Abstract
Mismatch Repair (MMR) is closely linked to DNA replication; however, other than the role of the replicative sliding clamp (PCNA) in various MMR functions, the linkage between DNA replication and MMR has been difficult to investigate. Here we use an in vitro DNA replication system based on simian virus 40, to investigate MMR recruitment to replicating DNA. Both DNA replication and MMR proteins are recruited to replicating DNA in an origin-dependent fashion. Primer synthesis is required for recruitment of both PCNA and MMR proteins, but not for recruitment of the single-stranded DNA-binding protein (RPA). Blocking PCNA recruitment to replicating DNA with a p21-based polypeptide blocks PCNA and MMR, but not RPA recruitment. Once PCNA and subsequent proteins required for replication are loaded onto DNA, addition of p21 leaves PCNA on the replicating DNA, but actively displaces MMR proteins. These findings indicate that the MMR machinery is recruited to replicating DNA through its interaction with PCNA, and suggests that this occurs via binding of the MMR proteins to the multi-protein interaction sites on PCNA. These studies demonstrate the utility of this system for further investigation of the role of DNA replication in MMR.
INTRODUCTION
Post-replicative Mismatch Repair (MMR) is crucial for repairing base–base mismatches and insertion/deletion loops (IDLs) generated primarily during DNA replication by nucleotide mis-incorporation and DNA slippage errors (1–4). Mutations in MMR genes are associated with various sporadic cancers as well as hereditary non-polyposis colorectal cancer (5–7). MMR is well studied in Escherichia coli where the entire repair reaction has been reconstituted in vitro and all the required MMR proteins have been identified and purified (8,9). Following DNA replication, mismatches on the DNA are recognized by MutS homodimer. The assembly of MutS–MutL complexes at mismatches activates the downstream endonuclease, MutH, the mispaired nucleotide(s) are removed and DNA is re-synthesized (10,11).
In E. coli, the association of MMR with newly synthesized DNA is due to the requirement for MutH. MutH is dependent on the transiently hemi-methylated nature of newly synthesized DNA for its function in MMR (8). Eukaryotes and bacteria other than gram-negatives, which have homologs for MutS and MutL, have no MutH homolog (12). While the eukaryotic MMR machinery is clearly present at DNA replication foci (13), showing its association with the DNA replication process, the mechanism behind the association of MMR with eukaryotic replication remains unclear. The interaction of the replicative sliding clamp (PCNA) with MutS complexes have been suggested to be a possible mechanism for recruiting MMR to replicating DNA (13–15); however, due to a lack of experimental systems that can concurrently investigate DNA replication and MMR, direct evidence showing PCNA plays a role in recruitment of MMR to replicating DNA has remained lacking.
PCNA has been shown to function in various steps of MMR. PCNA is a processivity factor for DNA polymerases δ and ε (16) and as such is required in the DNA re-synthesis step of MMR (17–20). PCNA has been shown to interact with exonuclease I (EXOI) and to colocalize with EXOI at DNA replication foci (21). PCNA is essential for 3′ to 5′ excision but not for 5′ to 3′ excision. These results suggest differential requirement for PCNA in 3′ to 5′ and 5′ to 3′ nick-directed excision pathways (22,23). However, it is unknown how PCNA functions differentially in the excision steps. PCNA has been shown to interact with MutSα, MutSβ, MutLα complexes in yeast as well as humans (15,24–27). Yeast and human MSH3 and MSH6 proteins contain a conserved PCNA-binding motif, known as the PCNA Interacting Protein motif (PIP box) (13,28,29). PCNA increases the specific binding of yeast MSH2–MSH6 complex to DNA mismatches as shown by gel-mobility shift assays (29), and has been proposed to possibly help target MMR to newly replicated DNA (14). However, there has been no direct data to support this model.
Studies herein utilize a Simian Virus 40 (SV40) in vitro DNA replication system to evaluate the role of PCNA in the recruitment of the MMR machinery to replicating DNA. We observe the recruitment of MutSα and MutSβ complexes along with MutL Homolog 1 (MLH1) to replicating SV40 DNA in a replication origin-dependent manner. This recruitment is dependent on PCNA; and moreover, is dependent on the availability of the conserved multi-protein interaction sites on PCNA. These results are the first direct demonstration that MMR is targeted to newly replicated DNA through its interactions with PCNA, and indicate that SV40 is a viable system for the study of the interaction between DNA replication and MMR.
MATERIALS AND METHODS
Antibodies
SV40-Tag antibodies, pAb419- and pAb101, and human-RPA70 antibody, Mab9 have been described previously (30,31). Anti-PCNA (pc10), -MutS Homolog 2 (MSH2) and –MutS Homolog (MSH3) antibodies were purchased from Santa Cruz. MLH1 antibody was purchased from BD biosciences. MSH6 antibody was ordered from Novus.
Chemicals
[α-32P]-dATP was purchased from GE Health Sciences. Aphidicolin was purchased from Sigma and dissolved in 95% (v/v) ethanol (final concentration 1 mg/ml).
Cell cultures
Human embryonic kidney 293 cells were grown in suspension at 37°C in Jolik's modified Eagle medium (ICN Chemicals) and 5% calf serum (Invitrogen). Hypotonic cell extracts were prepared as described (32). The cell extracts were further clarified by ultra-centrifugation at 100 000g for 30 min.
Plasmids and proteins
The SV40 replication plasmids pSV010 ori(+) and pSV010 ori(−) plasmids have been described previously (33,34). The plasmids were transformed and propagated in E. coli (JM109). Plasmid DNA was isolated by Qiagen DNA extraction kit as per the manufacturer's recommendations. Supercoiled DNA was further purified by velocity sedimentation in a 5–20% sucrose gradient. Tag was expressed by baculovirus infection of High-Five insect cells (Invitrogen) and purified by immunoaffinity chromatography using pAb101 monoclonal antibody as described earlier (35–37). The pGEX-p21C plasmid was a kind gift from Dr Anindya Dutta and pGEX-p21Cmut plasmid was generated by site-directed mutagenesis where amino acid residue 147 (methionine) was substituted to alanine using the Quickchange kit (Stratagene) as per the manufacturer's instructions. The plasmids were transformed into the E.coli expression strain BL21 (DE3) (38). Cells were grown to an OD of 0.6 and then induced with 0.4 mM isopropyl-1-thio-P-d-galactopyranoside (IPTG) for 4 h. The proteins were purified with glutathione-Sepharose beads (GE Biosciences) as described (39). Bacterially expressed PCNA was purified as described (40).
In vitro SV40 DNA replication reaction, gel filtration and protein recruitment assay
In vitro SV40 DNA replication was assembled as previously described (32). Plasmid DNA [pSV010 ori(+) or pSV010 ori(−)] measuring 450–675 ng was incubated with 2.25 µg of Tag, 0.1 mg/ml of bovine serum albumin (BSA), 9.375 µg of creatine phosphokinase (CPK), 450–750 µg of 293 cell extracts and Replication Buffer [30 mM HEPES (pH 7.5), 40 mM creatine phosphate, 7 mM MgCl2, 4 mM ATP, 200 µM CTP, 200 µM UTP, 200 µM GTP, 100 µM dCTP, 100 µM dGTP, 100 µM dTTP, 25 µM dATP and 0.5 mM DTT] in 150 µl. The reaction was incubated at 37°C for 30 min and subjected to gel filtration using a 6 ml Biogel A-50 m (Bio-Rad) or AF-02 (Sooner-Scientific) column in buffer A (25 mM Tris pH 7.5, 4 mM MgCl2, 50 mM NaCl, 1 mM DTT and 100 µM PMSF). The columns were developed with buffer A and 12 drops (∼400 µl) were collected for each fraction. Protein content was evaluated using the Bradford method (Bio-Rad). To evaluate the presence of specific proteins, the fractions were either precipitated using 5–20% trichloroacetic acid, or applied directly to SDS–PAGE, transferred to nitrocellulose and analyzed by immunoblotting. To evaluate DNA synthesis, [α-32P]-dATP was added to the DNA replication reaction. At the indicated times 2 µl was removed, dotted onto DE81 paper (Whatman) and the incorporated [α-32P]-dAMP evaluated after five washes in 0.5 M Na2HPO4 as described (41).
DNA synthesis capacity of isolated replicating complexes
To test the DNA synthesis capability of isolated replication complexes, Replication Buffer, CPK, BSA and radiolabeled [α-32P]-dATP were added to the collected fractions and incubated at 37°C for 45 min. The reaction was terminated by addition of stop buffer (20 mM Tris pH 7.5, 10 mM EDTA pH 8.0, 0.1% SDS, 1 µg/µl of Proteinase K). The DNA products were isolated, separated by 0.8% (w/v) agarose gel electrophoresis and analyzed by phosphorimage autoradiography.
RESULTS
Isolation of active replicating DNA–protein complexes
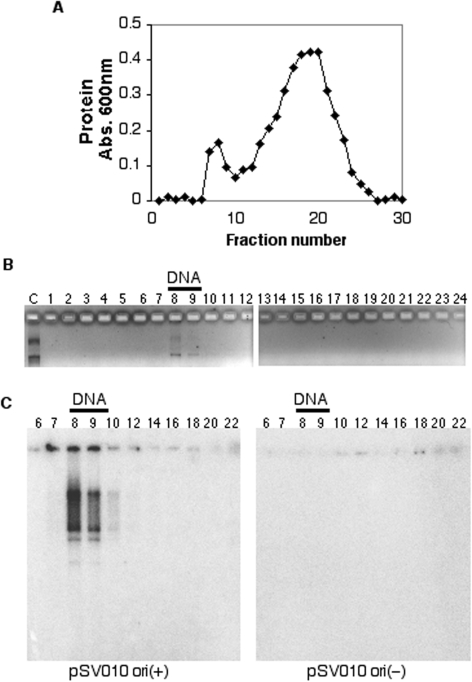
The in vitro SV40 replication system is useful for the study of eukaryotic DNA replication because it requires only one viral protein, large T-Antigen (Tag), and other replication factors are provided by the host cells (42,43). MMR has been shown to function on SV40 replicons in primate and human cells, and cell extracts lacking MMR activity have been shown to support SV40 DNA replication with decreased fidelity; these results clearly indicate that SV40 is subject to MMR both in vivo and in vitro (44,45). Here human cell extracts were used as a source of both DNA replication and MMR proteins. SV40 DNA replication reactions were assembled in vitro, incubated for 30 min to allow for assembly of the active replication complexes and subjected to gel filtration. The fractions collected were analyzed for protein and DNA. We observed two protein peaks corresponding to excluded and included fractions (Figure 1A). As expected, the plasmid DNA was also found to be excluded, in the same fractions as the excluded protein peak (Figure 1A and B). These fractions were analyzed for their ability to support DNA synthesis. The fractions were incubated with dNTPs, NTPs and [α-32P]-dATP to radiolabel any DNA products synthesized. The products were isolated and subjected to gel electrophoresis. DNA synthesis was observed in the plasmid-containing fractions but not in the free protein fractions (Figure 1C, left panel).
Figure 1.
Isolation of active SV40 DNA replication complexes. SV40 DNA replication reactions were assembled and subjected to gel filtration as described in Materials and Methods section. Fractions were collected and equal volumes were analyzed for comparative protein content using the Bio-Rad protein microplate assay, and the absorbance plotted (A). Fractions from a parallel separation were subjected to DNA extraction and agarose gel electrophoresis to detect the plasmid template (B). Fractions were also supplemented with [α-32P]-dATP, NTPs and dNTPs, and incubated at 37°C for 45 min. The extracted DNA products were subjected to agarose gel electrophoresis and autoradiography (C). The left panel shows synthesis in fractions from a separation of reactions assembled with a plasmid containing a functional origin of replication [pSV010 ori(+)]. The right panel shows synthesis in fractions from a separation of reactions assembled with a plasmid containing a mutated non-functional origin of replication [pSV010 ori(−)].
We addressed whether these actively replicating complexes were dependent on the presence of a functional replication origin on the plasmid template. As a control we used a plasmid DNA, pSV010 ori(−), whose origin of replication has been rendered non-functional by deletion of 4 bp within the core origin (33). As expected, this plasmid also fractionated in the excluded peak (as in Figure 1B, data not shown). When these fractions were analyzed for DNA synthesis, unlike the pSV010 ori(+) plasmid, we did not observe DNA synthesis in the pSV010 ori(−) plasmid-containing fractions (Figure 1C, right panel). Since, replication proteins might be bound to the pSV010 ori(−) plasmid, but would not be able to carry out DNA synthesis due to the non-functional origin of DNA replication, we analyzed these fractions for the ability to carry out DNA synthesis on an ori(+) template. Addition of pSV010 ori(+) plasmid and additional Tag to these fractions after gel filtration also did not result in DNA synthesis (data not shown), consistent with the absence of essential replication proteins in these fractions (see below). These results demonstrated that this procedure isolates active replicating complexes in an origin-dependent manner.
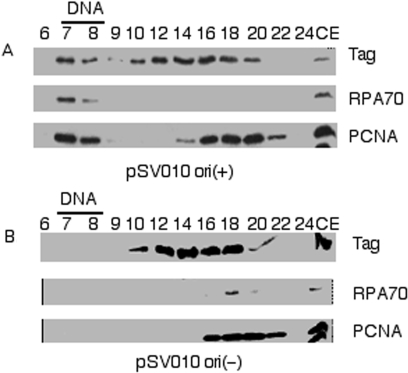
The fractions from the gel filtration separation were evaluated for the presence of known DNA replication proteins. The entire fractions were subjected to TCA precipitation and applied to SDS–PAGE, and subjected to immunoblotting for several known DNA replication proteins (Figure 2). The human DNA replication factors, RPA and PCNA, as well as SV40 Tag, were readily detectable in the fractions containing the origin-containing plasmid DNA, pSV010 ori(+) (Figure 2A). Conversely, these DNA replication proteins could not be detected in the plasmid-containing fractions from identical reactions assembled with plasmid DNA containing the mutated origin, pSV010 ori(−), which contains a 4 nt deletion that inactivates origin function (Figure 2B). We also showed that recruitment of the cellular replication factors was dependent on the addition of the viral replication protein, SV40 Tag (data not shown).
Figure 2.
Known replication proteins associate with replicating SV40 DNA in an origin-dependent manner After separation of replicating SV40 DNA complexes by gel filtration, proteins in the indicated fractions were precipitated and each entire fraction subjected to immunoblotting for large T-Antigen (Tag), Replication Protein A 70 kDa subunit (RPA70) and Proliferating Cell Nuclear Antigen (PCNA). Panel (A) shows immunoblots from a reaction assembled with pSV010 ori(+) and panel (B) shows immunoblots from a reaction assembled with pSV010 ori(−). As a positive control 5 µl of cell extract (10% of the proteins used in the entire reaction) was loaded in the last lane of both panel (A) and (B).
Association of MMR proteins with replicating DNA
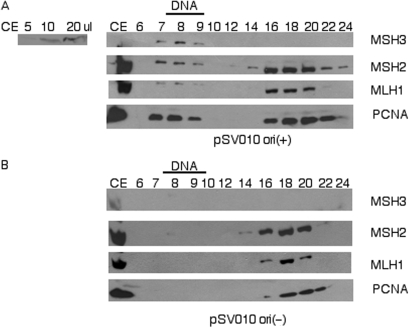
We then addressed whether MMR proteins associate with these SV40 replication complexes. We isolated replicating complexes as described above, and the fractions were analyzed for the presence of MMR proteins by immunoblotting. We observed the presence of MMR proteins in the fractions containing the replicating complexes assembled with pSV010 ori(+) DNA (Figure 3A), but not in the corresponding fractions from reactions assembled with pSV010 ori(−) DNA (Figure 3B). MutS Homolog 2 (MSH2), which is common to both the MutSα and MutSβ complexes, MutS Homolog 3 (MSH3) the other subunit of the MutSβ complex, and MutS Homolog 6 (MSH6, the other subunit of the MutSα complex) (46) were all found to be recruited to the SV40 replicating complexes, but not to the corresponding fractions produced with the mutant origin DNA template (Figure 3 and additional data not shown). Hence, both MutSα and MutSβ are recruited to replicating SV40 DNA complexes in a replication origin-dependent manner. [MSH3 was not detectable in the included protein fractions due to its relatively low levels in cell extracts (see inset to left of Figure 3A), and the fact that included proteins get widely spread across many fractions. However, when recruited to replicating complexes MSH3 could be detected because the recruited proteins get concentrated into only two or three plasmid-containing fractions, thereby maintaining sufficient concentration of MSH3 to be detectable by immunoblotting]. We also evaluated whether MutL complexes were recruited to the replicating complexes by evaluating the presence of MLH1, the common subunit of the MutL complexes (47). As with the MutS complexes, MLH1 was recruited to the replicating complexes in an origin-dependent manner (Figure 3). [In Figure 3A MLH1 appears to show a lower mobility on SDS–PAGE in the plasmid-containing fractions than in free protein-containing fractions. This is due to anomalous electrophoresis in this particular experiment, as can be seen by a similar apparent difference in PCNA mobility, and as no differences in mobility of MLH1 were seen in other separations (Supplemental Figure 1)].
Figure 3.
MMR proteins associate with replicating DNA in an origin-dependent manner. Precipitated protein fractions were analyzed by immunoblotting for MutS Homolog 3 (MSH3), MutS Homolog 2 (MSH2), MutL Homolog 1 (MLH1) and PCNA. Panel (A) shows reactions assembled with pSV010 ori(+) and panel (B) shows reactions assembled with pSV010 ori(−). As a positive control the first lane of both panel (A) and (B) contained 5 µl of cell extract, representing 10% of the proteins in the entire reaction. Inset to the left of Panel A shows a MSH3 immunoblot with increasing levels of cell extract as indicated.
Primer synthesis is required for recruitment of the MMR proteins
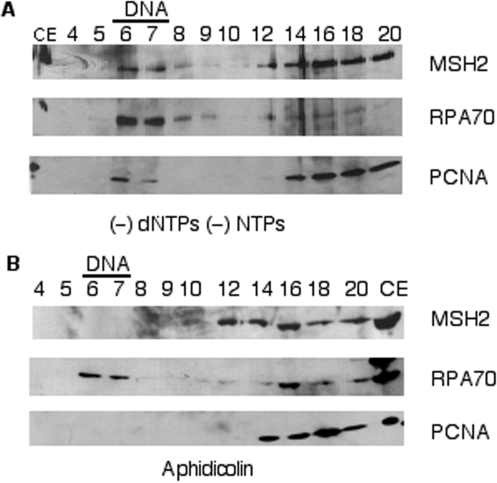
To further understand how MMR proteins are recruited to replicating DNA, we began to evaluate at which stage of DNA replication the MMR proteins are recruited. First, we investigated whether primer synthesis is required for recruitment of MMR proteins. We omitted addition of exogenous dNTPs and NTPs (other than ATP which is required for origin unwinding) and evaluated recruitment of MMR and replication proteins. We observed recruitment of RPA to the replicating DNA (Figure 4A), consistent with expected origin unwinding. We also observed recruitment of PCNA as well as MSH2, which we used as a common marker protein for MMR since it is a common subunit of both MutS complexes (Figure 4A). As primers are required for PCNA loading (48), this showed primer synthesis was occurring in the absence of exogenously added dNTPs and NTPs. This result can be explained by the presence of endogenous dNTPs and NTPs in human cell extracts, sufficient for primer synthesis. Primer synthesis by these extracts was verified by extensive labeling of short primers upon addition of [α-32P]-dATP alone to the reactions (Supplemental Figure 2). As dialysis was insufficient to remove enough nucleotides to prevent primer synthesis, we evaluated the requirement for primer synthesis for protein recruitment by using the DNA polymerase α-primase inhibitor, aphidicolin (49,50). Addition of 20 µM aphidicolin to the DNA replication reaction (which blocked dNMP incorporation better than withholding exogenous nucleotide, Supplemental Figure 2) prevented the recruitment of both MSH2 and PCNA (Figure 4B), consistent with the known requirement of primers for PCNA loading (48). Conversely, RPA was still recruited, showing that origin recognition and unwinding proceeded normally. These results are consistent with primer synthesis being required for MMR recruitment.
Figure 4.
Primer synthesis is required for MMR protein recruitment to replicating DNA. DNA replication reactions were subjected to gel filtration and the precipitated protein fractions were immunoblotted for MSH2, RPA70 and PCNA. In panel (A) exogenous dNTPs and NTPs except ATP were omitted. In panel (B) 20 µM aphidicolin was added to the SV40 DNA replication reaction. As a control for antibody recognition, lanes marked CE contained 5 µl of cell extract representing 7% of the proteins in the entire reaction.
Recruitment of MMR proteins is dependent on PCNA loading
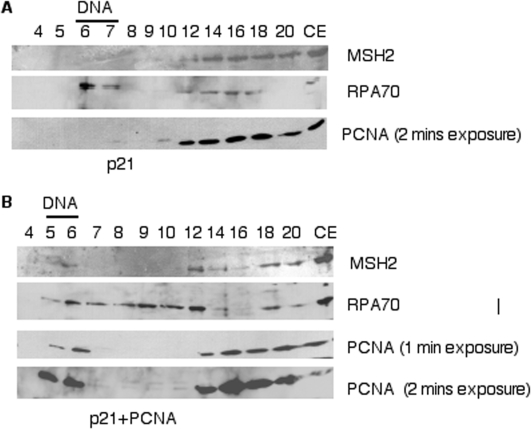
Since recruitment of both MMR proteins and PCNA appear to require primers, and MMR proteins are known to interact with PCNA, we addressed whether PCNA loading is required for MMR protein recruitment. This was tested by using a p21WAF1/CIP1 polypeptide that binds PCNA and inhibits interactions between PCNA and its interacting partner proteins (51,52). The carboxy-terminal of p21 contains a conserved PCNA Interacting Protein (PIP) motif, known as PIP box, shared by many PCNA interacting proteins (53). A GST-tagged carboxy-terminal p21 polypeptide consisting of amino-acids 87–164 (p21C) was added to cell extracts used for the in vitro DNA replication reactions (38). The effect of the p21 polypeptide on MMR protein recruitment was evaluated. RPA70 recruitment was observed in the presence of p21C. Conversely, we did not observe recruitment of PCNA or MSH2 to SV40 DNA in the presence of p21C (Figure 5A). This is consistent with p21 inhibiting loading of PCNA onto DNA by disrupting its interaction with RFC, which is required for PCNA loading (51,54). Addition of excess exogenous PCNA, which alleviates the inhibitory effect of p21C, restored recruitment of both PCNA and MSH2 (Figure 5B). Primer synthesis still occurs in the presence of p21C, as we observed labeling of small DNA products upon addition of labeled dATP (data not shown). This demonstrates that primer synthesis in the absence of PCNA is not sufficient for recruitment of MMR proteins to the replicating DNA. Rather, recruitment of MMR proteins to replicating DNA is dependent on PCNA loading, presumably due to either direct interaction of MMR proteins with PCNA, or due to steps in DNA replication subsequent to PCNA loading.
Figure 5.
p21 inhibits recruitment of MMR proteins and PCNA to replicating DNA. In panel (A) p21C (34 µg/ml) was added to the SV40 DNA replication reaction. In panel (B) p21C (34 µg/ml) and exogenous PCNA (30 µg/ml) were added to the SV40 DNA replication reaction. After gel filtration, precipitated protein fractions were analyzed by immunoblotting for MSH2, RPA70 and PCNA as above. As a control for antibody recognition, lanes marked CE contained 5 µl of cell extract representing 7% of the proteins in the entire reaction.
Continued access to the multi-protein binding sites on PCNA is required for the presence of MMR on replicating DNA
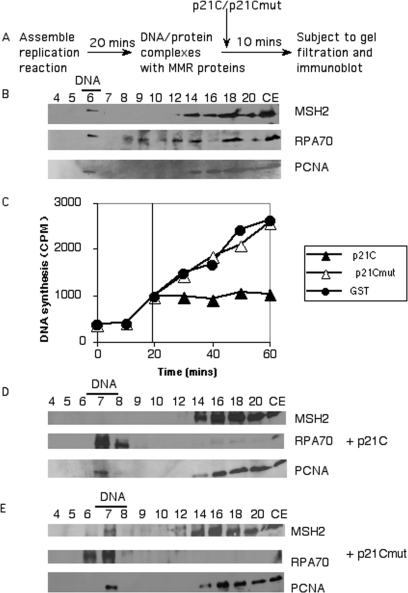
In the above experiments we inhibited DNA replication prior to PCNA loading and after primer synthesis. The absence of MMR recruitment in these cases could be due to the lack of later stages of DNA synthesis or the absence of DNA replication intermediates. We wanted to ascertain whether once DNA replication complexes are assembled, if the presence of MMR on replicating DNA becomes resistant to blocking of PCNA function. To address this question we carried out the experiment outlined in Figure 6A. This experiment was designed to evaluate whether MMR is still present on replicating DNA if p21C is added after assembly of active DNA replication/MMR complexes. As a control we verified that we could detect early recruitment of RPA, PCNA and MSH2 to replicating complexes after 20 min of incubation. We were able to clearly detect recruitment of these factors after 20 min (Figure 6B). We also needed to determine whether p21C could gain access to PCNA on the replication fork during the course of DNA replication. As p21 is known to inhibit SV40 DNA replication, we add p21C at 20 min and evaluated DNA synthesis. Figure 6C shows that addition of p21C during DNA replication rapidly inhibits ongoing SV40 DNA replication (within <5 min of addition of p21C). Conversely, addition of either GST alone or GST-p21C mutagenized to not bind to PCNA [methionine 147 mutated to alanine (55)] had no effect on DNA replication (Figure 6C). This demonstrated that p21C can readily gain access to PCNA in replicating complexes. Finally we addressed whether addition of p21C affected the presence of PCNA and MutS proteins on replicating complexes. The p21C polypeptide added at 20 min allowed recruitment of RPA and PCNA, but MSH2 was no longer detected (Figure 6D), whereas addition of the p21C mutant still allowed MSH2 to remain on the replicating complexes (Figure 6E). We conclude that addition of p21C to replicating complexes does not result in removal of PCNA, but does result in removal of MutS complexes. These results suggest that p21 is able to disrupt the interaction between PCNA and MutS complexes through competition for binding to the multi-protein-binding sites on PCNA. This is consistent with what is known about MutS complexes, as both MSH3 and MSH6 proteins have been shown to interact with PCNA through their PIP boxes (13,24,26,29). p21C also contains a conserved PIP box, and interacts with PCNA through the same motif (53,56). Together these findings indicate that MMR recruitment is PCNA-dependent, and is dependent on the availability of the multi-protein-binding sites on PCNA. This suggests that the MMR machinery is predominantly recruited to replicating DNA through the interaction of MMR proteins with PCNA.
Figure 6.
PCNA binding motif-dependent recruitment of MMR proteins to replicating DNA Panel (A) shows an outline of the experimental approach. After a 20 min incubation, to set up active DNA replication/repair complexes, either p21C or the p21C mutant is added. After 10 min, the complexes are subjected to gel filtration and immunoblotted as above. Panel (B) shows that at the 20 min time point that recruitment of RPA, PCNA and MSH2 to the replicating DNA can be readily detected via immunoblotting. In Panel (C) either GST-p21C, GST-p21C mutant or GST alone (34 µg/ml of each) were added to the replication reaction at 20 min. DNA incorporation was evaluated as described in Materials and Methods section and plotted as counts per minute (C.P.M.) incorporated at each of the indicated times. p21C (34 µg/ml; Panel D), or p21Cmut (34 µg/ml; Panel E), was added at 20 min during the SV40 DNA replication reaction. The reaction was further incubated for 10 min and then subjected to gel filtration. Precipitated protein fractions were analyzed by immunoblotting for MSH2, RPA70 and PCNA as above. As a control for antibody recognition, lanes marked CE contained 5 µl of cell extract representing 7% of the proteins in the entire reaction.
DISCUSSION
MMR is primarily a post-replicative DNA repair event. Mismatches or insertion/deletion loops generated during DNA replication are rapidly repaired by MMR. Earlier studies have shown co-localization of MMR proteins to DNA replication foci (13,57). How MMR associates with replicating DNA has been suspected to be via known interactions between the replicative sliding clamp, PCNA and MMR proteins; however, until now there is no direct evidence to support this hypothesis. In this study, we have developed a new system to investigate the linkage between DNA replication and MMR. This system uses the SV40 in vitro DNA replication system and DNA replication/repair competent human cell extracts. Using this system we showed that there is a physical association of MMR proteins with replicating SV40 DNA which is dependent on the presence of a functional origin of replication. This association is dependent on PCNA and the PCNA protein interaction motif. Earlier biochemical studies have shown interaction of MSH3 and MSH6 proteins with PCNA through a conserved PIP box, which interacts with the PCNA protein interaction motif (13,14,29). Using our system we show that the carboxyl-terminal half of p21, containing the conserved PIP box (56), was able to compete with MMR protein binding to PCNA on replicating SV40 DNA. This was dependent on a functional PIP box, as a single-point mutation in the PIP box known to destroy PIP box function, abrogated this effect. These results indicate that binding to PCNA, which is both present at the replication fork as well as transiently associated with DNA following replication fork passage, is the primary mechanism of MMR recruitment to replicating DNA.
Current knowledge of MMR shows that the pathway initiates through the interaction of MutS complexes with a mismatch (58–61), and then MutS complexes recruit other factors such as the MutLα complex, PCNA, and the DNA excision and re-synthesis machinery. Since addition of the p21 C-terminus can actively displace MutS complexes from replicating DNA, our results are consistent with mismatches not being required for the initial recruitment of the majority of MMR proteins to replicating DNA. Hence, our data are consistent with the model that PCNA may be responsible for initially recruiting the MMR machinery to actively replicating DNA, and actively participating in mismatch recognition (14,29). As PCNA is a sliding clamp that is capable of greatly increasing the processivity of DNA polymerases, such an interaction may assist MutS complexes to scan large regions of DNA more efficiently than it could alone (62). We have shown here that PCNA plays a role in recruitment of MMR to replicating DNA. Whether this role of PCNA is merely to increase the localized concentration of MutS complexes at replicating DNA to allow for more efficient MutS action in areas of newly replicated DNA, or to actually assist MutS in scanning the DNA for mismatches, remains unknown. However, results shown here clearly provide support for the model that MMR interaction with PCNA is the likely mechanism by which MMR acts predominantly in post-replicative manner.
Another aspect of MMR where PCNA is thought, but not yet proven, to have a critical function is that of strand discrimination. Our data, indicating that the MutS complex associates with replication forks via its interaction with PCNA, shows that PCNA is indeed present very early during MMR, likely often prior to mismatch recognition. Our results are therefore consistent with the model of PCNA providing the signal for strand discrimination at the earliest stages of MMR through PCNA's opposite polarity on the two newly synthesized daughter molecules. We anticipate that this new in vitro model system, which allows one to examine the relationship between DNA replication and MMR proteins, will provide new avenues to address the question of PCNA's role in MMR strand discrimination.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
National Institutes of Health funded grant CA89259 to T.M. We thank Dr Anindya Dutta for the p21C expression construct. We also thank Dr Jennifer Surtees for critiquing this manuscript, and Drs Joel Huberman and Dave Kowalski for helpful discussions. Thanks to Wan Zhang for purifying PCNA, Drs Jen-Sing Liu, Shu-Ru Kuo and John Fisk for experimental advice, and Dr Mark Sutton for much helpful advice, and assistance with the p21 mutagenesis. Funding to pay the Open Access publication charges for this article was provided by the University at Buffalo School of Medicine & Biomedical Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jiricny J. The multifaceted mismatch-repair system. Nat. Rev. Mol. Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 2.Jun SH, Kim TG, Ban C. DNA mismatch repair system. Classical and fresh roles. FEBS J. 2006;273:1609–1619. doi: 10.1111/j.1742-4658.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 3.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 4.Kunkel TA, Erie DA. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 5.Kolodner RD, Hall NR, Lipford J, Kane MF, Rao MR, Morrison P, Wirth L, Finan PJ, Burn J, et al. Human mismatch repair genes and their association with hereditary non-polyposis colon cancer. Cold Spring Harbor Symp. Quant. Biol. 1994;59:331–338. doi: 10.1101/sqb.1994.059.01.037. [DOI] [PubMed] [Google Scholar]
- 6.Kolodner RD. Mismatch repair: mechanisms and relationship to cancer susceptibility. Trends Biochem. Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 7.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N. Engl. J. Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 8.Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 9.Lu AL, Clark S, Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc. Natl Acad. Sci. USA. 1983;80:4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Au KG, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 11.Lahue RS, Modrich P. Methyl-directed DNA mismatch repair in Escherichia coli. Mutat. Res. 1988;198:37–43. doi: 10.1016/0027-5107(88)90037-1. [DOI] [PubMed] [Google Scholar]
- 12.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 13.Kleczkowska HE, Marra G, Lettieri T, Jiricny J. hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci. Genes Dev. 2001;15:724–736. doi: 10.1101/gad.191201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau PJ, Kolodner RD. Transfer of the MSH2.MSH6 complex from proliferating cell nuclear antigen to mispaired bases in DNA. J. Biol. Chem. 2003;278:14–17. doi: 10.1074/jbc.C200627200. [DOI] [PubMed] [Google Scholar]
- 15.Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 16.Kuriyan J, O’Donnell M. Sliding clamps of DNA polymerases. J. Mol. Biol. 1993;234:915–925. doi: 10.1006/jmbi.1993.1644. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Hays JB. Mismatch repair in human nuclear extracts. Time courses and ATP requirements for kinetically distinguishable steps leading to tightly controlled 5′ to 3′ and aphidicolin-sensitive 3′ to 5′ mispair-provoked excision. J. Biol. Chem. 2002;277:26143–26148. doi: 10.1074/jbc.M200358200. [DOI] [PubMed] [Google Scholar]
- 18.Holmes J, Jr, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc. Natl Acad. Sci. USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: reconstitution of a nick-directed bidirectional reaction. J. Biol. Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J. Biol. Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen FC, Jager AC, Lutzen A, Bundgaard JR, Rasmussen LJ. Characterization of human exonuclease 1 in complex with mismatch repair proteins, subcellular localization and association with PCNA. Oncogene. 2004;23:1457–1468. doi: 10.1038/sj.onc.1207265. [DOI] [PubMed] [Google Scholar]
- 22.Guo S, Presnell SR, Yuan F, Zhang Y, Gu L, Li GM. Differential requirement for proliferating cell nuclear antigen in 5′ and 3′ nick-directed excision in human mismatch repair. J. Biol. Chem. 2004;279:16912–16917. doi: 10.1074/jbc.M313213200. [DOI] [PubMed] [Google Scholar]
- 23.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol. Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 24.Gu L, Hong Y, McCulloch S, Watanabe H, Li GM. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 1998;26:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shell SS, Putnam CD, Kolodner RD. The N terminus of Saccharomyces cerevisiae Msh6 is an unstructured tether to PCNA. Mol. Cell. 2007;26:565–578. doi: 10.1016/j.molcel.2007.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SD, Alani E. Analysis of interactions between mismatch repair initiation factors and the replication processivity factor PCNA. J. Mol. Biol. 2006;355:175–184. doi: 10.1016/j.jmb.2005.10.059. [DOI] [PubMed] [Google Scholar]
- 27.Lau PJ, Flores-Rozas H, Kolodner RD. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol. Cell. Biol. 2002;22:6669–6680. doi: 10.1128/MCB.22.19.6669-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes. J. Biol. Chem. 2000;275:36498–36501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- 29.Flores-Rozas H, Clark D, Kolodner RD. Proliferating cell nuclear antigen and Msh2p-Msh6p interact to form an active mispair recognition complex. Nat. Genet. 2000;26:375–378. doi: 10.1038/81708. [DOI] [PubMed] [Google Scholar]
- 30.Gurney EG, Harrison RO, Fenno J. Monoclonal antibodies against simian virus 40 T antigens: evidence for distinct sublcasses of large T antigen and for similarities among nonviral T antigens. J. Virol. 1980;34:752–763. doi: 10.1128/jvi.34.3.752-763.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Din S, Brill SJ, Fairman MP, Stillman B. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 1990;4:968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- 32.Stillman BW, Gluzman Y. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol. Cell. Biol. 1985;5:2051–2060. doi: 10.1128/mcb.5.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stillman B, Gerard RD, Guggenheimer RA, Gluzman Y. T antigen and template requirements for SV40 DNA replication in vitro. EMBO j. 1985;4:2933–2939. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prelich G, Stillman B. Coordinated leading and lagging strand synthesis during SV40 DNA replication in vitro requires PCNA. Cell. 1988;53:117–126. doi: 10.1016/0092-8674(88)90493-x. [DOI] [PubMed] [Google Scholar]
- 35.Murphy CI, Weiner B, Bikel I, Piwnica-Worms H, Bradley MK, Livingston DM. Purification and functional properties of simian virus 40 large and small T antigens overproduced in insect cells. J. Virol. 1988;62:2951–2959. doi: 10.1128/jvi.62.8.2951-2959.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lanford RE. Expression of simian virus 40 T antigen in insect cells using a baculovirus expression vector. Virology. 1988;167:72–81. doi: 10.1016/0042-6822(88)90055-4. [DOI] [PubMed] [Google Scholar]
- 37.Simanis V, Lane DP. An immunoaffinity purification procedure for SV40 large T antigen. Virology. 1985;144:88–100. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Jackson PK, Kirschner MW, Dutta A. Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA. Nature. 1995;374:386–388. doi: 10.1038/374386a0. [DOI] [PubMed] [Google Scholar]
- 39.Chen G, Stenlund A. Characterization of the DNA-binding domain of the bovine papillomavirus replication initiator E1. J. Virol. 1998;72:2567–2576. doi: 10.1128/jvi.72.4.2567-2576.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fien K, Stillman B. Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol. Cell. Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melendy T, Sedman J, Stenlund A. Cellular factors required for papillomavirus DNA replication. J. Virol. 1995;69:7857–7867. doi: 10.1128/jvi.69.12.7857-7867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bullock PA. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 43.Hurwitz J, Dean FB, Kwong AD, Lee SH. The in vitro replication of DNA containing the SV40 origin. J. Biol. Chem. 1990;265:18043–18046. [PubMed] [Google Scholar]
- 44.Roberts JD, Izuta S, Thomas DC, Kunkel TA. Mispair-, site-, and strand-specific error rates during simian virus 40 origin-dependent replication in vitro with excess deoxythymidine triphosphate. J. Biol. Chem. 1994;269:1711–1717. [PubMed] [Google Scholar]
- 45.Brown TC, Jiricny J. Repair of base-base mismatches in simian and human cells. Genome/Natl Res. Council Canada. 1989;31:578–583. doi: 10.1139/g89-107. [DOI] [PubMed] [Google Scholar]
- 46.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc. Natl Acad. Sci. USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li GM, Modrich P. Restoration of mismatch repair to nuclear extracts of H6 colorectal tumor cells by a heterodimer of human MutL homologs. Proc. Natl Acad. Sci. USA. 1995;92:1950–1954. doi: 10.1073/pnas.92.6.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J. Biol. Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 49.Berger NA, Kurohara KK, Petzold SJ, Sikorski GW. Aphidicolin inhibits eukaryotic DNA replication and repair — implications for involvement of DNA polymerase alpha in both processes. Biochem. Biophys. Res. Commun. 1979;89:218–225. doi: 10.1016/0006-291x(79)90966-5. [DOI] [PubMed] [Google Scholar]
- 50.Krokan H, Wist E, Krokan RH. Aphidicolin inhibits DNA synthesis by DNA polymerase alpha and isolated nuclei by a similar mechanism. Nucleic Acids Res. 1981;9:4709–4719. doi: 10.1093/nar/9.18.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Podust VN, Podust LM, Goubin F, Ducommun B, Hubscher U. Mechanism of inhibition of proliferating cell nuclear antigen-dependent DNA synthesis by the cyclin-dependent kinase inhibitor p21. Biochemistry. 1995;34:8869–8875. doi: 10.1021/bi00027a039. [DOI] [PubMed] [Google Scholar]
- 52.Waga S, Hannon GJ, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 53.Warbrick E. PCNA binding through a conserved motif. Bioessays. 1998;20:195–199. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 54.Oku T, Ikeda S, Sasaki H, Fukuda K, Morioka H, Ohtsuka E, Yoshikawa H, Tsurimoto T. Functional sites of human PCNA which interact with p21 (Cip1/Waf1), DNA polymerase delta and replication factor C. Genes Cells. 1998;3:357–369. doi: 10.1046/j.1365-2443.1998.00199.x. [DOI] [PubMed] [Google Scholar]
- 55.Zheleva DI, Zhelev NZ, Fischer PM, Duff SV, Warbrick E, Blake DG, Lane DP. A quantitative study of the in vitro binding of the C-terminal domain of p21 to PCNA: affinity, stoichiometry, and thermodynamics. Biochemistry. 2000;39:7388–7397. doi: 10.1021/bi992498r. [DOI] [PubMed] [Google Scholar]
- 56.Warbrick E, Lane DP, Glover DM, Cox LS. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr. Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- 57.Smith BT, Grossman AD, Walker GC. Visualization of mismatch repair in bacterial cells. Mol. Cell. 2001;8:1197–1206. doi: 10.1016/s1097-2765(01)00402-6. [DOI] [PubMed] [Google Scholar]
- 58.Alani E, Chi NW, Kolodner R. The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and insertions. Genes Dev. 1995;9:234–247. doi: 10.1101/gad.9.2.234. [DOI] [PubMed] [Google Scholar]
- 59.Fishel R, Ewel A, Lescoe MK. Purified human MSH2 protein binds to DNA containing mismatched nucleotides. Cancer Res. 1994;54:5539–5542. [PubMed] [Google Scholar]
- 60.Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J. Biol. Chem. 1999;274:26668–26682. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 61.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr. Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 62.Kelman Z, Hurwitz J. Protein–PCNA interactions: a DNA-scanning mechanism? Trends Biochem. Sci. 1998;23:236–238. doi: 10.1016/s0968-0004(98)01223-7. [DOI] [PubMed] [Google Scholar]