contained the following samples: 1, Molecular mass markers (97, 66, 42, 30, 20 and 14 kDa); 2, ttRecF (37.8 kDa); 3, ttRecO (24.7 kDa); 4, ttRecR (21.2 kDa); 5, ttRecA (36.3 kDa); 6, ttSSB (29.8 kDa).
The physical interactions between the RecF pathway proteins
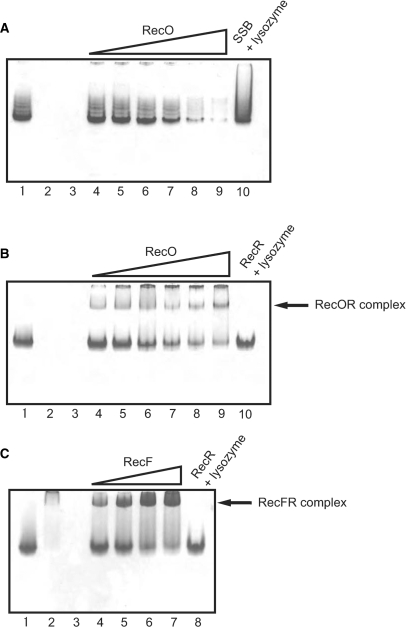
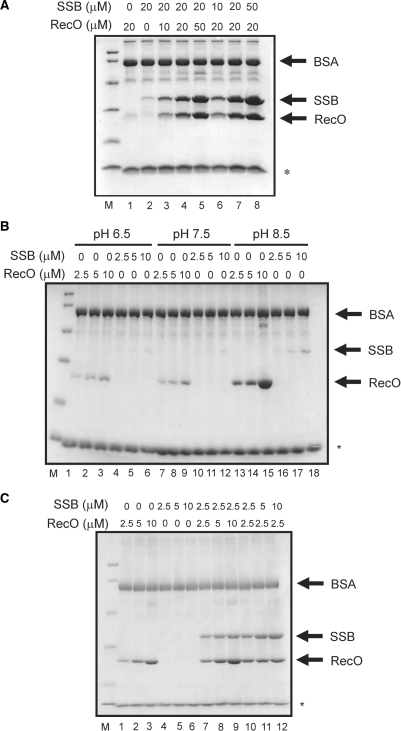
To examine and characterize the interactions between RecA, RecF, RecO, RecR and SSB, we performed native PAGE analysis (Figure 2). As expected, RecF (Figure 2C, lane 2) migrated slightly into the gel, whereas RecO (Figure 2A, lane 2) did not migrate into the gel at all because these proteins have a positive charge (pI = 9.09 and 10.7, respectively) in the gel running buffer. In contrast, SSB (Figure 2A, lane 1), RecR (Figure 2B, lane 1), and RecA (data not shown) migrated into the gel. The protein–protein interactions (complexes) were detected as a mobility shift and a disappearance of the corresponding single protein bands. As the concentration of RecO was increased, the band corresponding to SSB progressively disappeared (Figure 2A, lanes 4–9), becoming very faint when RecO and SSB were present at a molar ratio of 2:1 (Figure 2A, lane 8). As a control, when lysozyme (pI = 9.36) was added instead of RecO, the disappearance of the SSB band was not observed, even when the molar ratio of lysozyme:SSB was 5:1. These results indicated that RecO interacts specifically with SSB. A disappearance of the RecR band was also observed when the concentration of RecO was increased, while the control lysozyme had no effect (Figure 2B). This was accompanied by the appearance of a band corresponding to the RecOR complex (Figure 2B, lanes 4–9). These results indicated that RecO interacts with not only SSB but also RecR. In a similar set of experiments, RecR appeared to interact with RecF (Figure 2C). The SSB protein band was not affected by RecA, RecR or RecF, and none of the proteins affected the single RecA protein band (data not shown).
Figure 2.
Analysis of protein–protein interactions among RecF pathway proteins using native-PAGE. (A) Analysis of the interaction between RecO and SSB. SSB (10 μM) was incubated with 1, 2, 5, 10, 20 and 50 μM RecO, and the complexes were resolved by electrophoresis (lanes 4–9). SSB was also incubated with 50 μM lysozyme as a control (lane 10). 10 μM SSB, 10 μM RecO and 10 μM lysozyme were loaded in lanes 1, 2 and 3, respectively. (B) Analysis of the interaction between RecO and RecR. RecR (10 μM) was incubated with 1, 2, 5, 10, 20 and 50 μM RecO (lanes 4–9). RecR was also incubated with 50 μM lysozyme as a control (lane 10). 10 μM RecR, 10 μM RecO and 10 μM lysozyme were loaded in lanes 1, 2 and 3, respectively. (C) Analysis of the interaction between RecR and RecF. RecR (10 μM) was incubated with 1, 2, 5 and 7 μM RecF (lanes 4–7). RecR was also incubated with 10 μM lysozyme as a control (lane 8). 10 μM RecR, 10 μM RecF and 10 μM lysozyme were loaded in lanes 1, 2 and 3, respectively.
The ssDNA-binding properties of RecO and SSB
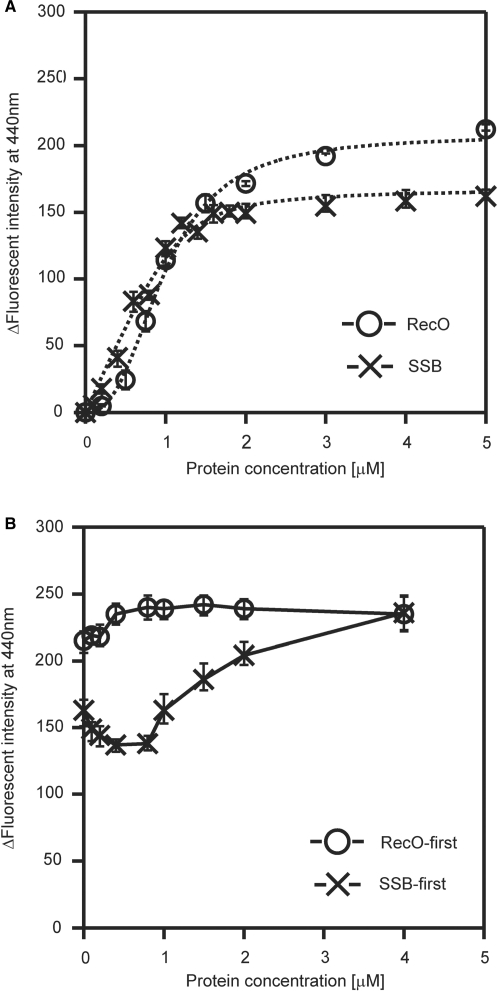
The analyses of the protein–protein interactions indicated that only RecO interacted with SSB, which suggests that RecO is an important factor in the displacement of SSB from the SSB-coated ssDNA. Since both RecO and SSB bind to ssDNA, we performed a set of binding experiments to determine whether RecO and SSB compete for binding to the ssDNA. We measured the change in fluorescence intensity (ΔF) of the etheno-modified ssDNA (εDNA) after protein binding (Figure 3A). In this analysis, the value of ΔF increases upon the addition of a protein to the εDNA solution. We found that the binding of RecO to εDNA resulted in a larger ΔF value compared to SSB. Also, the RecO-binding curve was sigmoidal, which suggests a cooperative binding of RecO to ssDNA. Although SSB had a similar binding curve, it was not sigmoidal. These results indicated that RecO and SSB have the ability to bind to the ssDNA, although RecO has a different mode of binding to ssDNA compared with SSB. The comparison of the binding curves of SSB and RecO suggested that RecO has a comparable ssDNA binding ability to that of SSB (Figure 3A).
Figure 3.
A ssDNA-binding assay using fluorophotometry and etheno-modified ssDNA. (A) Fluorescence spectral changes. The indicated concentrations of protein were incubated with 10 μM εDNA at 25°C for 5 min. The emission spectra (excitation wavelength of 305 nm) were measured using a 5 mm × 5 mm cell at 25°C. The ordinate axis represents the difference in fluorescence emission intensity at 440 nm between εDNA in the presence and absence of each protein. The data represents the average of three independent experiments. (B) Competition-binding experiments using RecO and SSB. 4 μM SSB or RecO was incubated with 10 μM εDNA at 25°C for 3 min prior to the addition of the competing protein. The indicated concentration of the competing protein was added to the solution for an additional 5 min before measuring the change in fluorescence.
Next, we carried out a set of competition experiments to examine the binding of RecO and SSB to ssDNA. RecO or SSB was first incubated with εDNA at a concentration that was sufficient to saturate the binding. The competing protein (either SSB or RecO) was then added to the solution. When RecO was incubated with the εDNA prior to adding SSB, the value of ΔF was approximately 220 and the addition of SSB had very little effect on this ΔF (Figure 3B, circle). In contrast, when RecO was added to the ssDNA that had been pre-incubated with SSB, the value of the ΔF increased with increasing protein concentration from about 150 to a final value of 230, which was the ΔF observed in the presence of 4 μM RecO (Figure 3B). In fact, in the presence of an excess of both SSB and RecO the ΔF was similar to that seen in response to the binding of RecO alone to the ssDNA, regardless of which protein was pre-incubated with the εDNA. These results suggested that RecO binds to ssDNA even if SSB is already bound to it and that RecO displaces SSB on the SSB-coated ssDNA.
The observation of the protein-ssDNA complexes by electron microscopy
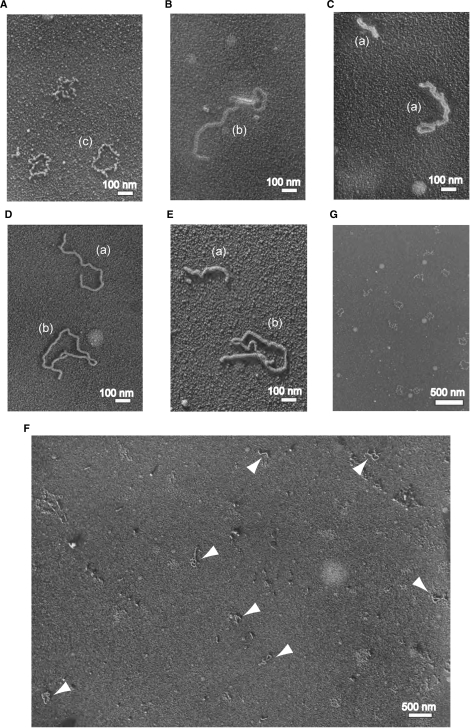
The results of the competitive ssDNA-binding experiments suggested that RecO displaces SSB on the SSB-coated ssDNA. To test this hypothesis, we observed the protein-ssDNA complex by electron microscopy and compared the images of the RecO-ssDNA complex with that of the SSB-ssDNA complex. The binding of SSB to M13 ssDNA formed a circular, rough-surfaced structure (Figure 4A and G), which was very similar to the published electron microscopy images of E. coli SSB-ssDNA complex (26). In contrast, the binding of RecO to M13 ssDNA formed a smooth-surfaced structure (Figure 4B and C). Almost all of the RecO-ssDNA complexes existed in linear, branched or twisted shapes, but were not circular. In addition, the complexes often contained more than two M13 ssDNAs, which looked like ssDNA molecules connected by RecO. The ssDNA annealing activity of RecO may relate to the formation of these structures.
Figure 4.
The observation of the protein-ssDNA complexes by electron microscopy. (A, G) The images of SSB in the presence of M13 ssDNA. (B, C) The images of RecO in the presence of M13 ssDNA. (D) The image of SSB-RecO in the presence of M13 ssDNA. M13 ssDNA was incubated with SSB for 10 min, and then RecO was added to the solution. (E) The image of RecO-SSB in the presence of M13 ssDNA. M13 ssDNA was incubated with RecO for 10 min, and then SSB was added to the solution. (F) The wide-ranging image of SSB-RecO in the presence of M13 ssDNA. The arrowheads indicate the protein-ssDNA complexes. Scales of each image are indicated by the white bars. Parentheses indicate the classified shapes in Table 1 (a, b and d).
Since the structural differences between the SSB-ssDNA and the RecO-ssDNA complexes were detectable, we attempted to observe the protein-ssDNA complexes in the presence of both SSB and RecO. M13 ssDNA was incubated with SSB and then RecO was added. Despite the pre-incubation of M13 ssDNA with SSB, we could only recognize RecO-ssDNA-like structures in the wide-field image (Figure 4F); no SSB-ssDNA specific structures were detected (Figure 4D, F). When RecO was incubated with M13 ssDNA prior to SSB, only RecO-ssDNA-like structures were also observed (Figure 4E). The observed complexes were classified according to their shapes; linear (including twist), circular, branch (including connected M13 ssDNA molecules) and SSB-type, and are summarized in Table 1. These results indicated that RecO can bind to the SSB-coated ssDNA and disrupt the SSB-ssDNA structure.
Table 1.
The number of classified shapes of the protein-ssDNA complex
| SSB + M13 | RecO + M13 | SSB +RecO + M13 | |
|---|---|---|---|
| Lineara,* | 0 | 46 | 85 |
| Circulard | 0 | 6 | 15 |
| Branchb,** | 0 | 10 | 63 |
| SSB-typec | 342 | 0 | 0 |
| Total | 342 | 62 | 163 |
Note: The 567 images were obtained from at least three independent experiments and were classified according to their shapes. The classification of the shapes is based on Figure 4A–E (a–c), and Supplementary Figure S1 (d).
*Including the twisted shape.
**Including the shape which looked like ssDNA molecules connected by RecO.
The inhibitory effect of SSB and RecO on the ssDNA-dependent ATPase activity of RecA
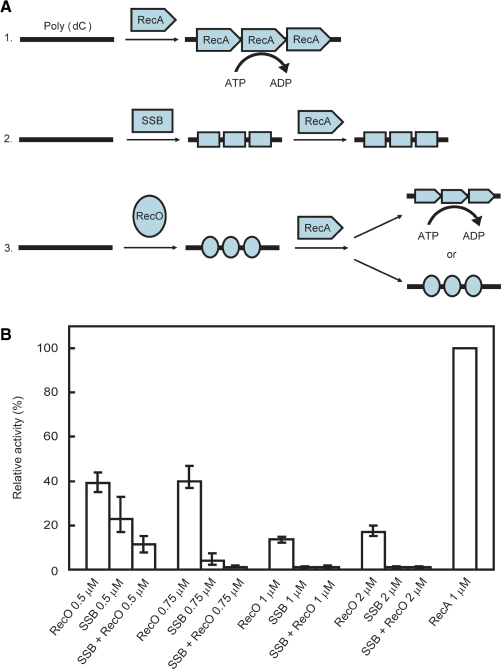
RecA hydrolyzes ATP in an ssDNA-dependent manner (Figure 5A-1) (27). However, when SSB binds to ssDNA prior to RecA, the ATPase activity of RecA is inhibited (Figure 5A-2). To date, it is not known whether the ATPase activity of RecA is affected by the binding of RecO to ssDNA prior to the binding of RecA (Figure 5A-3). Since the binding of RecO to the SSB-coated ssDNA disrupts the SSB-ssDNA structure, it may also have an effect on the inhibition of RecA by SSB. Therefore, we performed a set of experiments to determine the effect of RecO alone or in combination with SSB on the ATPase activity of RecA.
Figure 5.
The measurement of the ssDNA-dependent ATPase activity of RecA. (A) Schematic representation of the ATPase activity of RecA. 1. RecA hydrolyzes ATP in an ssDNA-dependent manner. 2. When SSB binds to the ssDNA prior to RecA, RecA ATPase activity is inhibited. 3. The effect of RecO binding to ssDNA prior to RecA on ATPase activity is not known. (B) The ssDNA-dependent ATPase activity of RecA in the presence of RecO and/or SSB. The ATPase activity of 1 μM RecA in the absence of RecO and SSB was defined as 100%. The ATPase activity of RecA in the presence of the indicated concentrations of SSB and/or RecO was measured. Each sample was incubated with a 340-mer poly (dC) prior to RecA, then 1 μM of RecA was added and the ATPase activity was measured. The data represent the average of three independent experiments.
SSB at a concentration of 0.5 µM reduced the ssDNA-dependent ATPase activity of 1 μM RecA to 20% of that observed in the absence of SSB. Concentrations of more than 0.75 μM of SSB almost completely inhibited the ATPase activity (Figure 5B, Supplementary Figure S2A). These results confirmed that SSB has a strong inhibitory effect on the ATPase activity of RecA.
When we examined the effect of RecO on the ATPase activity of RecA, we found that RecO also reduced the ssDNA-dependent ATPase activity of RecA, which was most likely due to the binding of RecO to the ssDNA. The ATPase activity of RecA was 40% of that of RecA alone even when the concentration of RecO was increased to 0.75 μM, a concentration at which SSB almost completely inhibited the ATPase activity. The degree of inhibition was less than that caused by SSB, even though RecO has a high affinity for ssDNA as shown in our earlier experiments (Figure 5B, Supplementary Figure S2A). The differences in the inhibitory effect of SSB and RecO may reflect the differences in the mode of binding of RecO and SSB to DNA.
The inhibitory effect of RecO on the ssDNA-dependent ATPase activity of RecA was less than that of SSB. Therefore, the degree of inhibition in the presence of both RecO and SSB would be less than that with SSB alone if SSB was completely dissociated from ssDNA by RecO. In the presence of both SSB and RecO, we observed a strong inhibitory effect on the ATPase activity of RecA, which was similar to that caused by SSB alone (Figure 5B, Supplementary Figure S2B). This result suggested that RecO did not dissociate SSB from the ssDNA. However, the results of the competition-binding experiments and the electron microscopic analysis suggested that RecO displaces SSB from the SSB-coated ssDNA. Since SSB can interact with RecO in solution, it is possible that SSB interacts with RecO, which is bound to the ssDNA. This would explain the strong inhibition of the ATPase activity of RecA in the presence of both SSB and RecO.
Co-localization of RecO and SSB on ssDNA
We performed a pull-down assay to confirm whether SSB co-localizes with RecO on ssDNA (Figure 6A). To prevent any non-specific interactions of RecO and SSB to the streptavidine sepharose, 0.15% Tween20 and 0.2 mg/ml BSA were added. Although the binding affinity of ssDNA was weakened in these conditions, a small amount of SSB and RecO were detected on the ssDNA-immobilized resin (Figure 6A, lanes 1 and 2, respectively). In the presence of an equal amount of SSB and RecO, these proteins were simultaneously present on the ssDNA (Figure 6A, lane 4). In addition, the amount of RecO and SSB that was pulled down was higher when the two proteins were added in combination compared with the addition of each protein alone. Moreover, when the concentration of RecO was progressively increased, the amount of SSB that was pulled down was increased relative to that of RecO (Figure 6A, lanes 3–5). Similar results were also obtained for RecO when the concentration of SSB was increased (Figure 6A, lanes 6–8). These results indicated that RecO did not dissociate SSB from the ssDNA and that these proteins co-localized on the ssDNA.
Figure 6.
The pull-down assay of the ssDNA-bound proteins. (A) The indicated concentrations of the proteins were incubated with ssDNA-immobilized streptavidine sepharose. After three washes, ssDNA-bound proteins were analyzed by SDS-PAGE. The asterisk indicates the band derived from the streptavidine sepharose. A band corresponding to BSA was observed in all lanes because it was present in the washing buffer. (B) Effect of pH on the ssDNA-binding properties of SSB and RecO. The buffers contained 50 mM MES-NaOH (pH 6.5, lanes 1–6), 50 mM Tris-HCl (pH 7.5, lanes 7–12) or 50 mM Tris-HCl (pH 8.5, lanes 13–18), together with 1 mM DTT, 0.15% Tween20 and 0.2 mg/ml BSA. The pull-down assays were performed as described in Figure 7. (C) The pull-down assay was performed in buffer containing 50 mM Tris-HCl (pH 8.5), 1 mM DTT, 0.15% Tween20 and 0.2 mg/ml BSA.
RecO-dependent localization of SSB on ssDNA
The salt concentration, length of DNA and pH often affect the DNA-binding properties of proteins that bind to DNA. To clarify whether SSB binds to ssDNA directly or through its interaction with RecO on ssDNA, the conditions under which RecO but not SSB binds efficiently to ssDNA were examined. Neither high KCl concentration (up to 1 M) nor ssDNA length within the range of 10–60 nucleotides affected the ssDNA-binding properties of these two proteins (data not shown). However, at a low concentration of MgCl2, the amount of pulled-down RecO was slightly increased whereas the amount of pulled-down SSB was unaffected (Figure 6B, Supplementary Figure S3). In addition, RecO bound to ssDNA more strongly than SSB at pH 8.5 (Figure 6B, Supplementary Figure S3A and S3C). Therefore, we performed a pull-down assay again at pH 8.5 in the absence of MgCl2 (Figure 6C). When the concentration of each protein was increased up to 10 μM, RecO was pulled down efficiently but SSB was not (Figure 6C, lanes 1–6). However, in the presence of both RecO and SSB, SSB was efficiently pulled down with RecO (Figure 6C, lanes 7–12) although SSB alone was hardly pulled down under these conditions (Figure 6C, lanes 4–6). In addition, the amount of pulled-down SSB increased as a function of the RecO concentration (Figure 6C, lanes 7–9). Similar results were also obtained in the presence of 1 and 10 mM MgCl2 at pH 8.5 (Supplementary Figure S3B and S3D). These results indicated that SSB bound to ssDNA indirectly through its interaction with the RecO-ssDNA complex.
Taken together, the above results indicated that, although SSB bound to ssDNA is displaced by RecO, it remains indirectly attached to ssDNA through its interaction with RecO in the RecO-ssDNA complex. One of the functions of RecO could be to rearrange SSB on ssDNA, resulting in the formation of a SSB-RecO-ssDNA complex. This complex would result in the strong inhibition of the ATPase activity of RecA (Figure 5B), indicating that the activity of RecA on SSB-coated ssDNA requires other factors, in addition to RecO.
The restoration of the ssDNA-dependent ATPase activity of RecA by RecR
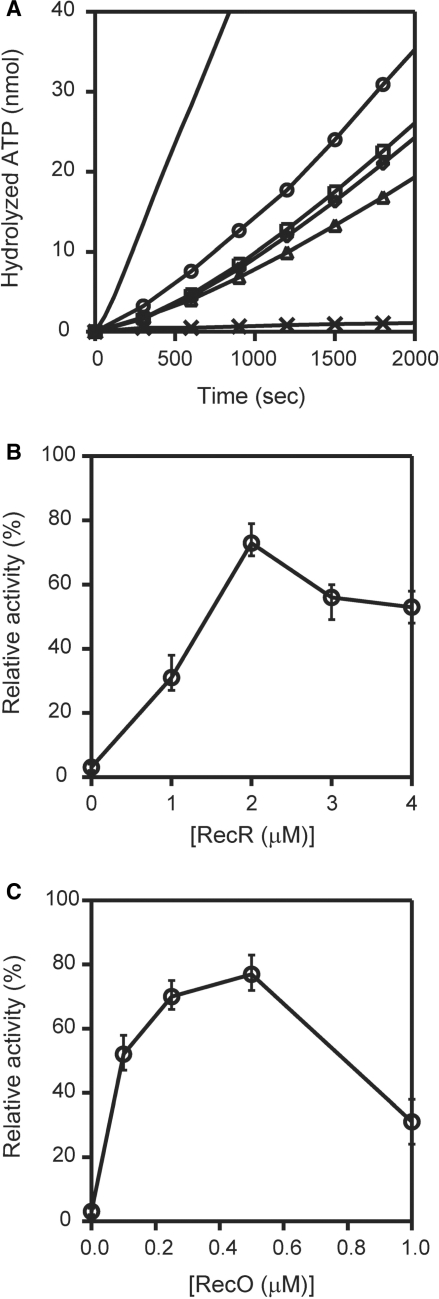
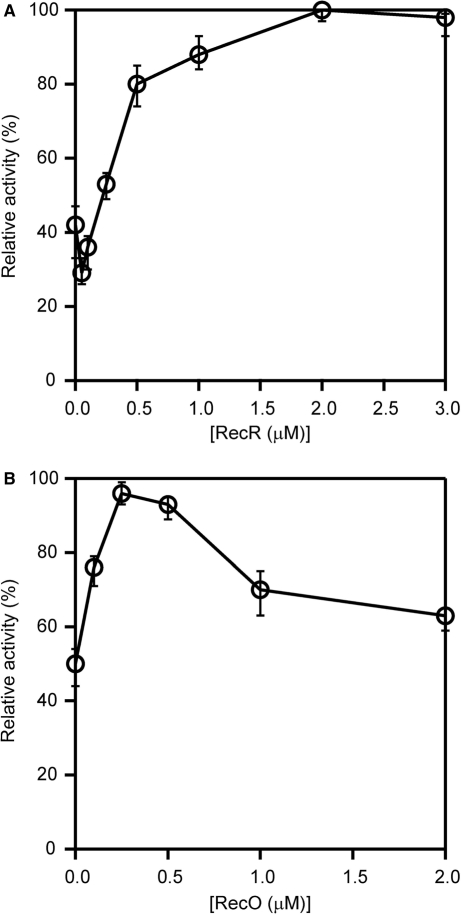
SSB inhibited the ssDNA-dependent ATPase activity of RecA, which was not restored by the addition of RecO. Since RecR interacts with RecO, we investigated whether RecR functioned as an additional factor in the displacement of SSB from the ssDNA. When we added RecR to a reaction mixture containing both RecO and SSB, in which ATPase activity was inhibited, we observed a dramatic restoration of the ATPase activity (Figure 7A), which was consistent with published results (17). This result indicated that the addition of RecR was necessary to restore the ATPase activity of RecA in the presence of SSB and RecO. Next, we determined the concentrations and the molar ratio of RecO and RecR in the presence of SSB that are required for the optimum ATPase activity of RecA. We added increasing concentrations of RecR (0–4 μM) to the reaction mixtures containing fixed concentrations of RecA, SSB and RecO (1 μM). We then compared the resulting ATPase activity to that of RecA alone and expressed it as a relative activity (Figure 7B). We restored a maximal ATPase activity (∼80%) at a concentration of 2 μM RecR. Interestingly, the ATPase activity decreased when the concentration of RecR exceeded 2 μM (to ∼60%). These results indicated that there is an optimum ratio of RecO and RecR (1:2) at which a maximum ATPase activity of RecA is restored in the presence of SSB. We also examined the relative ATPase activity of RecA in the presence of increasing concentrations of RecO (0–1 μM), maintaining the concentrations of RecA, SSB and RecR at a constant 1 μM. We observed a maximal ATPase activity (∼80%) when the concentration of RecO was 0.5 μM. These results indicated that the maximal restoration of the RecA ATPase activity in the presence of SSB occurs when RecO and RecR are present in a molar ratio of approximately 1:2, which suggests the formation of a functional RecO-RecR complex with a stoichiometry of 1:2. Under normal conditions, RecR interacts with RecO but not ssDNA, therefore these results also suggested that either the interaction of RecR weakens the ssDNA-binding affinity of RecO or the interaction of RecR with RecO causes a functional modulation of RecO to relieve the inhibition of RecA.
Figure 7.
The removal of inhibition of RecA by RecR. (A) The ordinate axis is the amount of hydrolyzed ATP. The ssDNA-dependent ATPase activity of RecA was measured in the presence of 1 μM RecA (line), 1 μM SSB and 1 μM RecO (cross); 1 μM RecA, 1 μM SSB, 1 μM RecO and 1 μM RecR (triangle); 1 μM RecA, 1 μM SSB, 1 μM RecO and 2 μM RecR (circle); 1 μM RecA, 1 μM SSB, 1 μM RecO and 3 μM RecR (square); and 1 μM RecA, 1 μM SSB, 1 μM RecO and 4 μM RecR (diamond). Proteins were incubated with 340-mer poly (dC) in following order: SSB, RecO and RecR; RecA was then added to the mixture and the ATPase activity was measured. The data represent the averages of three independent experiments. (B) The proteins were incubated with a 340-mer poly (dC) in the following order: 1 μM SSB, 1 μM RecO and the indicated concentrations of RecR and then 1 μM RecA was added to the mixture, and ssDNA-dependent ATPase activity was measured. The activity relative to the activity of RecA alone (defined as 100%) was plotted against RecR concentration. (C) The proteins were incubated with a 340-mer poly (dC) in the following order: 1 μM SSB, the indicated concentrations of RecO, and 1 μM RecR, and then 1 μM RecA was added to the mixture and the ssDNA-dependent ATPase activity was measured. The ATPase activity was plotted against RecO concentration as described in (B).
The effect of RecR on the ssDNA-binding properties of RecO
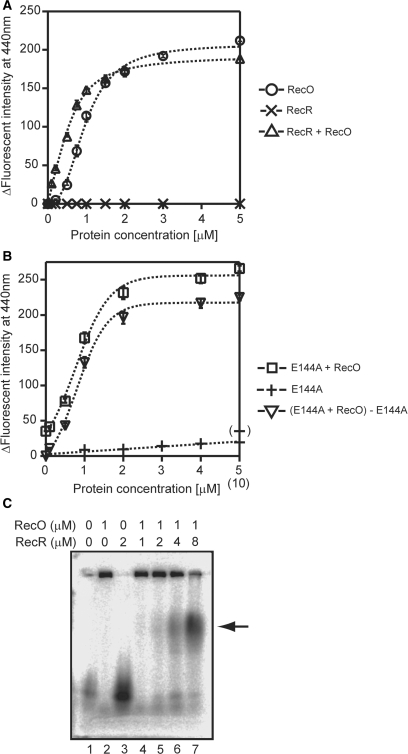
We examined the effect of RecR on the ssDNA-binding activity of RecO to determine whether or not RecR dissociates RecO from ssDNA using fluorophotometry and εDNA. The ΔF in the presence of RecR alone was undetectable (Figure 8A, cross), which suggested that the binding affinity of RecR for ssDNA was very weak; thus, any value of ΔF obtained in the presence of both RecO and RecR would reflect the binding of RecO to the ssDNA. We found that the binding ability of the RecO-RecR complexes did not differ dramatically from that of RecO (Figure 8A, triangle). However, the sigmoidal binding curve of RecO changed to a common curve in the presence of RecR. These results suggested that the interaction of RecR with RecO does not result in a dissociation of RecO from ssDNA, but changes the mode of binding of RecO to ssDNA.
Figure 8.
The effect of RecR on the ssDNA-binding activity of RecO. (A) The indicated concentrations of RecO were incubated with 10 μM εDNA at 25°C for 5 min in the absence (circle) or presence (triange) of 10 μM RecR. The effect of RecR alone is represented by a cross. The measurement conditions were the same as described for Figure 4. (B) The indicated concentrations of RecO were incubated with 10 μM εDNA at 25°C for 5 min in the presence of 10 μM RecR_E144A (square). The effect of RecR_E144A alone (0–5 µM) is represented by a plus, and the effect of 10 μM RecR_E144A is shown by a bracketed plus. The reverse triangle represents the binding curve of RecO to ssDNA with the ΔF of the 10 μM RecR_E144A alone (∼35) subtracted. (C) An agarose gel retardation assay of the RecO-RecR complex. A 5′-33P labeled 66-mer ssDNA was incubated with the indicated concentrations of RecO and RecR for 5 min at 50°C, and then the samples were analyzed by electrophoresis on a 2.5% agarose gel.
To clarify whether the interaction of RecR with RecO modulated the ssDNA-binding properties of RecO, we employed a RecR_E144A (E144A) mutant protein. Previously, we showed that E144A did not bind to RecO (17). In this study, we found that E144A slightly bound to the ssDNA, probably due to the removal of a negative charge, and increased the ΔF to ∼35 (Figure 8B, plus); therefore, this value was subtracted from the binding curve of RecO (Figure 8B, reverse triangle). The original binding curve of RecO in the presence of 10 μM E144A was also depicted (Figure 8B, square). The binding curves of RecO in the presence of E144A were sigmoidal and were similar to that of RecO alone. This result indicated that the interaction of RecR with RecO was necessary for the modification of ssDNA-binding properties of RecO.
The effect of RecR on the ssDNA-binding properties of RecO was also confirmed by an agarose gel retardation assay. When RecO was incubated with ssDNA, a large complex was formed and was trapped in the loading well (Figure 8C, lane 2). RecR alone did not show clear band shift because of its weak affinity to ssDNA (lane 3). In the presence of an equal concentration of RecO and RecR, a small amount of a new band appeared (indicated by an arrow) in addition to the trapped complexes in the loading well (lane 4). The intensity of this new band dramatically increased with increasing concentrations (1 to 4 µM) of RecR (lanes 4–7). In contrast, the intensity of the trapped complexes gradually decreased. Therefore, the new band was thought to be the RecR-RecO-ssDNA complex. Taken together, these results suggested that the interaction of RecR with the RecO-ssDNA complex modulated the ssDNA-binding properties of RecO.
The effect of RecR and RecO on the ssDNA-dependent ATPase activity of RecA
In the absence of SSB, RecO alone inhibited the ssDNA-dependent ATPase activity of RecA, probably because it binds to ssDNA (Figure 5B). We were interested in whether RecR affected the inhibition of the ATPase activity by RecO. In the absence of RecR, 0.25 μM RecO inhibited the ssDNA-dependent ATPase activity of RecA to about 40% of that of RecA alone (Figure 9A). In the presence of 0.5 μM RecR, a drastic recovery of inhibition was observed, and the recovery rate became saturated. These results suggested that RecO and RecR function at a 1:2 molar ratio. The addition of the higher concentrations of RecR resulted in nearly 100% recovery, indicating that excess RecR does not inhibit ATPase activity in the presence of RecO.
Figure 9.
The effect of changing the molar ratio of RecO and RecR on the activity of RecA in the absence of SSB. (A) The proteins were incubated with a 340-mer poly (dC) in the following order: 0.25 μM RecO, the indicated concentrations of RecR and then 1 μM RecA was then added to the mixture and the ssDNA-dependent ATPase activity was measured. The RecA activity relative to that of RecA alone (defined as 100%) was plotted against the concentration of RecR. The data represent the average of three independent experiments. (B) The proteins were incubated with a 340-mer poly (dC) in the following order: the indicated concentrations of RecO, 1 μM of RecR and then 1 μM RecA was added to the mixture and the ssDNA-dependent ATPase activity was measured. RecA activity was plotted as described in (A).
Although the binding affinity of RecR for ssDNA is very weak, RecR at a concentration of 1 µM alone inhibited the ssDNA-dependent ATPase activity of RecA to about 50% (Figure 9B). The addition of RecO up to a concentration of 0.5 μM overcame this inhibition and restored the ATPase activity of RecA to almost 100%. However, a further increase in the concentration of RecO had an inhibitory effect. These results also indicated that the optimum molar ratio of RecO and RecR is approximately 1:2. The inhibitory effect of the high concentrations of RecO may be caused by the binding of excess RecO to ssDNA. Earlier results showed that at concentrations above 1 μM, RecO inhibited the ssDNA-dependent ATPase activity of RecA to about 20% (Figure 5B). In the presence of RecR, however, the activity did not fall below 60% even when the concentration of RecO was increased to 2 μM (Figure 9B). Excess RecR did not inhibit activity in the presence of RecO (Figure 9A). These results indicated that RecOR complexes can overcome the inhibitory effect of an excess of either protein on the activity of RecA. The RecOR complexes may stabilize the active RecA filaments.
DISCUSSION
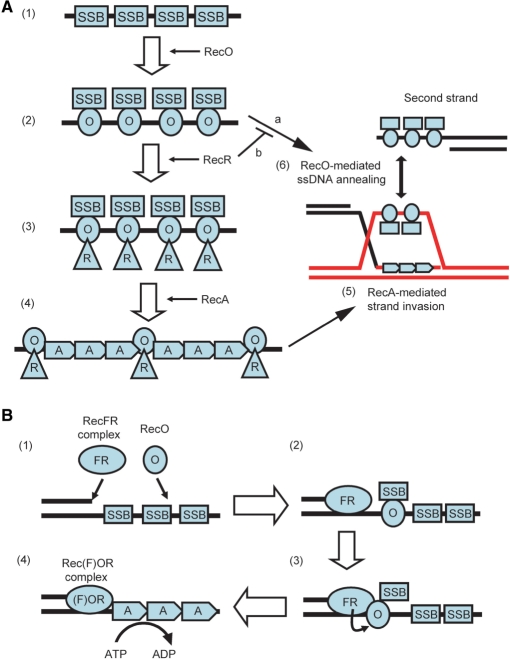
In this study, we examined the mechanism of RecA loading onto the SSB-coated ssDNA by the T. thermophilus RecO and RecR. Based on the results of this study, we propose a model for the function of RecO and RecR, which is depicted in Figure 10A. In this model, the first step of RecA loading onto the SSB-coated ssDNA would require that RecO disrupt the existing SSB-ssDNA complex and then bind to the ssDNA (Figure 10A-2). Although this step requires the high-affinity binding of ssDNA by RecO, our data indicate that the ssDNA-binding affinity of RecO is comparable to that of SSB and that RecO preferentially binds to the ssDNA in the presence of SSB (Figure 3A and B). Moreover, we demonstrated by using electron microscopy that the RecO-ssDNA-like structure was predominantly formed even though SSB was bound to the ssDNA prior to RecO (Figure 4D and F).
Figure 10.
A model for the displacement of SSB by RecO, RecR and RecA. (A): (1) ssDNA regions are coated by SSB immediately. (2) RecO interacts with SSB, remodels the SSB-ssDNA complex and binds to ssDNA directly. The displaced SSB interacts with RecO in the RecO-ssDNA complex. If a complementary ssDNA strand exists, the ssDNAs are annealed by RecO (a). (3) RecR interacts with RecO in the SSB-RecO-ssDNA complex. Then RecR modulates the ssDNA-binding mode of RecO and inhibits the ssDNA annealing activity of RecO (b). At this time, SSB may be released from RecO. (4) RecO and RecR remain on ssDNA as the RecOR complex, where they assist the formation of the RecA filament. RecA makes nucleoprotein filament on ssDNA, resulting in SSB dissociation. The RecOR complex may stabilize the RecA filament probably by binding to the end of the RecA filament. (5) RecA-mediated strand invasion occurs. The strand which is displaced by RecA is captured by SSB and RecO like in (1) and (2). (6) The displaced strand is annealed with a second strand by SSB and RecO. (B): (1) When dsDNA-ssDNA junctions are generated as a consequence of DNA damage, the RecFR complex binds to the dsDNA-ssDNA junction, and RecO interacts with the SSB-coated ssDNA. (2) RecO displaces SSB on the SSB-coated ssDNA. The released SSB remains on the ssDNA indirectly via its interaction with RecO. (3) RecR, which is in a RecFR complex at the dsDNA-ssDNA junction site, interacts with RecO that attaches SSB to the ssDNA at this site. (4) The RecOR complex, which may also associate with RecF, facilitates the displacement of SSB from the ssDNA region and the loading of RecA onto the ssDNA. This may be enhanced by extension of RecA filaments.
RecO inhibited the ssDNA-dependent ATPase activity of RecA, similar to SSB, which was probably because of its high-affinity ssDNA-binding activity (Figure 5B); however, the magnitude of inhibition by RecO was much less than that caused by SSB. Therefore, if RecO causes a complete dissociation of SSB from ssDNA the ATPase activity of RecA should be restored to levels seen in the presence of RecO alone. However, in the presence of both RecO and SSB, the ATPase activity of RecA was strongly inhibited (Figure 5B). In addition, the results of the pull-down assay indicated that RecO and SSB co-localize on ssDNA (Figure 6A) and that the localization of SSB on ssDNA is strongly depended on the presence of RecO (Figure 6C, Supplementary Figure S3B and S3D). These results indicated that SSB binds to RecO on the ssDNA (Figure 10A-2). Thus, we have demonstrated that RecO remodels the SSB-ssDNA structure during the first step of RecA loading onto SSB-coated ssDNA.
In relation to this observation, it has been reported that RecO mediates annealing of SSB-coated ssDNA (6), which also suggests the remodeling of the SSB-ssDNA structure by RecO. In eukaryotes, the functional homologue of RecO, Rad52, can also anneal RPA-coated ssDNA (6). This activity is involved in the single strand annealing (SSA) pathway, which is one of the DSB repair pathways. Therefore, the ssDNA-annealing activity of RecO may be involved in an SSA-like pathway in bacteria, although the pathway, including the mechanism of RecO involvement, has not been defined yet (Figure 10A-6). In this study, we have shown that RecR modulates the ssDNA-binding mode of RecO (Figure 8). In relation to this result, it has been reported that RecR inhibits the ssDNA-annealing activity of RecO (6) (Figure 10A-2, line b). Since the SSA pathway is sometimes mutagenic, the ssDNA-annealing activity of RecO may be inhibited until RecA-mediated strand invasion occurs. In this case, RecO would be involved in two repair pathways like Rad52; the RecA-independent SSA-like pathway (Figure 10A, arrow a) and the RecA-dependent strand invasion pathway (Figure 10A-2, 3, 4, 5), and the selection of the operative pathway might depend on the interaction of RecR with RecO (Figure 10A-2, 3).
This notion is supported by observations made by us and others. We observed a dramatic restoration of the ATPase activity of RecA after the addition of RecO and RecR when linear ssDNA was used as a substrate (Figure 7A). This result is consistent with a previous study by Shan and Bork et al. (14,16). They also demonstrated that RecR at least is associated with the end of the RecA nucleoprotein filament, and suggested that the RecOR complex preferentially is associated with the end of the RecA filament and stabilized RecA filament to prevent dissociation of the RecA monomer from the end of the RecA filament, as depicted in Figure 10A-4. In this study, we also showed that the interaction between RecR and RecO did not dissociate RecO from ssDNA (Figure 8A) and that the addition of RecO and RecR at an appropriate ratio could overcome the inhibitory effect of excess RecO or RecR on the ATPase activity of RecA (Figure 9). These results suggested that the interaction of RecR with RecO stabilizes the formation of the active RecA filament and enhances the activity. Therefore, it is plausible that the interaction of RecR with the RecO-SSB complex induces the displacement of SSB from the RecO-ssDNA complex (see below) and that the RecO-RecR complex then functions to facilitate the nucleation of RecA on ssDNA.
Once a stable RecA filament is formed, RecA-mediated strand invasion can occur (Figure 10A-5). It has been reported that the strand displaced by Rad51-mediated strand invasion is captured by RPA and annealed to the second ssDNA strand by Rad52 (7). This phenomenon was also demonstrated by using bacterial RecO and SSB instead of Rad52 and RPA (7). Therefore, the ssDNA-annealing activity of RecO could be exerted after RecA-mediated strand invasion (Figure 10A-6). Recently, it has been reported that BRCA2 homologues, Brh2 and CeBRC-2, can also anneal RPA-coated ssDNA (8,9). These results suggested that the ssDNA annealing activity of the mediator protein is conserved from bacteria to eukaryotes. Since RecO and BRCA2 homologues also have some structural similarity, the mechanism through which these two factors remodel SSB/RPA-ssDNA structure may also be conserved (see below).
RecF is not necessary to relieve the inhibition of RecA activity by SSB when linear ssDNA is used as a substrate. When gapped DNA is used as a substrate, however, the addition of RecF is important for the efficient recovery of RecA activity (13), suggesting that RecF functions to allow the dsDNA-ssDNA junction-specific loading of RecA. Therefore, we propose a model of junction-specific loading of RecA by RecO, RecR and RecF (Figure 10B). We previously reported that RecR preferentially forms a RecFR complex in the presence of RecF, RecO and RecR (17), and others reported that the RecFR complex of E. coli binds to the dsDNA-ssDNA junctions (13). Therefore, RecR of the RecFR complex at the dsDNA-ssDNA junction site could interact with RecO in the SSB-RecO complex in this region (Figure 10B-3). In our previous study, we identified an acidic cluster region of RecR as the RecO-binding site (17). In accordance with this result, the crystal structure of the Deinococcus radiodurans (dr) RecOR complex has been reported recently, in which the acidic cluster of drRecR interacts with the positively charged OB-fold of drRecO (28). In contrast to RecO (pI = 10.7), both RecR (pI = 5.3) and SSB (pI = 5.1) are acidic proteins. Therefore, it is plausible that the acidic cluster region of RecR competes with SSB for binding to RecO. In support of this idea, the C-terminal region of SSB is especially acidic and has been reported to be the most plausible RecO-binding site (29). Thus, RecR would affect the interaction between RecO and SSB through its interaction with RecO, which would facilitate both the displacement of SSB from RecO (Figure 10B-3) and the nucleation of RecA on the ssDNA (Figure 10B-4). In general, SSB strongly inhibits the nucleation of RecA. However, once RecA nucleates on ssDNA, SSB stimulates the extension of the RecA filament. Thus, filament extension of RecA would cause further displacement of SSB from ssDNA (Figure 10B-4).
In the fluorescent DNA-binding assay, a decrease in ΔF was observed after the addition of <1 μM RecO (Figure 3B). The ssDNA-binding properties of drSSB have been analyzed in detail (30,31). These reports indicated that the site size of drSSB on ssDNA is ∼50 nucleotides (nt). Since D. radiodurans is related to T. thermophilus, the site size of ttSSB would be similar to that of drSSB. In contrast to SSB, the site size of RecO on ssDNA has not been determined. However, an analysis of the drRecO crystal structure suggests that the site size of RecO on DNA is much smaller than that of SSB (∼15 nt) (32). Therefore, RecO binding to ssDNA may cause the release of a large amount of ssDNA from the SSB-ssDNA complex, resulting in a decrease in ΔF.
This is the first article that has demonstrated the structure of the RecO-ssDNA complex by electron microscopy. RecO covered the entire ssDNA region and formed a filament-like structure (Figure 4B and C). Most of the RecO-ssDNA complexes formed linear structures, although we used circular M13 ssDNA (Table 1). We also observed branched shaped structures in which the complexes contained more than two M13 ssDNAs. This may relate to the ssDNA annealing activity of RecO, which is expected to anneal a partial complementary sequence in M13 ssDNA. In this case, the intramolecular annealing would result in a linear structure that partially contains dsDNA. Intermolecular annealing would connect two of the M13 ssDNAs to form larger complexes. In contrast to the RecO-ssDNA complexes, the SSB-ssDNA complex formed a bead-like, circular structure (Figure 4A, Table 1). Therefore, the structures of the RecO-ssDNA and SSB-ssDNA complexes were quite different and were easily distinguished.
When M13 ssDNA was incubated with both SSB and RecO, no SSB-ssDNA complexes were observed, even though SSB was incubated with ssDNA prior to the addition of RecO. The observed complexes were similar to the RecO-ssDNA complexes (Figure 4D–F). This result suggests that RecO can bind to SSB-coated ssDNA and rearrange SSB on ssDNA. This is further supported by the results of the pull-down assay, which indicated that displaced SSB still binds to ssDNA indirectly through its interaction with RecO on ssDNA (Figure 6A and B). In addition, we observed that these proteins bound to the ssDNA more stably than each protein alone (Figure 6A and B, Supplementary Figure S3B and S3D). Thereby, the ATPase activity of RecA was inhibited strongly in the presence of both SSB and RecO (Figure 5B). We searched for an intermediate structure that would contain both SSB and RecO on one M13 ssDNA molecule. However, no intermediate structures were observed. RecO may rearrange SSB very efficiently and once the rearrangement occurs, SSB may not be able to bind to the ssDNA again. Thus, the complexes shown in Figure 4D–F must be SSB-RecO-ssDNA complexes. Although remarkable differences were not observed between the SSB-RecO-ssDNA and RecO-ssDNA complexes, minor structural changes may have occurred that were eclipsed by the platinum coat. Further investigations are required to determine how SSB interacts with RecO on the ssDNA.
One of the functions of RecO seems to be the rearrangement of SSB on ssDNA for subsequent RecA nucleation with RecR. The structural similarity of RecO and SSB supports this rearrangement function of RecO. The crystal structures of drRecO and drSSB have been determined (32–34). Interestingly, both drRecO and drSSB have an oligonucleotide-/oligosaccharide-binding fold (OB-fold) that participates in ssDNA binding (35). And while one could hypothesize that the OB-fold of RecO plays a role in the binding of ssDNA, there is currently no evidence for this. However, in eukaryotes RPA and BRCA2 also have OB-folds (36). In addition, BRCA2 has been reported to stimulate the loading of Rad51onto the RPA-coated ssDNA (3,9,36). BRCA2 has structural and functional features that are similar to RecO and RecR, which supports our hypothesis and suggests that the fundamental mechanism of loading of RecA (or Rad51) onto SSB (or RPA)-coated ssDNA may be conserved from prokaryotes to higher eukaryotes. It is unknown whether or not Rad52 also has an OB-fold because the entire structure of Rad52 has not yet been determined. However, the undetermined C-terminal region is involved in the interaction of RPA (37), raising the possibility that Rad52 has an OB-fold. If this were true, our hypothesis would be more conclusive.
In this study, to analyze RecA loading onto SSB-coated ssDNA by RecOR, we measured the ATPase activity of RecA at steady state (last 5 min of the measurement) and examined which protein bound to ssDNA directly after at least 5-min incubation. In contrast, the initial state of RecA loading by RecOR has been analyzed in detail by Hobbs et al. (29). Based on kinetic studies of ATP hydrolysis by RecA in the presence of RecO, RecR and SSB or a SSB variant that lacks 8 amino acids from its C-terminus, they proposed that RecO associates with the C-terminal region of SSB on ssDNA. Their report indicated that the recovery of ATPase activity of RecA by RecOR was accompanied by a lag phase. This lag might be due to the inhibition of ssDNA binding of RecOR by SSB since ssDNA binding of the RecOR complex seems to be necessary for the recovery of RecA activity. In fact, a lag was also observed in our study (Figure 7A). Therefore, we speculate that RecO primarily associates with the acidic C-terminal region of SSB on ssDNA and then remodels the SSB-ssDNA complex with RecR to facilitate filament formation of RecA on ssDNA. As this process would require some time, it might be responsible for observed lag phases.
Both SSB and RecO inhibited the ssDNA-dependent ATPase activity of RecA (Figure 5B). However the inhibitory effect of RecO on the ATPase activity of RecA was weaker than that of SSB, even though RecO had a comparable ssDNA-binding affinity to SSB. The differences in the DNA-binding mode between RecO and SSB may explain this discrepancy. ttSSB has two OB-folds per polypeptide chain (Satoh and coworkers, PDB ID: 2CWA) and forms a stable dimer. Thus, the SSB functional unit has four OB-folds. In contrast, E. coli SSB has one OB-fold per polypeptide chain and forms a stable tetramer (38). Although the oligomeric state of ttSSB and E. coli SSB are different, the tertiary structures of the ttSSB dimer and the E. coli SSB tetramer are quite similar. The crystal structure of the E. coli SSB-ssDNA complex revealed that the ssDNA wraps around the four OB-folds of the E. coli SSB tetramer (39). In contrast to SSB, RecO exists as a monomer in solution and binds to the ssDNA via a single OB-fold. These differences in the mode of DNA binding may reflect the degree of inhibition of the ssDNA-dependent ATPase activity of RecA.
The inhibition of the ssDNA-dependent ATPase activity of RecA by SSB or RecO was efficiently removed when RecO and RecR were present in a 1:2 molar ratio (Figures 7 and 9). This result suggested that RecO and RecR functions at a stoichiometry of 1:2. In accordance with this result, drRecO and drRecR formed a 1:2 complex in the crystal (6). When the ratio of RecO to RecR deviated from 1:2, the inhibition of the ATPase activity of RecA was again observed (Figures 7 and 9B). This could be explained as an excess of protein competing with RecA for binding to the ssDNA, since the inhibitory effect of RecO was higher than that of RecR. It has been reported that RecR can bind to ssDNA, although the binding affinity is very weak (40,41). In contrast, RecO exhibited a high affinity for ssDNA binding, which was comparable to that of SSB. These facts indicate that the ssDNA-binding affinity of RecR is much weaker than that of RecO; therefore, an excess of RecR may not exhibit a strong inhibitory effect in the presence of RecO (Figures 7B and 9A). In the absence of SSB, the inhibition of the ssDNA-dependent ATPase activity of RecA was almost completely removed by RecO and RecR (Figure 9). In contrast, the activity was only restored to 80% in the presence of SSB (Figure 7). Even if SSB is completely displaced from the ssDNA by the sequential action of the RecF pathway proteins, it may re-associate with ssDNA or RecO and the differences in restoring the ATPase activity of RecA may be related to the presence of the displaced SSB.
The shape of the RecO-binding curve for ssDNA was sigmoidal, suggesting that the binding of RecO to the ssDNA is cooperative. RecO exists as a monomer and does not associate in solution. However, the results of the electron microscopy indicated that RecO bound to the entire ssDNA and formed a filament like structure (Figure 4B and C). Therefore, several RecO molecules may bind cooperatively to the ssDNA and form filaments through protein–protein interactions, similar to RecA. It is clear that further investigations are required to determine whether or not such RecO filaments are formed. Alternatively, the cooperative binding of RecO on the ssDNA may relate to binding mode of RecO to ssDNA. RecO can anneal two ssDNAs (6,42). The surface of the RecO protein also has two distinct basic regions that are predicted to participate in DNA binding (32). Therefore, it is possible that RecO binds two ssDNAs simultaneously. In this way, the resulting binding curve would be sigmoidal if the two binding sites have apparent differences in their binding affinities. This idea is consistent with the results of the gel retardation assay, in which a large ssDNA-RecO complex was trapped in the loading well (Figure 8C, lane 2). In addition, some connected RecO-M13 ssDNA complexes were observed in the electron microscopic analysis, although we employed a very low protein concentration to prevent the formation of large RecO-ssDNA complexes. Interestingly, when RecR was added to the large RecO-ssDNA complexes, a protein band corresponding to a new complex was detected (Figure 8C, lanes 4–7). Since the position of this band was different than that of the RecR-ssDNA or RecO-ssDNA complexes, it may represent a RecO-RecR-ssDNA complex. These results indicated that the association state of the RecO-ssDNA complex was changed by the interaction of RecR with RecO. Since drRecR interacted with the OB-fold of drRecO in the RecOR crystal structure, RecR may cover one DNA-binding site of RecO and prevent RecO forming a RecO-ssDNA network. In addition, the ssDNA annealing activity of RecO was inhibited by RecR (6), which also suggests that RecR covers at least one of the DNA-binding sites of RecO. In support of this idea, we found that the sigmoidal binding curve of RecO changed to a common curve in the presence of RecR (Figure 8A). In addition, when we used a RecR mutant whose RecO interaction was disrupted, this phenomenon was not observed (Figure 8B). These results implicate RecR in the alteration of the RecO ssDNA-binding activity. Furthermore, the change in the RecO-binding curve induced by RecR seems to reflect the stoichiometric binding of RecOR. If this were true, RecR would significantly enhance the affinity of RecO for ssDNA. Therefore, RecR may cover only a weak ssDNA-binding site of RecO and enhance the net affinity of RecO for ssDNA. We have previously reported that the combination of RecO and the RecR mutant could not restore the inhibition of the ATPase activity of RecA by SSB (17). A RecR-induced functional change in RecO may be important for the subsequent nucleation of RecA on ssDNA and stabilization of the active RecA filaments.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Professor Seiki Kuramitsu for providing the expression plasmids of the T. thermophilus RecF pathway proteins used in this study. This work was supported in part by the Grants-in-Aid from the Japan Society for the Promotion of Science (JSPS) to T.S. and T.M. Funding to pay the Open Access publication charges for this article was also provided by JSPS.
REFERENCES
- 1.Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 3.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 4.Song B, Sung P. Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J. Biol. Chem. 2000;275:15895–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- 5.Sugiyama T, Kowalczykowski SC. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J. Biol. Chem. 2002;277:31663–31672. doi: 10.1074/jbc.M203494200. [DOI] [PubMed] [Google Scholar]
- 6.Kantake N, Madiraju MV, Sugiyama T, Kowalczykowski SC. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proc. Natl Acad. Sci. USA. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugiyama T, Kantake N, Wu Y, Kowalczykowski SC. Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. Embo. J. 2006;25:5539–5548. doi: 10.1038/sj.emboj.7601412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazloum N, Zhou Q, Holloman WK. DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry. 2007;46:7163–7173. doi: 10.1021/bi700399m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petalcorin MI, Sandall J, Wigley DB, Boulton SJ. CeBRC-2 stimulates D-loop formation by RAD-51 and promotes DNA single-strand annealing. J. Mol. Biol. 2006;361:231–242. doi: 10.1016/j.jmb.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 10.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 11.Tseng YC, Hung JL, Wang TC. Involvement of RecF pathway recombination genes in postreplication repair in UV-irradiated Escherichia coli cells. Mutat. Res. 1994;315:1–9. doi: 10.1016/0921-8777(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 12.Rocha EP, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005;1:e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 14.Bork JM, Cox MM, Inman RB. The RecOR proteins modulate RecA protein function at 5′ ends of single-stranded DNA. Embo. J. 2001;20:7313–7322. doi: 10.1093/emboj/20.24.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Webb BL, Cox MM, Inman RB. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell. 1997;91:347–356. doi: 10.1016/s0092-8674(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 16.Shan Q, Bork JM, Webb BL, Inman RB, Cox MM. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- 17.Honda M, Inoue J, Yoshimasu M, Ito Y, Shibata T, Mikawa T. Identification of the RecR Toprim domain as the binding site for both RecF and RecO. A role of RecR in RecFOR assembly at double-stranded DNA–single-stranded DNA junctions. J. Biol. Chem. 2006;281:18549–18559. doi: 10.1074/jbc.M512658200. [DOI] [PubMed] [Google Scholar]
- 18.Umezu K, Chi NW, Kolodner RD. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl Acad. Sci. USA. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- 20.Hegde SP, Qin MH, Li XH, Atkinson MA, Clark AJ, Rajagopalan M, Madiraju MV. Interactions of RecF protein with RecO, RecR, and single-stranded DNA binding proteins reveal roles for the RecF-RecO-RecR complex in DNA repair and recombination. Proc. Natl Acad. Sci. USA. 1996;93:14468–14473. doi: 10.1073/pnas.93.25.14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inoue J, Shigemori Y, Mikawa T. Improvements of rolling circle amplification (RCA) efficiency and accuracy using Thermus thermophilus SSB mutant protein. Nucleic Acids Res. 2006;34:e69. doi: 10.1093/nar/gkl350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honda M, Rajesh S, Nietlispach D, Mikawa T, Shibata T, Ito Y. Backbone 1H, 13C, and 15N assignments of a 42 kDa RecR homodimer. J. Biomol. NMR. 2004;28:199–200. doi: 10.1023/B:JNMR.0000013821.02982.01. [DOI] [PubMed] [Google Scholar]
- 23.Shigemori Y, Mikawa T, Shibata T, Oishi M. Multiplex PCR: use of heat-stable Thermus thermophilus RecA protein to minimize non-specific PCR products. Nucleic Acids Res. 2005;33:e126. doi: 10.1093/nar/gni111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuramitsu S, Hiromi K, Hayashi H, Morino Y, Kagamiyama H. Pre-steady-state kinetics of Escherichia coli aspartate aminotransferase catalyzed reactions and thermodynamic aspects of its substrate specificity. Biochemistry. 1990;29:5469–5476. doi: 10.1021/bi00475a010. [DOI] [PubMed] [Google Scholar]
- 25.Menetski JP, Kowalczykowski SC. Interaction of recA protein with single-stranded DNA. Quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 1985;181:281–295. doi: 10.1016/0022-2836(85)90092-0. [DOI] [PubMed] [Google Scholar]
- 26.Chrysogelos S, Griffith J. Escherichia coli single-strand binding protein organizes single-stranded DNA in nucleosome-like units. Proc. Natl Acad. Sci. USA. 1982;79:5803–5807. doi: 10.1073/pnas.79.19.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cox MM, Lehman IR. Enzymes of general recombination. Annu. Rev. Biochem. 1987;56:229–262. doi: 10.1146/annurev.bi.56.070187.001305. [DOI] [PubMed] [Google Scholar]
- 28.Timmins J, Leiros I, McSweeney S. Crystal structure and mutational study of RecOR provide insight into its mode of DNA binding. Embo. J. 2007;26:3260–3271. doi: 10.1038/sj.emboj.7601760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobbs MD, Sakai A, Cox MM. SSB protein limits RecOR binding onto single-stranded DNA. J. Biol. Chem. 2007;282:11058–11067. doi: 10.1074/jbc.M611007200. [DOI] [PubMed] [Google Scholar]
- 30.Witte G, Urbanke C, Curth U. Single-stranded DNA-binding protein of Deinococcus radiodurans: a biophysical characterization. Nucleic Acids Res. 2005;33:1662–1670. doi: 10.1093/nar/gki310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eggington JM, Kozlov AG, Cox MM, Lohman TM. Polar destabilization of DNA duplexes with single-stranded overhangs by the Deinococcus radiodurans SSB protein. Biochemistry. 2006;45:14490–14502. doi: 10.1021/bi061178m. [DOI] [PubMed] [Google Scholar]
- 32.Leiros I, Timmins J, Hall DR, McSweeney S. Crystal structure and DNA-binding analysis of RecO from Deinococcus radiodurans. Embo. J. 2005;24:906–918. doi: 10.1038/sj.emboj.7600582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makharashvili N, Koroleva O, Bera S, Grandgenett DP, Korolev S. A novel structure of DNA repair protein RecO from Deinococcus radiodurans. Structure. 2004;12:1881–1889. doi: 10.1016/j.str.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Bernstein DA, Eggington JM, Killoran MP, Misic AM, Cox MM, Keck JL. Crystal structure of the Deinococcus radiodurans single-stranded DNA-binding protein suggests a mechanism for coping with DNA damage. Proc. Natl Acad. Sci. USA. 2004;101:8575–8580. doi: 10.1073/pnas.0401331101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murzin AG. OB(oligonucleotide/oligosaccharide binding)-fold: common structural and functional solution for non-homologous sequences. Embo J. 1993;12:861–867. doi: 10.1002/j.1460-2075.1993.tb05726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang H, Jeffrey PD, Miller J, Kinnucan E, Sun Y, Thoma NH, Zheng N, Chen PL, Lee WH, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 37.Park MS, Ludwig DL, Stigger E, Lee SH. Physical interaction between human RAD52 and RPA is required for homologous recombination in mammalian cells. J. Biol. Chem. 1996;271:18996–19000. doi: 10.1074/jbc.271.31.18996. [DOI] [PubMed] [Google Scholar]
- 38.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc. Natl Acad. Sci. USA. 1997;94:6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 40.Alonso JC, Stiege AC, Dobrinski B, Lurz R. Purification and properties of the RecR protein from Bacillus subtilis 168. J. Biol. Chem. 1993;268:1424–1429. [PubMed] [Google Scholar]
- 41.Lee BI, Kim KH, Park SJ, Eom SH, Song HK, Suh SW. Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. Embo. J. 2004;23:2029–2038. doi: 10.1038/sj.emboj.7600222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luisi-DeLuca C, Kolodner R. Purification and characterization of the Escherichia coli RecO protein. Renaturation of complementary single-stranded DNA molecules catalyzed by the RecO protein. J. Mol. Biol. 1994;236:124–138. doi: 10.1006/jmbi.1994.1123. [DOI] [PubMed] [Google Scholar]