Abstract
Escherichia coli DEAD-box protein A (DbpA) is an ATP-dependent RNA helicase with specificity for 23S ribosomal RNA. Although DbpA has been extensively characterized biochemically, its biological function remains unknown. Previous work has shown that a DbpA deletion strain is viable with little or no effect on growth rate. In attempt to elucidate a phenotype for DbpA, point mutations were made at eleven conserved residues in the ATPase active site, which have exhibited dominant-negative phenotypes in other DExD/H proteins. Biochemical analysis of these DbpA mutants shows the expected decrease in RNA-dependent ATPase activity and helix unwinding activity. Only the least biochemically active mutation, R331A, produces small colony phenotype and a reduced growth rate. This dominant slow growth mutant will be valuable to determine the cellular function of DbpA.
INTRODUCTION
DbpA is an Escherichia coli DEAD-box protein that binds tightly and specifically to 23S rRNA and stimulates rapid ATP hydrolysis (1,2). Extensive truncation (2–4) and footprinting (5) experiments have shown that DbpA binds to a 153 nt region (153-mer) of 23S rRNA consisting of helices 89 through 93, which are all part of the peptidyltransferase center (PTC) of the ribosome. Shorter fragments that contain only hairpin 92 bind to DbpA with similar affinity as the 153-mer, demonstrating this hairpin as the primary binding site (3). In the presence of RNA duplexes containing hairpin 92, DbpA demonstrates weakly processive 3′ to 5′ RNA helicase activity (6). However, mature ribosomes are poor substrates for DpbA because hairpin 92 is buried within the PTC (4).
The N-terminal 381 amino acids of DbpA contains the ATPase core, 9 conserved motifs spanning two domains that define DExD/H proteins and are responsible for the ATP binding and hydrolysis activities. The C-terminal 76 amino acids form a basic domain that forms an RNA recognition motif (RRM) and is involved in RNA-binding specificity (7). The isolated C-terminal domain of YxiN, the Bacillus subtilis ortholog of DbpA, can bind hairpin 92 tightly and specifically (5). However, the isolated N-terminal domain of YxiN binds RNA weakly and nonspecifically but does show ATPase activity, thus demonstrating the functional modularity of this DExD/H protein (5).
Despite this extensive biochemical characterization, relatively little is known about the function of DbpA in E. coli. DbpA mRNA levels are found in low amounts in logarithmic cells, but they increase upon acid shock, addition of ciprofloxacin and at stationary phase (8). Several groups have deleted the dbpA gene using different methods, however no growth defects were observed in different culture media and growth conditions (9,10) (Tsu,C. and Uhlenbeck,O.C., unpublished data). Because DbpA is activated by 23S rRNA in vitro, it was hypothesized that DbpA is either involved in the ribosome assembly pathway or in some specialized aspect of translation (2). Under many growth conditions, DbpA is not essential and therefore, either one of the other E. coli DExD/H proteins can substitute for its function or it is only required under certain growth conditions.
The goal of this article is to identify a dominant negative mutant of DbpA that can be used to better understand its cellular function. DbpA is a good candidate for creating a dominant negative mutant because its binding affinity and specificity lies almost completely in the CTD, while its ATPase and helicase activity are determined by the conserved ATPase core (5). These modular characteristics make it possible to inactivate DbpA by mutating catalytic residues without affecting the ability of the CTD to bind substrate RNA. Unlike a knockout mutation of DbpA, a site-directed mutation would produce a protein that could compete with wild-type DbpA for the rRNA-binding site and subsequently block action at the normal site of function. Therefore, several point mutations were made in the conserved sequence motifs of DbpA that were identical to those showing dominant negative properties in other DExD/H proteins, including eIF-4A, Prp16, Prp22, Prp28, Prp43, Rok1p, Sub2p, Ded1p and Dbp8p (11–19).
MATERIALS AND METHODS
Site-directed mutagenesis and purification of His-tagged DbpA mutations
The gene coding for DbpA with an N-terminal Met-His6 sequence was cloned into the pET-3a vector between the NdeI and BamHI sites (20). Site-directed mutagenesis was performed on this plasmid using QuikChange® XL Site-Directed mutagenesis kit (Stratagene). DbpA-containing plasmids were transformed into BL21(DE3) pLysS cells (Stratagene) and purified as previously described (20) with the following changes. Cell lysate was purified by FPLC over a nickel column or by batch purification over Ni-NTA beads (Qiagen) in buffer A (500 mM NaCl, 20 mM MOPS pH 7, 1 mM β-mercaptoethanol, 10% glycerol) and 10 mM imidazole. His6-DbpA was eluted in buffer A with 300 mM imidazole and further purified using a Pharmacia Superdex75 sizing column in buffer A.
Preparation of RNA
Native E. coli rRNA (16S and 23S) was from Roche Diagnostics or purified from E. coli MRE600 cells by the following method. The cell pellet was thawed on ice and resuspended in ribosome lysis buffer (50 mM Tris pH 7.5, 10 mM MgCl2, 100 mM NH4Cl, 0.5 mM EDTA pH 8, 6 mM DTT) and RQ1 DNase (Promega). Cells were lyzed by a Thermo IEC french press and cell debris removed by centrifugation for 20 min at 11 000 r.p.m. in an Eppendorf table top centrifuge. Ribosomes were collected by centrifugation for 1 h at 40 000 r.p.m. in a Ti45 rotor in a Beckman Optima LE-80K ultracentrifuge. The pellet was cleared by rinsing with TE buffer and then resuspended in SDS–Urea extraction buffer (6 M Urea, 0.1% SDS, 10 mM Tris pH 7.8, 10 mM EDTA pH 8, 350 mM NaCl). Ribosomal RNA was purified by phenol–chloroform extraction, ethanol precipitation, LiCl precipitation, a second ethanol precipitation and elution over NAP-25 columns (Amersham Biosciences).
The 9-mer and 32-mer RNA were purchased from Dharmacon. A 153-mer segment of domain V of 23S rRNA was prepared by in vitro transcription. The 153-mer DNA template was PCR amplified from p2454 to 2606 (4) and purified with the QIAquick Gel Extraction kit (Qiagen). Transcription of 153-mer RNA was carried out in transcription buffer (80 mM Tris pH 7.5, 20 mM DTT, 4 mM Spermidine, 0.1% TritonX, 12 mM MgCl2, 0.004 U/ml pyrophosphatase) with 4 mM NTP's, 15 ng/μl template DNA and 0.1 mg/ml T7 RNA polymerase at 37°C for 3 h, concentrated by ethanol precipitation and then purified by 10% PAGE. RNA was extracted from the gel by passive elution (0.5 M NaOAc and 1 mM EDTA pH 8) overnight at 4°C and then concentrated by ethanol precipitation. The concentration of the final product was calculated using ɛ260 = 1.3 × 106 L/mol cm. RNA substrates were 5′-32P labeled with polynucleotide kinase and γ-32P ATP and purified by 10% denaturing PAGE. Labeled RNA was extracted from the gel by passive elution overnight at 4°C and then precipitated with ethanol.
Assays
The rate of ATP hydrolysis of DbpA in the presence of 23S + 16S rRNA was measured using the previously described high-throughput coupled spectroscopic assay (5). All reported values are the average of at least three data sets and are all within 2-fold error.
The binding of DbpA to a 153-mer portion of 23S rRNA was determined based on the previously described gel shift assay (21). Binding reactions were carried out in binding buffer (50 mM HEPES pH 7.5, 5 mM MgCl2, 100 mM KCl, 100 μM DTT, 100 μg/ml BSA and 5% (v/v) glycerol) with 200 pM 5′-32P RNA, 70 μM polyA and increasing amounts of DbpA protein. Samples (20 μl) were equilibrated at 24°C for 30 min and then an 18 μl aliquot was loaded on a 5% native acrylamide gel (29 : 1 acrylamide:bis) running at 200 V in 1/3 × TBE buffer. Gels were run for 2 h, dried and exposed to a phosphor screen. The fraction of bound protein was determined from the ratio of counts in the bound RNA band relative to the total counts of the free and bound RNA. The fraction of bound RNA versus DbpA concentration was plotted and fit to simple binding equilibria to determine the dissociation constant (Kd) (21). The binding affinity was also evaluated in the presence of a non-hydrolyzable ATP analog, AMPPNP. Reactions were carried out as above with 50 pM 5′-32P RNA and the addition of 4 mM AMPPNP. All reported values are the average of at least three data sets and are corrected for percent activity as measured by a stoichiometric binding assay (described below). The errors are less than 2.5-fold for all measurements.
The stoichiometry of RNA binding for each wild-type and mutant protein was measured using the gel shift binding assay in the presence of 500 nM 5′-32P 153-mer and varying protein concentrations (21). The fraction of bound RNA versus protein concentration was plotted and fit linearly (data not shown). The linear slope is the percent active protein, which is then multiplied by the average value of Kd to obtain final values corrected for percent protein activity.
Helicase activity was measured using a 5′-32P labeled 9-mer (*9-mer) annealed 5′ to 32-mer RNA containing hairpin 92 (Figure 4A) using the assay described in Diges et al. (22) with the following modifications. A total of 1.2 μM 32-mer and 0.6 μM *9-mer were annealed by mixing in 50 mM HEPES pH 7.5 and 50 mM KCl and then heating to 95°C for 1 min, 80°C for 1 min, 75°C for 2 min, 70°C for 3 min. At 70°C, 5 mM MgCl2 was added and the reaction was cooled to 24°C for 15 min and then to 4°C and used immediately. Helicase assays in the presence of excess protein were based on the previously described method in Ref. (22) and carried out at 24°C in binding buffer (as above). Seventy-five microliter aliquots of the RNA mixture (5 nM annealed substrate with 70 μM polyA) and protein mix (0.6 μM DbpA, 2 mM ATP, 2 mM MgCl2) were combined to initiate unwinding. At various times, 9 μl of the reaction mixture was removed and mixed with 3 μl of quench dye [2.5 mM EDTA pH 8, 0.025% SDS (w/v), 1.25% glycerol (v/v), pinch xylene cyanol]. Then 9 μl of the quenched reaction was immediately loaded onto a 10% native acrylamide gel (29 : 1 acrylamide:bis) running at 150 V in 1/3× TBE buffer at 24°C. Gels were run at 200 V for 2 h, dried and exposed to a phosphor screen. The fraction of bi-substrate (*9-mer + 32-mer) was determined from the ratio of counts in the displaced RNA band (*9-mer alone) relative to the total counts of the annealed and displaced RNA bands. The fraction of duplex RNA was plotted versus time and fit to a single exponential curve. All reported rates of unwinding represent the average of at least three measurements and the error is within 2-fold.
Figure 4.
(A) An annealed duplex consisting of 32-mer containing hairpin 92 and 5′-32P labeled 9-mer, is unwound upon addition of DbpA and saturating ATP (2 mM). (B) Helicase assays for wild-type DbpA (circles), S184A (triangles) and R334A (squares) performed in binding buffer in the presence of 600 nM protein, 5 nM duplex RNA and 2 mM ATP at 24°C. Lines are the best fit of the data and an end point using a single exponential equation.
Western blots
XL10-Gold® (Stratagene) or Tuner™ (DE3)pLacI (Novagen) modified to be RecA− (a gift from M.E. Saks) cells were transformed with wild-type or mutant plasmids. Overnight cultures were diluted in LB, grown to OD600 ∼ 0.3–0.4, then the cells were harvested, quick-cooled by pouring over an equal volume of ice and centrifuged. Cells were lyzed as described by J. Choralis et al. (23) except the lysis buffer contained 20 mM HEPES pH 7.5, 30 mM NH4Cl, 10 mM MgCl2 and 4 mM β-mercaptoethanol. Clarified lysates were diluted in lysis buffer plus 5 × SDS load dye (85 mM Tris–HCl pH 6.8, 4% SDS, 40% glycerol, 10% β-mercaptoethanol, bromophenol blue) and then 10–25 μl were loaded on a 15% Tris–HCl Ready gel (Bio-Rad). Gels were transferred to PVDF (Bio-Rad) membrane in transfer buffer, 25 mM Tris–HCl pH 8.3, 192 mM glycine, 20% methanol and 0.1% SDS, at 100 V for 1 h at 4°C. PVDF membranes were treated according to the Li-Cor Odyssey® manual, probed with a 1 : 10 000 dilution of rabbit anti-DbpA (provided by F. Fuller-Pace) and a 1 : 5000 dilution of secondary antibody Alexa Fluor® 680 goat anti-rabbit IgG (H+L) (Invitrogen) and then scanned on the Odyssey® Infrared Imaging System, which can report relative intensities of each band on the blot. The number of moles of each protein was estimated using a standard curve based on the band intensities for a serial dilution of purified DbpA protein. Molecules per cell were estimated using the number of moles divided by the approximate amount of cells (according to the volume collected and lyzed) and loaded on the blot. Due to the small amount of endogenous DbpA, 40 times more of wild-type Tuner™ (DE3)pLacI RecA− cells were loaded compared to those containing DbpA and mutant plasmids.
RESULTS
DbpA mutant design
Mutations were focused in regions of the protein expected to be involved in ATP binding and hydrolysis in order to disrupt the activity of DbpA without affecting its substrate binding. Although the complete structure of DbpA is not available, the structures of the C-terminal half of the ATPase core and the CTD of the B. Subtilis ortholog, YxiN, are known (7,24). The C-terminal half of the ATPase core forms a parallel αβ-fold as in other RNA helicase structures (25–29) and RecA (24,27,30), while the C-terminal domain (CTD) structure as mentioned above forms a RRM-fold and is responsible for RNA binding and specificity (7). The structure of a closely related DEAD-box family member, Drosophila Vasa, in complex with single-stranded RNA and a non-hydrolyzable ATP analog, helps to identify the likely critical catalytic residues of DbpA (Figure 1). In addition, extensive biochemical characterization of ATPase core mutations in human eIF-4A and yeast Prp16, Prp22 and Prp43 have been reported (31,32). Finally, several mutations in the ATPase active sites of yeast eIF-4A, Prp16, Prp22, Prp28, Prp43, Rok1p, Sub2p, Ded1p and Dbp8p were previously found to have dominant negative phenotypes (11–19). Therefore, mutational analysis of DbpA was designed according to the structures, focusing on residues that contact ATP, and based on the other RNA helicase mutations that caused dramatic biochemical differences or dominant negative phenotypes.
Figure 1.
(A) Conserved ATPase core motifs with conserved DbpA sequences shown below the corresponding motif. Residues mutated in this study are in bold. (B) Drosophila Vasa RNA Helicase ATP-binding pocket structure (PDB : 2db3) showing only the residues that were mutated in DbpA (E. coli DbpA numbering). Colors coordinate to the motifs are as in Figure 1A; motif I is purple, motif II is red, motif III is yellow, motif VI is cyan and AMPPNP is green. The white sphere is the Mg2+ that is coordinated to the AMPPNP. Important contacts defined in the Vasa structure are indicated with a dashed line.
Eleven DbpA residues that span four conserved motifs (I, II, III, VI) were mutated to alanine. Motifs I, II and III are in the N-terminal RecA-like domain of the catalytic core, (N-Rec) and motif VI lies in the C-terminal RecA-like domain of the core (C-Rec) (Figure 1). In the GSGKT sequence of motif I, also known as the Walker A motif, the conserved lysine (K53) hydrogen bonds with the β and γ phosphates of ATP, whereas the threonine (T54) coordinates a Mg2+ ion that also interacts with the β and γ phosphates (26,30). In the DEAD sequence of motif II, the Walker B motif, the highly conserved aspartate (D153) and glutamate (E154) residues also coordinate the Mg2+ ion that interacts with the β and γ phosphates of ATP (26,30) and are responsible for hydrolysis (33). The last aspartate (D156) makes interdomain contacts with motif III (26,30). The serine (S184), alanine (A185) and threonine (T186) residues that form motif III do not interact directly with ATP, but are thought to coordinate ATP binding and helicase activities (31). In the HRIGRTAR sequence, motif VI, the second arginine (R331) interacts with the γ phosphate of ATP, while the third arginine (R334) interacts with the α, β and γ phosphates (26,30,33). One of the arginines that contacts the γ phosphate is proposed to be an ‘arginine finger’ that is involved in transition state stabilization during hydrolysis and may be critical for coupling the ATPase cycle with helicase activity (26,30,33,34).
RNA binding
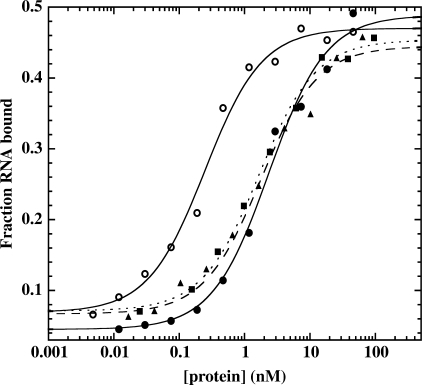
The majority of DbpA's RNA-binding affinity is derived from the interactions of the CTD (5), therefore mutants in the ATPase motifs are not expected to significantly affect binding. A native gel electrophoretic mobility shift assay was used to evaluate the binding affinity (Kd) of wild-type DbpA and the eleven mutants for the 153-nt fragment of 23S rRNA (21). In agreement with other experiments (21), the binding affinity for wild-type DbpA is 1.7 nM and, as expected, most of the mutant proteins bind to substrate RNA with similar affinities (Figure 2 and Table 1). Surprisingly, the Kd for two of the mutants, D153A and R328A is approximately 10-fold weaker than wild-type (Table 1). As with all the mutants, the CTD is likely to remain unchanged, so these two mutations must affect the overall structure of DbpA thereby, causing the decrease in binding affinity.
Figure 2.
RNA-binding curves for wild-type DbpA (circles and solid line), H327A (triangles and dashed line), R334A (squares and dotted line) and wild-type DbpA with 4 mM AMPPNP (open circles and solid line) derived from gel mobility shift assays. All experiments are performed with 200 pM 5′-32P labeled 153-mer RNA in binding buffer [50 mM HEPES pH 7.5, 5 mM MgCl2, 100 mM KCl, 100 μM DTT, 100 μg/ml BSA, 5% (v/v) glycerol] and 70 μM polyA at 24°C.
Table 1.
Binding affinities for wild-type and the eleven DbpA mutants to 153- mer RNA in the absence and presence of 4 mM AMPPNP
| Motif | Protein | Kd RNA (nM) | Kd RNA + AMPPNP (nM) |
|---|---|---|---|
| wild-type | 1.7 | 0.2 | |
| I | K53A | 2.4 | 0.4 |
| T54A | 2.8 | 4.0 | |
| II | D153A | 14 | 14 |
| E154A | 5.0 | 3.4 | |
| D156A | 1.9 | 0.1 | |
| III | S184A | 2.8 | 1.5 |
| T156A | 1.8 | 0.9 | |
| VI | H327A | 1.5 | 0.7 |
| R328A | 18 | 18 | |
| R331A | 2.3 | 0.8 | |
| R334A | 1.1 | 0.4 |
Dissociation constants (Kd) were determined from data such as those presented in Figure 2. Kd RNA values are corrected for percent active protein and are averages of at least three determinations which vary by less than 2.5-fold.
In the presence of saturating amounts of non-hydrolyzable ATP (AMPPNP), DbpA's binding affinity for 153-mer RNA is 8.5-fold tighter (Figure 2) (21). This additional RNA-binding affinity is thought to be due to interactions with the catalytic domains that only form when their structure is pre-organized by the binding of ATP (21). The RNA-binding affinity of each mutant was measured in the presence of 153-mer and AMPPNP to determine whether they have this additional affinity (Table 1). Most of the mutants displayed little or no increase in affinity when AMPPNP was present (Table 1) indicating that their ATP-binding site is either partially or fully disrupted thus preventing the additional contacts with the RNA. Only two of the mutants, K53A and D156A, show a greater than 5-fold increase in RNA-binding affinity in the presence of AMPPNP, suggesting that their ATP-binding site remains nearly fully intact (Table 1).
RNA-dependent ATPase activity
The ATPase activity of each mutant protein was initially assayed in the presence of 250 nM 23S rRNA and 5 mM ATP which are saturating for wild-type DbpA. For each mutation, the measured hydrolysis rate was plotted versus concentration of protein to determine the specific activity, kobs. For all of the mutants, kobs values were significantly reduced compared to wild-type, with the activities ranging from <5% to ∼70% active (Table 2). The mutants with high specific activity (D156A, S184A, T186A and H327A) make interdomain contacts but do not contact the ATP directly (Figure 1B), suggesting that they are not directly involved in hydrolysis (30). The two mutants with the lowest specific activity, R331A and K53A, both contact the phosphates of the ATP directly (26). This lysine is invariant in all ATPases, and when mutated usually abolishes hydrolysis and unwinding (Figure 1B) (33). As expected, the K53A mutant of DbpA shows very weak ATPase activity (kobs = 0.7 min−1). Interestingly, K53A still demonstrates increased RNA-binding affinity in the presence AMPPNP suggesting that the mutation affects catalysis without affecting ATP binding (Tables 1 and 2). It has been proposed that one of the arginines in motif VI acts as an ‘arginine finger’ that stabilizes the transition state of the hydrolysis reaction and connects domain two to the ATP bound in domain one (30). In DbpA, R331 is the likely ‘arginine finger’ due to the very low ATPase activity (kobs = 0.4 min−1) of the mutant. Alanine substitutions of the other two arginines in motif VI of DbpA, R328 and R334, have more ATPase activity than R331A and thus are less likely to serve as the arginine finger. In addition, R328 does not seem to bind the ATP in the crystal structure of Vasa (Figure 1B) (26).
Table 2.
ATPase activities of wild-type and the eleven DbpA mutants in the presence of 23S rRNA substrate and ATP
| Motif | Protein | kobs (min−1) | kmax (min−1) | Kapp,RNA (nM) | kcat (min−1) | Km,atp (µM) |
|---|---|---|---|---|---|---|
| wild-type | 120 | 140 | 33 | 160 | 400 | |
| I | K53A | 0.7 | – | – | – | – |
| T54A | 6.0 | – | – | – | – | |
| II | D153A | 1.7 | – | – | – | – |
| E154A | 2.0 | – | – | – | – | |
| D156A | 28 | 31 | 98 | 19 | 64 | |
| III | S184A | 53 | 68 | 44 | 59 | 410 |
| T156A | 79 | 97 | 64 | 87 | 1000 | |
| VI | H327A | 50 | 82 | 210 | 58 | 2600 |
| R328A | 1.1 | – | – | – | – | |
| R331A | 0.4 | – | – | – | – | |
| R334A | 14 | – | – | – | – |
The kobs is the slope of a plot of the rate of ATP hydrolysis versus protein concentration at constant ATP (5 mM) and RNA (250–500 nM) at 24°C. The Kapp,RNA, kmax, KM,ATP and kcat for D156A, S184A, T186A and H327A were determined from data such as presented in Figure 3. Values are averages of at least three data sets and are within 2-fold error.
Four mutants, D156A, S184A, T186A and H327A, have sufficient ATPase activity to allow the determination of the apparent RNA-binding constant, Kapp, RNA, the maximum catalytic rate, kmax and the Michaelis–Menten constants, KM, ATP and kcat. Here, Kapp, RNA and kmax were measured with 5 mM ATP and RNA concentration between 0 and 500 nM (Figure 3A), whereas the Michaelis–Menten values are measured at 250–500 nM RNA with ATP concentration between 0 and 4 mM ATP (Figure 3B). As had been determined previously, wild-type DbpA has a very tight Kapp, RNA of 33 nM (Table 2) (4). The mutants D156A, S184A and T186A all have apparent binding constants within 2.5-fold of wild-type (Table 1 and Figure 2), indicating that they bind substrate RNA equally well. This conclusion is consistent with the gel mobility shift data for these mutants, except for H327A, which has a 6-fold higher Kapp, RNA even though the Kd is similar to wild-type (Tables 1 and 2). This discrepancy could be due to the weaker ATP binding of this mutant or just an artifact of the indirect measurement of RNA binding using the ATPase assay.
Figure 3.
ATPase activities for wild-type DbpA (circles), D156A (diamonds), S184A (upside down triangles), T186A (squares) and H327A (triangles) at (A) saturating ATP (5 mM) and RNA concentration between 0 and 500 nM and (B) saturating RNA (250–500 nM) and ATP concentration between 0 and 4 mM. All experiments were carried out in 50 mM HEPES pH 7.5, 10 mM MgCl2, 50 mM KCl, 100 μM DTT, 200 μM NADH, 1 mM phospho(enol)pyruvate, 10 mM phosphate kinase/lactate dehydrogenase buffer at 24°C. Lines are the best fit of the data using the Michaelis–Menten equation.
The Michaelis–Menten constant for wild-type DbpA (KM, ATP = 400 μM) is consistent with a previous measurement (Table 2 and Figure 3B) (4). KM, ATP values for T186A and H327A are moderately weaker than wild-type DbpA, while mutant D156A shows a slightly tighter KM, ATP than wild-type, suggesting that it may bind ATP better (Figure 3B, Tables 1 and 2). These data are all consistent with gel mobility shift data in the presence of AMPPNP described earlier. The two mutants that fail to bind ATP in this assay do not increase in affinity for RNA when AMPPNP is present in the gel shift assay. The D156A mutant has a tighter KM, ATP, which is also reflected by the 20-fold increase in binding affinity when AMPPNP is present compared with the 10-fold increase for wild-type DbpA (Table 1). The only inconsistency between the two assays is in the case of S184A, which binds ATP equally well as wild-type (KM, ATP = 410 μM) but does not show an expected increase in affinity for RNA in the presence of AMPPNP in gel shift assays (Tables 1 and 2).
RNA helicase assay
The RNA substrate used in the helicase assay contains a 32 nt oligomer containing hairpin 92 annealed to a 5′-32P labeled 9-mer (Figure 4A) (22). In the presence of saturating (2 mM) ATP, wild-type DbpA unwinds the substrate at a rate of 2.2 min−1 (Figure 4B). As expected, the majority of the mutants that hydrolyze ATP poorly; K53A, D153A, E154A, R328A and R331A are unable to unwind the duplex RNA substrate (Table 3). However, T54A, D156A, S184A, T186A and H327A, which have kobs greater than ∼6 min−1, can unwind the RNA duplex at varying rates. Interestingly, R334A has a higher kobs of 14 min−1 (∼12% active), but cannot unwind the duplex (Figure 4B, Tables 1 and 3). In the structure of Drosophila Vasa, the equivalent Arginine (R582) seems to contact all phosphates of the AMPPNP (26). It is possible that this residue is more involved in release of ATP and coordination of the hydrolysis to helicase action, which would result in this uncoupling of activities.
Table 3.
Unwinding rates of wild-type and the eleven DbpA mutants
| Motif | Protein | Unwinding rate (min−1) |
|---|---|---|
| wild-type | 2.2 | |
| I | K53A | <0.1 |
| T54A | 0.1 | |
| II | D153A | <0.1 |
| E154A | <0.1 | |
| D156A | 0.4 | |
| III | S184A | 0.8 |
| T156A | 0.2 | |
| VI | H327A | 0.3 |
| R328A | <0.1 | |
| R331A | <0.1 | |
| R334A | <0.1 |
Unwinding rates were determined from data such as those presented in Figure 4 and are the averages of at least three separate determinations that vary by less than 2-fold.
Growth phenotype
The wild-type dbpA gene was cloned into the pET-3a system (Novagen) where the target genes are under the control of the T7 RNA polymerase promoter (20). Eleven point mutations were made using this dbpA-containing plasmid and Stratagene's QuikChange® XL Site-Directed mutagenesis kit. Plasmids were initially transformed into the T7-lacking E. coli XL10-Gold strain (Stratagene). When mutant DbpA plasmids were transformed into XL10-Gold, all eleven strains behaved the same as wild-type DbpA and wild-type XL10-Gold controls. In order to produce protein for biochemical studies, the same eleven plasmids were transformed into E. coli BL21(DE3) pLysS (Stratagene), which harbors a T7 RNA polymerase gene under control of the lac repressor. During the course of this experiment, we noticed that strains containing the R331A mutant plasmid consistently produced a small colony phenotype while the other ten strains showed colony sizes similar to controls [BL21(DE3) pLysS with wild-type DbpA plasmid and BL21(DE3) pLysS expressing endogenous levels of DbpA]. This small colony phenotype for R331A is only seen in cells containing the T7 RNA polymerase gene and is likely the result of ‘leaky’ expression of the mutant from the plasmid (35). Presumably, the nonfunctional R331A protein from the multicopy plasmid competes with the endogenous DbpA to produce the small colony phenotype. It is striking that none of the other DbpA mutations show the small colony phenotype, even when cells are plated at 24°C instead of 37°C because mutations in DExD/H proteins are often cold sensitive (23,36,37). This may reflect the fact that of the eleven mutations tested, R331A shows the lowest amount of ATPase activity (Table 1). To confirm that the small colony phenotype was a consequence of the R331A point mutation, the mutation was made a second time with identical results.
Two additional growth experiments support the idea that the small colony phenotype results from competition between the R331A protein and the endogenous wild-type DbpA. First, when the R331A plasmid is transformed into E. coli BL21(DE3) cells, even smaller colonies are observed, consistent with an even greater amount of ‘leaky’ plasmid expression in the absence of the pLysS gene encoding the T7 lysozyme, which inhibits T7 RNA polymerase (38). Second, when the R331A plasmid is transformed into an E. coli BL21(DE3) strain with a deletion of the DbpA gene (provided by I. Iost), even smaller colonies were observed, which is expected when the competing wild-type protein is unavailable. Finally, if T7 RNA polymerase is induced by the addition of 1 mM IPTG to any of the strains discussed above, there is no significant change in the results. Presumably, the relatively large amount of ‘leaky’ expression from the plasmid-encoded gene is more than sufficient to compete with the wild-type protein.
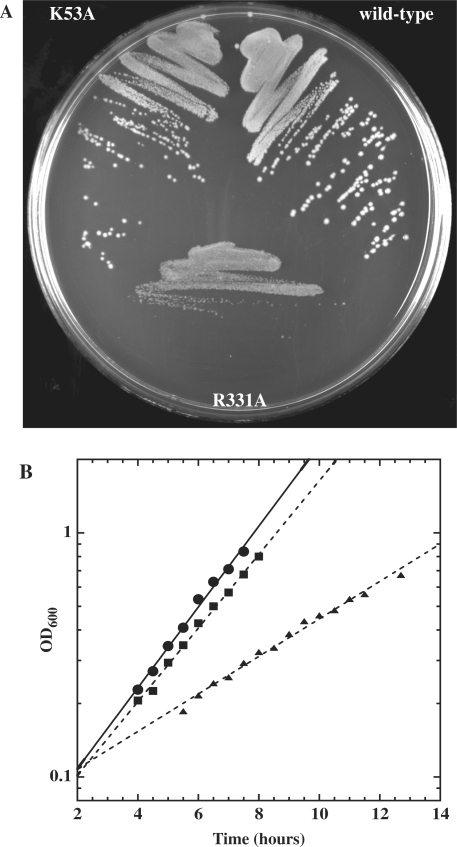
Further analysis of the R331A mutant in BL21(DE3) pLysS cells was complicated by the fact that when cells were grown in liquid culture for many hours, and then plated, the colony size was no longer homogenously small. Instead, a significant fraction of larger colonies was observed. This phenotypic instability is probably the result of recombination between the mutant plasmid gene and the genomic copy of the DbpA gene. Thus, the R331A plasmid and several other mutations were transformed into a Tuner™ (DE3) pLacI RecA− strain. In this case, the small colony phenotype of R331A still occurs (Figure 5A) and is much more stable at both 22°C and 37°C. When the R331A mutation in Tuner™ (DE3) pLacI RecA− was grown in liquid culture at both temperatures, it had a modestly slower growth rate (1.6-fold at 37°C, 2.2-fold at 22°C) than either the wild-type DbpA or the empty vector control (Figure 5B). In addition, at both temperatures, cells harboring the R331A mutation took significantly longer to exit stationary phase and enter log phase (Figure 5B).
Figure 5.
(A) Wild-type DbpA and mutants K53A and R331A in Tuner™ (DE3) pLacI RecA− cells at 37°C. (B) Growth rates and doubling times at 22°C for DbpA (circles, 113 min), K53A (squares, 134 min) and R331A (triangles, 251 min) in Tuner™ (DE3) pLacI RecA− cells (M.E. Saks). Doubling times are an average of 3–4 growth experiments.
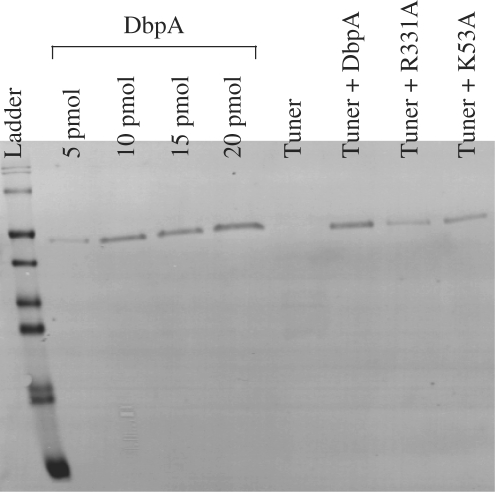
To compare the expression of uninduced R331A mutant with that of endogenous DbpA, semi-quantitative western blot experiments using a polyclonal antibody against DbpA (provided by F. Fuller-Pace) were performed on the R331A mutant and wild-type DbpA expressed in Tuner™ (DE3) pLacI RecA− along with K53A in Tuner™ (DE3) pLacI RecA− and wild-type Tuner™ (DE3) pLacI RecA− controls (Figure 6). The amount of endogenous DbpA present in wild-type cells cannot be detected even when large amounts of lysate are analyzed. Based upon the detection limit of the DbpA antibody, we estimate that there are less than 1000 molecules of DbpA per cell. However, when plasmids containing wild-type DbpA, R331A or K53A are present, much larger amounts of protein were observed. Based on three separate experiments ∼75 000 copies per cell could be estimated for all three proteins. This high level of expression is observed as the result of ‘leaky’ expression from T7 RNA polymerase (35) compounded by the multiple copies of the plasmid. Because the amount of R331A protein is not significantly different from the others, its small colony phenotype is not due to the presence of a larger or smaller amount of protein. Furthermore, the 75-fold excess of R331A mutant protein over endogenous DbpA protein means that it can effectively compete for the available 23S rRNA hairpin 92 binding site, despite the fact that in the presence of ATP it binds less well than wild-type DbpA.
Figure 6.
Western blot of cell lysates. A serial dilution of purified DbpA was used to calibrate the sensitivity of the blot and allows an estimate of the amount of DpbA present in a given cell.
DISCUSSION
The mutations in the active site of DbpA possess similar biochemical properties as those observed in several other DExD/H proteins including, human eIF-4A, yeast Prp16, Prp22, Prp43 (12,31,32). The classic ATPase motifs I and II, also known as the Walker A and B motifs, are highly conserved and important in the ATP hydrolysis cycle (30,33). In DbpA, alanine substitution of the strictly conserved residues of these motifs (K53, T54, D153 and E154) causes drastic reductions in ATP hydrolysis rates and helicase activities as expected. Residue K53 of motif I is expected to make contacts with the β and γ-phosphates of ATP and, when mutated in DbpA, kobs is low (0.7 min−1) and helicase activity is abolished. According to crystal structures and other mutagenesis data, motif II residue E154 coordinates a Mg2+ ion while the D153 residue binds to the ATP through that Mg2+ and is proposed to catalyze hydrolysis of the β−γ phosphate bond (30,33). Similar mutation of all these residues to alanine in yeast Prp22, Prp16 and Prp43, as well as more conservative mutation to other residues in human eIF-4A, also show little to no ATPase and no RNA helicase activity (12,31,32). In contrast with the Walker A and B residues, crystal structures of Vasa show that the last residue of motif II, D156, does not contact the ATP but instead makes interdomain contacts with the H327 of motif VI (26). In DbpA, D156A is the only mutant from these two motifs that has significant ATPase activity and moderate helicase activity as also shown with similar mutations of this last residue (D/H) in HCV NS3, human eIF-4A, and yeast Prp16 and Prp43 (12,31,32,39).
Motif III does not contact ATP directly but makes important interdomain contacts, i.e. S184 and T186 contact D156 in motif II and T186 contacts H327 in motif VI, which most likely assists in domain closure (26). Individual mutations of S184 and T186 to alanine in DbpA only moderately decrease helicase and ATPase activity similar to Prp16 and Prp43 (12,32). However, simultaneous mutations of the serine and threonine in motif III of human eIF-4A causes a small decrease in ATP hydrolysis while almost completely abolishing unwinding, suggesting this motif is coupling the ATPase and helicase activities (31).
Motif VI, unlike motifs I, II and III, is found in domain 2, along the ATP-binding pocket at the interface of the two domains (Figure 1B). As mentioned above, H327 makes contacts to domain 1 and to motif III (26) and its mutation to alanine in DbpA, like the other interdomain contacting mutants, (D156A, S184A and T186A) only moderately affects the ATPase and helicase activities. However, individual mutations of each of the three arginines all have severe effects on ATPase and helicase activity as seen with yeast Prp16, Prp 22 and human eIF-4A (21,32). Two of these arginines in the Vasa structure, R331 and R334, make contacts with the ATP (Figure 1B) (26) and when mutated to alanine in DbpA, kobs decreases to <12% of wild-type for R334A and <0.5% of wild-type for R331A (Table 2). The first arginine of motif VI, R328, does not contact the ATP but also has severely decreased ATPase activity when mutated in DbpA (Figure 1B and Table 2). The reduced binding affinity of the R328A mutant might explain the low kobs (Tables 1 and 2) but it is unclear why the binding is affected. One of the arginines in motif VI is thought to act as an ‘arginine finger’, important in transition state stabilization of the ATP hydrolysis cycle (30,33,34). Out of the three arginine mutations in DbpA, the R331A mutant has the most dramatic effect, with no helicase activity, and the lowest kobs of 0.4 min−1 (Tables 2 and 3). Therefore, we propose that R331 is most likely to act as the ‘arginine finger’ in DbpA.
The DbpA R331A mutant is also the only one of the eleven ATPase mutants that displays a dominant negative phenotype (Figure 5). It is interesting to compare its biochemical properties to the K53A mutation, which does not display a phenotype. In other DExD/H helicases, yeast Prp16, Prp22, Prp43 and human and yeast eIF-4A, mutations in either of the amino acids corresponding to K53 or R331 result in lethal or dominant negative phenotypes and greatly reduced ATPase and helicase activities (11–14,31,32). In DbpA, the severely decreased ATPase activity for R331A and K53A are nearly the same (0.4 and 0.7 min−1, respectively) and both show no helicase activity (Tables 2 and 3). They both can bind substrate well (Table 1) and therefore both should block the binding and activity of the wild-type protein in vivo. Why then does only the R331A mutation show a phenotype? The only major biochemical difference between the two mutant proteins is that K53A binds RNA substrate 6-fold tighter in the presence of AMPPNP, similar to wild-type DbpA, where the R331A-binding affinity for RNA is only 3-fold tighter when AMPPNP is present (Table 1). This would suggest that the K53A mutation would be more effective than the R331A mutation in competing with wild-type protein in vivo where ATP is present. Thus a more likely explanation of this caveat lies in the specific activities of the two mutant proteins. It could be that the slightly higher residual ATPase activity of the K53A mutation is sufficient to permit the catalytic cycle and substrate rearrangement to occur and thus avoid the phenotype. In contrast, the R331A mutation eliminates the ‘arginine finger’, which thereby removes the ATP-dependent interdomain motion that is critical for helicase function (34).
Until now, there has not been an available DbpA phenotype. This dominant slow growth mutation will be extremely useful to further investigate the molecular function of DbpA (2). From what is known thus far, DbpA is expected to be important in some role of ribosome biogenesis or translation because it interacts with the 23S rRNA at the peptidyltransferase center. Initially, it will be interesting to study the ribosome populations in the mutant strains to hopefully begin to understand the actual function of DbpA in vivo.
ACKNOWLEDGEMENTS
The project described above was supported by R01-GM60268-08 from the National Institutes of Health, and 1C06RR018850-01from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. The authors would like to thank Dr Ivelitza Garcia, Dr Isabelle Iost and Dr Margaret E. Saks for helpful discussions and support. The DbpA deletion strain was gererously supplied by Dr Isabelle Iost and the DbpA antibody by Dr Frances Fuller-Pace. Funding to pay the Open Access publication charges for this article was provided by R01–GM60268.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicol SM, Fuller-Pace FV. The ‘DEAD Box’ protein DbpA interacts specifically with the peptidyltransferase center in 23S rRNA. Proc. Natl Acad. Sci. USA. 1995;92:11681–11685. doi: 10.1073/pnas.92.25.11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsu CA, Kossen K, Uhlenbeck OC. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsu CA, Uhlenbeck OC. Kinetic analysis of the RNA-dependent adenosinetriphosphatase activity of DbpA, an Escherichia coli DEAD protein specific for 23S ribosomal RNA. Biochemistry. 1998;37:16989–16996. doi: 10.1021/bi981837y. [DOI] [PubMed] [Google Scholar]
- 5.Karginov FV, Caruthers JM, Hu Y, McKay DB, Uhlenbeck OC. YxiN is a modular protein combining a DExD/H core and a specific RNA-binding domain. J. Biol. Chem. 2005;280:35499–35505. doi: 10.1074/jbc.M506815200. [DOI] [PubMed] [Google Scholar]
- 6.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck OC, McKay DB. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA. 2006;12:959–967. doi: 10.1261/rna.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, et al. The Complete Genome Sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 9.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iost I, Dreyfus M. DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 2006;34:4189–4197. doi: 10.1093/nar/gkl500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hotz H-R, Schwer B. Mutational analysis of the Yeast DEAD-box splicing factor Prp16. Genetics. 1998;149:807–815. doi: 10.1093/genetics/149.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of Lariat-intron from the spliceosome. J. Biol. Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 13.Schmid SR, Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol. Cell. Biol. 1991;11:3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwer B, Meszaros T. RNA helicase dynamics in pre-mRNA splicing. EMBO J. 2000;19:6582–6591. doi: 10.1093/emboj/19.23.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang TH, Latus LJ, Liu Z, Abbott JM. Genetic interactions of conserved regions in the DEAD-box protein Prp28p. Nucleic Acids Res. 1997;25:5033–5040. doi: 10.1093/nar/25.24.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daugeron M-C, Linder P. Characterization and mutational analysis of yeast Dbp8p, a putative RNA helicase involved in ribosome biogenesis. Nucleic Acids Res. 2001;29:1144–1155. doi: 10.1093/nar/29.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 18.Oh JY, Kim J. ATP hydrolysis activity of the DEAD box protein Rok1p is required for in vivo ROK1 function. Nucleic Acids Res. 1999;27:2753–2759. doi: 10.1093/nar/27.13.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang M, Green MR. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes Dev. 2001;15:30–35. doi: 10.1101/gad.851701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karginov FV, Uhlenbeck C. Interaction of Escherichia coli DbpA with 23S rRNA in different functional states of the enzyme. Nucleic Acids Res. 2004;32:3028–3032. doi: 10.1093/nar/gkh640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 22.Diges CM, Uhlenbeck OC. Escherichia coli DbpA is a 3′ –> 5′ RNA helicase. Biochemistry. 2005;44:7903–7911. doi: 10.1021/bi050033x. [DOI] [PubMed] [Google Scholar]
- 23.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 24.Caruthers JM, Hu Y, McKay DB. Structure of the second domain of the Bacillus subtilis DEAD-box RNA helicase YxiN. Acta Crystallograph. Sect. F Struct. Biol. Cryst. Commun. 2006;62:1191–1195. doi: 10.1107/S1744309106044642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl Acad. Sci. USA. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 27.Story RM, Steitz TA. Structure of the recA protein-ADP complex. Nature. 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 28.Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, Weber PC. Structure of the hepatitis C virus RNA helicase domain. Nat. Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 29.Zhao R, Shen J, Green MR, MacMorris M, Blumenthal T. Crystal structure of UAP56, a DExD/H-box protein involved in pre-mRNA splicing and mRNA export. Structure. 2004;12:1373–1381. doi: 10.1016/j.str.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 31.Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider S, Hotz H-R, Schwer B. Characterization of dominant-negative mutants of the DEAH-box splicing factors Prp22 and Prp16. J. Biol. Chem. 2002;277:15452–15458. doi: 10.1074/jbc.M112473200. [DOI] [PubMed] [Google Scholar]
- 33.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 34.Dittrich M, Hayashi S, Schulten K. ATP hydrolysis in the betaTP and betaDP catalytic sites of F1-ATPase. Biophys. J. 2004;87:2954–2967. doi: 10.1529/biophysj.104.046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 36.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noble SM, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Studier FW. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 39.Kim DW, Kim J, Gwack Y, Han JH, Choe J. Mutational analysis of the hepatitis C virus RNA helicase. J. Virol. 1997;71:9400–9409. doi: 10.1128/jvi.71.12.9400-9409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]