Abstract
Aichi virus, a member of the family Picornaviridae, encodes a leader (L) protein of 170 amino acids (aa). The Aichi virus L protein exhibits no significant sequence homology to those of other picornaviruses. In this study, we investigated the function of the Aichi virus L protein in virus growth. In vitro translation and cleavage assays indicated that the L protein has no autocatalytic activity and is not involved in polyprotein cleavage. The L-VP0 junction was cleaved by 3C proteinase. Immunoblot analysis showed that the L protein is stably present in infected cells. Characterization of various L mutants derived from an infectious cDNA clone revealed that deletion of 93 aa of the center part (aa 43 to 135), 50 aa of the N-terminal part (aa 4 to 53), or 90 aa of the C-terminal part (aa 74 to 163) abolished viral RNA replication. A mutant (Δ114-163) in which 50 aa of the C-terminal part (aa 114 to 163) were deleted exhibited efficient RNA replication and translation abilities, but the virus yield was 4 log orders lower than that of the wild type. Sedimentation analysis of viral particles generated in mutant Δ114-163 RNA-transfected cells showed that the mutant has a severe defect in the formation of mature virions, but not in that of empty capsids. Thus, the data obtained in this study indicate that the Aichi virus L protein is involved in both viral RNA replication and encapsidation.
Aichi virus, which is associated with human acute gastroenteritis (41), is a member of the genus Kobuvirus of the family Picornaviridae (31, 42). This virus was first isolated in 1989 from a stool specimen from a patient with oyster-associated nonbacterial gastroenteritis in Aichi, Japan (41), and the complete genome sequence was determined in 1998 (42). The single-stranded, positive-sense RNA genome of Aichi virus consists of 8,280 nucleotides (nt) and a poly(A) tract and encodes a polyprotein of 2,432 amino acids (aa) (34, 42). One of the features of the genome organization of Aichi virus is that it encodes a leader (L) polypeptide upstream of the capsid coding region.
Among picornaviruses besides Aichi virus, aphtho-, cardio-, erbo-, and teschoviruses (10) and porcine enterovirus 8 (18) encode L proteins. The L protein of foot-and-mouth disease virus (FMDV), an aphthovirus, is produced in two forms, Lab and Lb, through initiation of translation at two in-frame AUG codons located 84 nt apart (3). Both forms of the FMDV L protein are papain-like thiol proteinases that cleave at their own C termini (23, 30, 36). In addition, the FMDV L protein cleaves a eukaryotic translation initiation factor, eIF4G (9), contributing to the shutoff of host cap-dependent protein synthesis. The shutoff of host protein synthesis leads to inhibition of alpha/beta interferon production. The L deletion mutant of FMDV can replicate and spread in non-interferon-responsive baby hamster kidney (BHK) cells (29). However, it can replicate but cannot spread in secondary cells from susceptible animals, such as bovine, ovine, and porcine cells (8). This is associated with the inability of the L deletion mutant to inhibit cap-dependent alpha/beta interferon mRNA translation (7, 8). The erbovirus L protein also has autocatalytic activity but does not cleave eIF4GI (15).
The cardiovirus L proteins have neither the amino acid sequence motif characteristic of picornavirus proteases nor autocatalytic activity. They are released from a polyprotein by viral protease 3C (27, 33). The L protein of the GDVII strain of Theiler's murine encephalomyelitis virus (TMEV) has been shown to be essential for neurovirulence in mice (6). In addition, it has been reported that L mutants of TMEV and mengovirus exhibit distinct growth properties depending on the host cells: certain L mutants of TMEV and mengovirus can spread in BHK cells but not in mouse L929 cells (1, 40, 43). For the GDVII strain of TMEV, it has been shown that the L deletion mutant has a defect in the assembly of virions in L929 cells (1), which mainly causes the inability of the mutant to spread in L929 cells. On the other hand, for the DA1 strain of TMEV and mengovirus, the L proteins have been reported to be involved in the inhibition of alpha/beta interferon production by L929 cells (40, 44). Furthermore, in the case of encephalomyocarditis virus, it has been suggested that the L protein is involved in internal ribosome entry site-mediated translation of viral RNA (11, 17).
The Aichi virus L protein does not exhibit significant homology to other picornavirus L proteins in either the amino acid sequence or the sequence motif found in picornavirus proteases (42). In this study, we investigated the function of the L protein in virus replication, using various mutants derived from a full-length infectious cDNA clone. Deletion of 93 aa of the center part, 50 aa of the N-terminal part, or 90 aa of the C-terminal part of the L protein abolished viral RNA replication. Deletion of 50 aa of the C-terminal part did not affect RNA replication ability but, remarkably, reduced plaque-forming ability. Sedimentation analysis of viral particles generated in mutant Δ114-163 RNA-transfected cells showed that the mutant Δ114-163 has a severe defect in the formation of mature virions. Thus, the results obtained in this study indicate that the Aichi virus L protein is involved in both viral RNA replication and encapsidation.
MATERIALS AND METHODS
Cell culture.
Vero cells were grown in Eagle's minimal essential medium (MEM) containing 5% fetal calf serum (growth medium) at 37°C.
Plasmids.
pAV-FL is an infectious cDNA clone of Aichi virus strain A846/88 (34). The Aichi virus sequences used for the construction of plasmids in this study were derived from pAV-FL.
(i) pL-VP0.
A fragment containing the T7 promoter sequence and a part of the Aichi virus sequence (nt 1 to 2364) was amplified by PCR with primers M13-RV, which anneals to the vector sequence upstream of the T7 promoter sequence in pAV-FL, and VP0-Stop-M (5′ CATTACTGTGGGGCGAGGTAGC 3′; minus-strand sequence; nt 2348 to 2364; the stop codon is underlined), using pAV-FL as the template. The derived fragment was ligated into pUC118, and then the nucleotide sequence of the fragment was confirmed.
(ii) p3A-3D.
The Aichi virus sequence spanning from nt 5755 to the poly(A) tract was amplified by PCR with primers 5755-P (5′ GATCTCCTGGAAGCCATGG 3′; plus-strand sequence; nt 5755 to 5773) and M13-20, which anneals to the vector sequence downstream of the poly(A) tract in pAV-FL, using pAV-FL as the template, and ligated into the pGEM-T vector (Promega), generating p3A-3D. The nucleotide sequence of the cloned fragment was confirmed. In p3A-3D, the Aichi virus sequence was under the control of the SP6 promoter.
(iii) pL-VP0(Q170P).
An EcoRI-SmaI fragment (nt 386 to 1384) of pAV-FL was subcloned into the EcoRI-SmaI sites of pUC118, yielding pU-ES. A mutation changing the adenine at nt 1253 to a cytocine was introduced into pU-ES by PCR with primers 1254-P (5′ AGGCAACTCGGTCACCAACATC 3′; plus-strand sequence; nt 1254 to 1275) and A1253C-M (5′ GGCCGTTGGAGGTTAGTGGT 3′; minus-strand sequence; nt 1234 to 1253; the mutated nucleotide is underlined), and the amplified fragment was self-ligated. The nucleotide sequence between the SpeI (nt 403) and SmaI sites of the derived clone was confirmed, and then this fragment was ligated into pL-VP0, which had been digested with SpeI and SmaI, generating pL-VP0(Q170P).
(iv) p5′-Bgl.
Two oligonucleotides, BglStop-P (5′ GATCTAAGAGAGAGA 3′; the stop codon is underlined) and HindStop-M3 (5′ AGCTTCTCTCTCTTA 3′; the stop codon is underlined), were annealed with each other. The derived double-stranded oligonucleotide adapter, which has protruding single-stranded BglII and HindIII ends, was ligated into the BglII (nt 3914)-HindIII (just downstream of the poly(A) tail) sites of pAV-FL to introduce a stop codon at nt 3918. The generated plasmid was called p5′-Bgl.
(v) pΔP1-Xho.
An EcoRI-StuI fragment (nt 387 to 4644) of pAV-FL was subcloned into the EcoRI-SmaI sites of pUC118. The P1 region (nt 1276 to 3771) of this subclone was deleted by PCR with primers Sac-3772P (5′ AAGAGCTCACAGTGGCCGTGATCAAGCA 3′; the Aichi virus sequence, nt 3772 to 3791, is underlined) and Xba-1275 M (5′ TTCTAGAGATGTTGGTGACCGAGTTGCCT 3′; the sequence complementary to the Aichi virus sequence, nt 1254 to 1275, is underlined). The PCR product was self-ligated, and the nucleotide sequence between the KpnI (nt 663) and BglII (nt 3914) sites of the derived clone was confirmed. The KpnI-BglII fragment of pAV-FL was replaced with the KpnI-BglII fragment with deletion of nt 1276 to 3771, generating pAV-FLΔP1. To introduce a stop codon at nt 5335, two oligonucleotides, XhoStop-P (5′ TCGACTAAGAGAGAGA 3′; the stop codon is underlined) and HindStop-M1 (5′ AGCTTCTCTCTCTTAG 3′; the stop codon is underlined), were annealed with each other, and the derived double-stranded oligonucleotide adapter with protruding single-stranded XhoI and HindIII ends was ligated into the XhoI (nt 5329)-HindIII [just downstream of the poly(A) tail] sites of pAV-FLΔP1. The generated plasmid was called pΔP1-Xho.
(vi) pΔP1ΔBglStu-Csp.
pAV-FLΔP1 was digested with BglII (nt 3914) and StuI (nt 4644 and 5056), and the resultant largest fragment was blunt ended and then self-ligated, yielding pΔP1ΔBglStu. To introduce a stop codon at nt 6487, two oligonucleotides, CspStop-P (5′ CGAATAAGAGAGAGA 3′; the stop codon is underlined) and HindStop-M2 (5′ AGCTTCTCTCTCTTATT 3′; the stop codon is underlined), were annealed with each other, and the resultant double-stranded oligonucleotide adapter was ligated into the Csp45I (nt 6481)-HindIII [just downstream of the poly(A) tail] sites of pΔP1ΔBglStu. The derived plasmid was called pΔP1ΔBglStu-Csp.
(vii) Deletion mutants.
A deletion was introduced into pU-ES by PCR using the primers shown in Table 1. The PCR product was self-ligated, and the nucleotide sequence between the SpeI (nt 403) and SmaI (nt 1384) sites of the derived plasmid was confirmed. This SpeI-SmaI fragment harboring the deletion was ligated into pAV-FL, which had been digested with SpeI and SmaI.
TABLE 1.
Primers used for construction of mutants
| Mutant | Primer | Sequence | Position (nt) |
|---|---|---|---|
| pΔ43-135 | 1147P | 5′ CAAGCAAAGACCCCGGAGGAGCGT | 1147-1170 (plus strand) |
| 870M | 5′ AGAGAGATGGGGGTTGGTGTT | 850-870 (minus strand) | |
| pΔ4-53 | 901P | 5′ TGCCCGTTCTGTCAGTACGACG | 901-922 (plus strand) |
| 753-M | 5′ TGCAGCCATGACTCCGACATAAAAGG | 728-753 (minus strand) | |
| pΔ74-163 | 1234P | 5′ ACCACTAACCTCCAACGGCA | 1234-1253 (plus strand) |
| 963M | 5′ CTCGCCACGGAGAGACTCTGGGGA | 940-963 (minus strand) | |
| pΔ114-163 | 1234P | 5′ ACCACTAACCTCCAACGGCA | 1234-1253 (plus strand) |
| 1083M | 5′ GTATGTTTCAGGATAGAACCAGGAT | 1060-1083 (minus strand) | |
| pLfs | 766P | 5′ CGATCTGTGCTCGCTGCTGT | 766-785 (plus strand) |
| 764M | 5′ GAAACCCGTGTTGCAGCCATG | 764-744 (minus strand) | |
| 1234P | 5′ ACCACTAACCTCCAACGGCA | 1234-1254 (plus strand) | |
| Ains-1233M | 5′ TAGGGAGTGTGAGAGGGGGGGTGGAa | 1210-1233 (minus strand) |
The inserted nucleotide is underlined.
(viii) pLfs.
A deletion mutation of the adenine at nt 765 was first introduced into pU-ES by PCR using primers 766P and 764M (Table 1), and the amplified DNA fragment was self-ligated. After the nucleotide sequence between the SpeI and SmaI sites of the derived plasmid had been confirmed, the second PCR was performed with primers 1234P and Ains-1233M (Table 1) using this plasmid as the template, with an adenine inserted between nt 1233 and 1234. The PCR fragment was self-ligated, and the nucleotide sequence between the SpeI and SmaI sites of the derived plasmid was confirmed again. Then, this SpeI-SmaI fragment with the two mutations was ligated into pAV-FL, which had been digested with SpeI and SmaI.
In vitro transcription-translation. In vitro translation using rabbit reticulocyte lysate (RRL) was carried out with a TNT quick-coupled transcription-translation system (Promega). Each plasmid was reacted in the reaction mixture at a concentration of 25 μg/ml with or without biotinylated lysyl-tRNA (Promega) or [35S]methionine-cysteine (Promix L-[35S] in vitro cell-labeling mix; Amersham) at 30°C for various times. One microliter of the reaction mixture was subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE). Biotin-labeled translation products were detected by chemiluminescence using a BM chemiluminescence blotting kit (biotin-streptavidin) (Roche Molecular Biochemicals) as described previously (34). For the detection of 35S-labeled products, the gel was dried and then exposed to a Fujifilm imaging plate. Radioactive signals were detected with a phosphorimager (FUJIX BAS1000).
Cleavage assay.
The in vitro translation reaction mixture (10 μl) of pL-VP0 or pL-VP0(Q170P), which had been reacted in the presence of biotinylated lysyl-tRNA, was mixed with 2 μl of the reaction mixture of p3A-D, which had been reacted in the absence of labeled amino acids, and 1 μl of an RNase A solution (10 μg/ml), followed by incubation at 37°C. In the indicated case, 1 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg of aprotinin/ml, 1 μg of pepstatin/ml, and 2 mM EDTA were added to the reaction mixture. After incubation for various times, samples were taken from the cleavage reaction mixture, and 1 μl of each sample was analyzed by SDS-PAGE. Biotin-labeled proteins were detected as described above.
Expression of the L protein in E. coli.
The L coding region was amplified by PCR with primers Nde-L-P (5′ AACATATGGCTGCAACACGGGTTTCAC 3′; plus-strand sequence; nt 745 to 766; the NdeI site is underlined) and Hind-L-M (5′ TTAAGCTTTTGCCGTTGGAGGTTAGTGGTA 3′; minus-strand sequence; nt 1233 to 1254; the HindIII site is underlined), using pAV-FL as the template. The PCR product was cloned into the pGEM-T vector, and the L coding region of the derived clone was sequenced. The NdeI-HindIII fragment of the clone containing the L coding sequence was ligated into the same sites of pET-21a (Novagen), which provided a C-terminal polyhistidine tag (His tag) on the L protein, and the derived plasmid was called pET-L. Escherichia coli BL21(DE3) transformed with pET-L was grown at 37°C in Luria-Bertani broth containing ampicillin until the optical density at 600 nm reached 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to the culture to a final concentration of 1 mM, and the cells were cultured for 2 h. The cells were harvested, and inclusion bodies were purified using BugBuster protein extraction reagent (Novagen) according to the manufacturer's protocol. The inclusion bodies were lysed with 6 M guanidine-HCl, and the His-tagged protein was purified with a HisTrap kit (Pharmacia) under denaturing conditions according to the manufacturer's recommendations. Since SDS-PAGE analysis of the purified fraction revealed two proteins, a major 20-kDa protein and a minor 18-kDa one, the 20-kDa protein, whose molecular mass corresponded to that expected for the L protein plus a His tag, was eluted from the gel electrophoretically. A guinea pig was immunized with the eluted protein in Freund's incomplete adjuvant three times at 3-week intervals.
Synthesis of RNA transcripts.
pAV-FL and the mutants were linearized by digestion with HindIII and then transcribed with T7 RNA polymerase using a T7 RiboMAX express large-scale RNA production system (Promega). The integrity of the synthesized RNAs was confirmed by agarose gel electrophoresis.
Immunoblotting.
Vero cells were transfected with RNA transcripts by electroporation as described previously (34), and at various times after electroporation, cells were lysed in 200 μl of lysis buffer (20 mM Tris-HCl [pH 7.4], 1% sodium deoxycholate, 1% Triton X-100, 0.1% SDS). The lysates (1.5 μl) were analyzed by SDS-PAGE, and then the proteins were blotted onto a polyvinylidene difluoride membrane. Immunoblotting analysis was performed with guinea pig antiserum raised against the Aichi virus L protein, as described previously (34).
Titration of viable viruses generated from transcripts.
Vero cell monolayers in 35-mm-diameter dishes were transfected with 1 μg of in vitro transcripts using Lipofectamine Plus reagent (Life Technologies). After incubation at 37°C for 3 h, 1.5 ml of growth medium was added to each dish, and then the cells were incubated for 72 h. The cells were lysed by three consecutive freeze-thaw cycles, and the lysates were used for the plaque assay. The number of plaques was determined 72 h after infection.
Dot blot hybridization.
Vero cells were transfected with RNA transcripts by electroporation. Total RNA was extracted from the transfected cells using Trizol reagent (Life Technologies) at various times after electroporation. Using these total-RNA samples, dot blot hybridization was carried out to investigate the accumulation of viral positive-strand RNAs in cells as described previously (34).
Analysis of protein synthesis in cells transfected with RNA transcripts.
Vero cells were electroporated with RNA transcripts and then cultured in several 35-mm-diameter dishes with growth medium. After 2 or 5 h, the medium in each dish was replaced with 0.7 ml of methionine-free MEM, and then a total of 40 μCi of [35S]methionine-cysteine was added. After incubation for 1 h, the cells were washed with phosphate-buffered saline and then lysed with 200 μl of lysis buffer. The lysates (1.5 μl) were analyzed by SDS-PAGE. The gel was dried, and radioactive signals were detected with a phosphorimager as described above. The intensity of each band was quantified with the NIH Image program.
Preparation of radiolabeled viruses and sucrose gradient analysis.
The methods used for the preparation of radiolabeled viruses and sucrose gradient analysis were described previously (35). Briefly, Vero cells were electroporated with RNA transcripts and then cultured in growth medium for 2 h. The medium was replaced with MEM without methionine, and then [35S]methionine-cysteine was added. After 4 h, the cells were lysed, and the viral particles were precipitated through a sucrose cushion. The pelleted viral particles were resuspended in phosphate-buffered saline and then sedimented through a 10 to 30% sucrose gradient. The gradients were fractionated, and the radioactivity in each fraction was counted with a liquid scintillation counter. A part of the fraction containing virions or empty capsids was analyzed by SDS-PAGE, and radioactive signals were detected with a phosphorimager.
RESULTS
Aichi virus L protein exhibits no autocatalytic activity.
Analysis of the N-terminal sequence of VP0 was not successful, probably due to its blocked N terminus (42). However, a Gln-Gly pair, which is the typical recognition site of picornavirus 3C proteinase, is present at aa 170 and 171. In addition, the glycine residue at aa 171 constitutes a myristoylation motif (GXXXT), which is found at the N termini of the VP0 sequences of most picornaviruses (Fig. 1A). Based on these observations, Yamashita et al. (42) predicted that the Aichi virus L protein would be released from a polyprotein through cleavage by 3C at the Gln170-Gly171 pair.
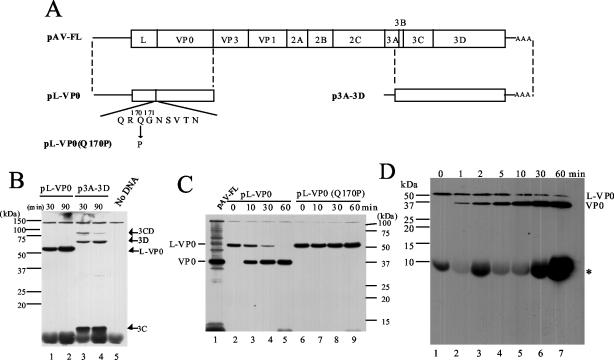
FIG. 1.
Analysis of cleavage at the L-VP0 junction. (A) Diagrams of an infectious cDNA clone of Aichi virus, pAV-FL, and the constructs used for in vitro translation and cleavage assays. The open box and lines indicate coding and noncoding regions, respectively. Below the diagram of pL-VP0, the predicted amino acid sequence around the L-VP0 junction is shown with the myristoylation motif. In pL-VP0(Q170P), a glutamine residue at aa 170 is mutated to a proline. In pAV-FL, pL-VP0, and pL-VP0(Q170P), the Aichi virus sequence is located downstream of the T7 promoter sequence, and in p3A-3D, the Aichi virus sequence is under the control of the SP6 promoter. (B) In vitro transcription-translation of pL-VP0 and p3A-3D in RRL. The plasmids were subjected to in vitro transcription-translation reactions in the presence of biotinylated lysyl-tRNA for 30 or 90 min at 30°C. Proteins were analyzed by SDS-PAGE and then blotted onto a polyvinylidene difluoride membrane. The biotin-labeled translation products were detected by chemiluminescence. The positions of translation products or cleavage products and molecular mass markers are indicated on the right and left, respectively. (C) In vitro cleavage assay. Biotin-labeled translation products of pL-VP0 and pL-VP0(Q170P) were incubated with unlabeled translation products of p3A-3D for various times at 37°C in the presence of RNase A. Then, biotin-labeled proteins were analyzed by SDS-PAGE and detected by chemiluminescence. As markers, in vitro transcription-translation products of pAV-FL were used. The positions of L-VP0 and VP0 and molecular mass markers are indicated on the left and right, respectively. (D) In vitro cleavage assay in the presence of protease inhibitors. The biotin-labeled translation product of pL-VP0 was reacted with the unlabeled translation product of p3A-3D in the presence of 1 mM PMSF, 2 μg of aprotinin/ml, 1 μg of pepstatin/ml, and 2 mM EDTA. Aliquots were taken at the indicated times and analyzed by SDS-15% PAGE. The asterisk indicates nonspecific signal, which was also observed in the sample containing no DNA (Fig. 1B, lane 5). The bromophenol blue dye front corresponds to the bottom of the blot.
First, we examined whether the Aichi virus L protein has autocatalytic activity. The L-VP0 region (Fig. 1A; pL-VP0) was translated in RRL in the presence of biotinylated lysyl-tRNA. As a control, p3A-3D harboring the sequence encoding the C-terminal three-fourths of 3A to 3D (aa 1676 to 2432) was used (Fig. 1A). When the 3A-3D region was translated, three products that were thought to be 3CD, 3D, and 3C were found (Fig. 1B, lanes 3 and 4). In contrast, pL-VP0 produced a 58-kDa protein after a transcription-translation reaction at 30°C for 90 min; this molecular mass corresponded to that of L plus VP0 (lane 2). These results indicate that the Aichi virus L protein has no autocatalytic activity.
Next, we examined whether 3C cleaves the L-VP0 protein at the Gln170-Gly171 pair. A plasmid, pL-VP0(Q170P), was constructed by introducing a mutation that changes the glutamine at aa 170 to a proline in pL-VP0 (Fig. 1A). pL-VP0 and pL-VP0(Q170P) were subjected to a transcription-translation reaction in RRL containing biotinylated lysyl-tRNA. This labeling method was chosen because the predicted amino acid sequences of L and VP0 include seven and six lysines, respectively. Then, each reaction mixture was incubated with the unlabeled p3A-3D translation products for various times. The pL-VP0(Q170P) product was not cleaved on incubation with the unlabeled p3A-3D translation products even after incubation for 1 h at 37°C (Fig. 1C, lanes 6 to 9). In contrast, cleavage of the pL-VP0 product was observed after incubation for only 10 min (lane 3), and after 1 h of incubation, almost all L-VP0 had been cleaved (lane 5). However, we could detect only a cleavage product with mobility corresponding to that of VP0, i.e., not one corresponding to L (lanes 2 to 5). This suggests the possibility that the cleaved L is rapidly degraded. Protease inhibitors (1 mM PMSF, 2 μg of aprotinin/ml, 1 μg of pepstatin/ml, and 2 mM EDTA) were added to the cleavage reaction mixture, and aliquots of the reaction mixture were taken at various times, including 1, 2, and 5 min, and subjected to SDS-15% PAGE. L-VP0 was cleaved in the presence of protease inhibitors, but L or its smaller cleavage products were not detected even after short incubations (Fig. 1D). The predicted amino acid sequence of the L protein includes one methionine and five cysteines. We performed the same experiment with [35S]methionine-cysteine-labeled L-VP0. Similarly, a cleavage product corresponding to VP0 was detected, but we failed to detect products corresponding to L or its smaller cleavage products (data not shown). The cause of the failure to detect L is unknown. However, these results suggest that L-VP0 is cleaved by 3C at the Gln170-Gly171 pair.
The Aichi virus L protein is not involved in polyprotein cleavage.
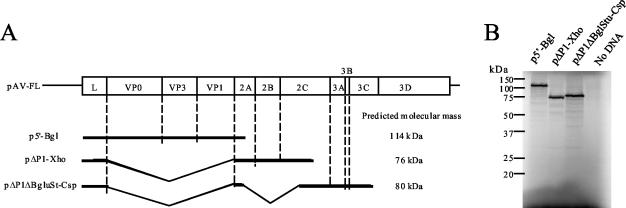
To examine whether the Aichi virus L protein is involved in polyprotein cleavage, we constructed three plasmids, p5′-Bgl, pΔP1-Xho, and pΔP1ΔBglStu-Csp (Fig. 2A). p5′-Bgl harbors the sequence spanning from nt 1 to 3919, encoding L to the N-terminal one-third of 2A. pΔP1-Xho contains the sequence spanning from nt 1 to 5334 with an in-frame deletion of the entire P1 coding region except for the N-terminal 7 amino acids of VP0 and the C-terminal 7 amino acids of VP1. pΔP1ΔBglStu-Csp harbors the sequence from nt 1 to 6486, encoding L to the N-terminal two-thirds of 3C with two in-frame deletions, almost the entire P1-coding region and the region encompassing 2A to 2C. As shown in Fig. 2A, the three plasmids cover all predicted cleavage sites except the 3C-3D site, which was shown to be cleaved by 3C, as described above (Fig. 1B). These three plasmids were subjected to transcription-translation reactions in RRL containing [35S]methionine-cysteine at 30°C for 90 min. Each plasmid produced one major protein with the expected molecular mass (Fig. 2B), indicating that the L protein has no ability to cleave the polyprotein.
FIG. 2.
Investigation of the ability of the Aichi virus L protein to process polyproteins. (A) Diagrams of pAV-FL and deletion mutants. In pAV-FL, the open box and lines indicate coding and noncoding regions, respectively. In deletion mutants, only the regions to be translated are shown as thick lines. Internal deletions are represented by angled lines. The predicted molecular masses of products of deletion mutants are shown. (B) In vitro transcription-translation of p5′-Bgl, pΔP1-Xho, and pΔP1ΔBglStu-Csp in RRL. The plasmids were subjected to in vitro transcription-translation reactions in the presence of [35S]methionine-cysteine for 90 min at 30°C. The translation products were analyzed by SDS-PAGE, and the gel was dried. Radioactive signals were detected with a phosphorimager. The positions of molecular mass markers are indicated on the left.
Expression of the L protein in infected Vero cells.
As shown in Fig. 1C and D, we failed to detect the L protein in the in vitro translation-cleavage experiment. One possible explanation for this failure is that the L protein is unstable. We investigated whether the L protein is stably present in infected Vero cells.
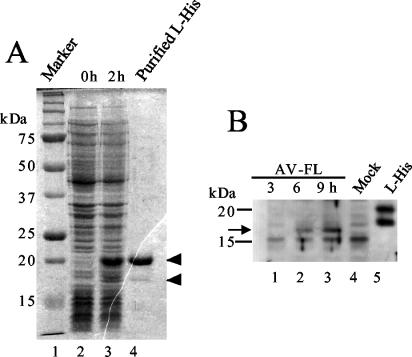
The C-terminally His-tagged L protein was expressed in E. coli. The predicted molecular mass of the His-tagged L protein is 20.9 kDa. After induction, the expression of a 20-kDa protein was observed (Fig. 3A, lane 3). In addition, a protein of 18 kDa was expressed at a lower level. Since the fractions purified with an Ni affinity column contained this minor protein in addition to the major 20-kDa one (Fig. 3A, lane 4), the 20-kDa protein was eluted from the gel and used to immunize a guinea pig.
FIG. 3.
(A) Expression of the His-tagged L protein in E. coli and its purification. Lysates of E. coli cells transformed with pET-L were prepared at 0 and 2 h after induction with IPTG and then analyzed by SDS-PAGE. A fraction purified using an Ni affinity column was also analyzed (lane 4). After electrophoresis, the gel was stained with Coomassie blue. The positions of the 20- and 18-kDa proteins are indicated by arrowheads. (B) Synthesis of the L protein in Vero cells transfected with the AV-FL RNA. At the indicated times after transfection by electroporation, cell lysates were prepared and subjected to SDS-PAGE, and then the L protein was detected by Western blotting using antiserum raised against the purified His-tagged L protein. A fraction purified using an Ni affinity column was also analyzed (lane 5). The positions of molecular mass markers are indicated on the left. An arrow indicates the position of the 17-kDa protein.
The RNA transcript synthesized from an infectious cDNA clone of Aichi virus, pAV-FL, was transfected into Vero cells by electroporation. At 3, 6, and 9 h after electroporation, cell lysates were prepared and analyzed by immunoblotting with anti-L antiserum. The antiserum reacted with a 17-kDa protein, which accumulated with the passage of time (Fig. 3B). Since the predicted molecular mass of the intact L protein is 19.4 kDa, it is possible that a small part of the N or C terminus of the L protein was removed in the cells. Thus, the result indicates that the L protein, or a large part of the L protein, is stable in infected cells.
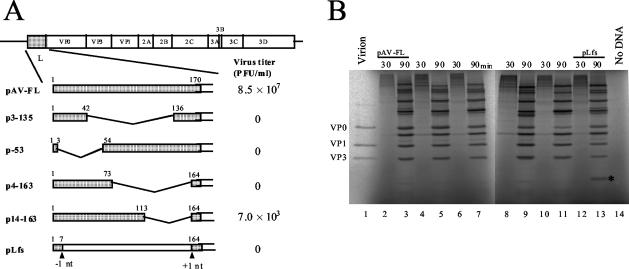
Abilities of L mutants to produce viable viruses.
To investigate whether the L protein is required for virus multiplication in Vero cells, various deletions were introduced into the L coding region of an infectious cDNA clone of Aichi virus, pAV-FL (Fig. 4A). Vero cells were transfected with AV-FL and mutant RNAs by lipofection, and then the virus yields 72 h after transfection were determined by means of plaque assay (Fig. 4A). Deletion of 93 aa of the center part (Δ43-135), 50 aa of the N-terminal part (Δ4-53), and 90 aa of the C-terminal part (Δ74-163) abolished plaque-forming ability. While a mutant, Δ114-163, in which 50 aa of the C-terminal part of the L coding region were deleted, generated viable viruses, the virus titer was 4 log orders lower than that of the wild type. These results indicate that the Aichi virus L protein is essential for virus growth. In addition, it was found that aa 114 to 163 are not essential for virus viability but that the deletion of these amino acids markedly affects viability.
FIG. 4.
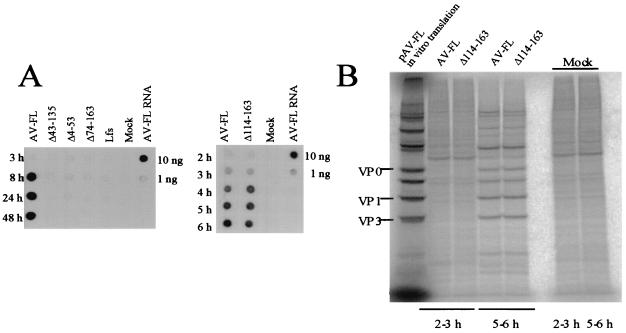
(A) Schematic diagrams of pAV-FL and the L mutants and their abilities to produce viable viruses. The open boxes and lines indicate coding and noncoding regions, respectively. Vertical lines within the box represent putative cleavage sites for viral proteinase. L protein sequences are shaded, and deleted regions are represented by angled lines. Amino acid numbers are indicated above the boxes. In pLfs, the positions where a nucleotide was deleted (−1 nt) and added (+1 nt) are indicated. RNA transcripts synthesized from these plasmids were transfected into Vero cells by lipofection, and the virus titers in the cells 72 h after transfection were determined by plaque assay. The number of plaques was determined 72 h after infection. (B) In vitro transcription-translation of pAV-FL and the L mutants in RRL. The plasmids were reacted in an in vitro transcription-translation system in the presence of [35S]methionine-cysteine for 30 or 90 min at 30°C. The translation products were analyzed by SDS-PAGE, and the gel was dried. Radioactive signals were detected with a phosphorimager. To show the mobility of capsid proteins, 35S-labeled virions were analyzed. The asterisk indicates the unique protein produced from pLfs.
It has been reported that picornavirus genomes contain an internal cis-acting RNA replication element (cre) (13, 14, 20, 21, 22), and cre is located within the coding region in most cases. In order to examine whether the L protein or the L-coding RNA sequence is required for virus growth, pLfs was constructed. In this mutant, the adenine at nt 765 was deleted and an adenine was added between nt 1233 and 1234, so this mutant should express a protein encoded in a different reading frame from the L coding frame (Fig. 4A). This mutant, Lfs, did not generate viable viruses (Fig. 4A), suggesting that the L protein, not the L-coding RNA sequence, is required for virus viability.
The Aichi virus L protein is required for viral RNA replication.
We tried to clarify which stage of virus replication is affected by the mutations in the L mutants. First, the mutant plasmids were subjected to in vitro transcription-translation reactions using RRL. No apparent decrease in translation efficiency was observed in the mutants compared to the wild type (Fig. 4B). This suggests that the mutants do not have a significant defect in protein synthesis. However, in pΔ114-163 the amount of VP0 was reduced, whereas the amounts of other proteins were similar in the wild type and Δ114-163 (lanes 3 and 11). Densitometric quantitation revealed that the VP0/VP1 ratio in Δ114-163 was 35% of that in the wild type. The reduction in VP0 observed in Δ114-163 was probably due to inefficient cleavage at the L-VP0 site caused by the deletion upstream of the cleavage site. We will further investigate below whether the inefficient cleavage at the L-VP0 site is the major cause of the decrease in viable virus produced by Δ114-163.
Next, the RNA replication abilities of the mutants were examined. Vero cells were electroporated with the mutant RNAs, and then total RNAs were extracted from the cells at various times after electroporation and used for dot blot hybridization (Fig. 5A). No replication of the Δ43-135, Δ4-53, Δ74-163, and Lfs RNAs was detected (Fig. 5A, left). While the Δ114-163 RNA replicated as efficiently as the wild type (Fig. 5A, right). These results indicate that the L protein is required for viral RNA replication and that aa 114 to 163 are dispensable for RNA replication.
FIG. 5.
(A) RNA replication of AV-FL and the L mutants. Vero cells were electroporated with the RNA transcripts, and then total RNAs were extracted from the cells at the indicated times after electroporation. The total-RNA samples were dotted and probed with digoxigenin-labeled negative-sense viral RNA (nt 4790 to 5253). As controls, 10 and 1 ng of AV-FL transcripts were dotted. (B) Protein synthesis in Vero cells electroporated with the AV-FL or Δ114-163 RNA. At 2 or 5 h after electroporation, cells were labeled with [35S]methionine-cysteine for 1 h. The cells were lysed, and then the proteins were analyzed by SDS-PAGE. Radioactive signals were detected with a phosphorimager. As markers, in vitro transcription-translation products of pAV-FL were used. Mock, mock transfected.
The Aichi virus L protein is involved in viral RNA encapsidation.
Mutant Δ114-163 exhibited efficient RNA replication ability (Fig. 5A) but produced a 4-log-order-lower level of viable viruses than the wild type (Fig. 4A). As described above, in vitro translation analysis (Fig. 4B) suggested that the inefficient cleavage at the L-VP0 site might be the cause of the defect of Δ114-163 in generating viable viruses. To address this issue further, we investigated protein synthesis in Δ114-163 RNA-transfected cells by pulse-labeling cells with [35S]methionine-cysteine. Between 5 and 6 h after transfection, similar levels of viral protein synthesis were observed in cells transfected with the Δ114-163 RNA and the wild-type RNA (Fig. 5B). Importantly, similar amounts of VP0 were accumulated in the wild-type RNA- and Δ114-163 RNA-transfected cells. This indicates that Δ114-163 has no defect in protein synthesis or cleavage at the L-VP0 site in cells.
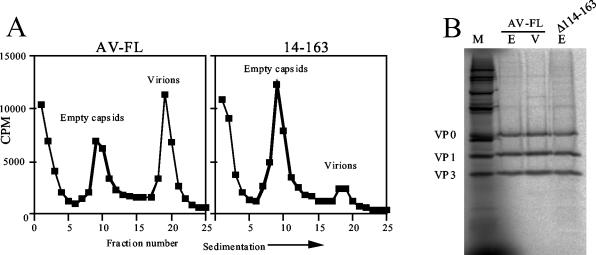
For the reduction in virus production by Δ114-163, we postulated another possible cause, i.e., that Δ114-163 is defective in the formation of virus particles. To examine this possibility, we carried out sedimentation analysis of viral particles generated in RNA-transfected cells. Vero cells were electroporated with the wild-type or Δ114-163 RNA and then labeled with [35S]methionine-cysteine. At 6 h after transfection, the labeled particles were sedimented through a 10 to 30% sucrose gradient, and the gradient was fractionated. As shown in Fig. 6A, both virions and empty capsids accumulated in cells transfected with the wild-type RNA. In contrast, in cells transfected with the Δ114-163 RNA, empty capsids accumulated efficiently but the number of virions was remarkably decreased. To confirm that the uncleaved L-VP0 is not present in empty capsids generated in Δ114-163 RNA-transfected cells, the fraction containing empty capsids from Δ114-163 was analyzed by SDS-PAGE. Empty capsids generated in the Δ114-163 RNA-transfected cells, as well as wild-type virions and empty capsids, were composed of properly cleaved VP0, VP1, and VP3 (Fig. 6B). These results show that Δ114-163 has a defect in RNA encapsidation and, therefore, indicate that the Aichi virus L protein is involved in RNA encapsidation.
FIG. 6.
(A) Sedimentation analysis of viral and subviral particles generated in Vero cells electroporated with AV-FL or Δ114-163 RNA. Vero cells transfected with the RNAs were labeled with [35S]methionine-cysteine. At 6 h after electroporation, the labeled viral and subviral particles were collected and centrifuged through a 10 to 30% sucrose gradient. The gradient was fractionated, and the radioactivity in each fraction was counted with a liquid scintillation counter. (B) Proteins constructing virions and empty capsids. Parts of fractions containing virions (lane V) of the AV-FL virus and empty capsids (lane E) of the AV-FL virus and the Δ114-163 virus were analyzed by SDS-PAGE, and 35S-labeled proteins were visualized with a phosphorimager. As markers, in vitro transcription-translation products of pAV-FL were used (lane M).
DISCUSSION
Aichi virus encodes an L protein of 170 aa which exhibits no significant sequence homology to those of other picornaviruses, aphtho-, cardio-, erbo-, or teschoviruses or porcine enterovirus 8. In this study, we investigated the function of the Aichi virus L protein in virus growth. In vitro translation analysis and cleavage assays indicated that the L protein has no autocatalytic activity and is not involved in polyprotein cleavage. The L-VP0 protein was cleaved by 3C at Gln170-Gly171. Characterization of various L mutants derived from an infectious cDNA clone revealed that the L protein is involved in both viral RNA replication and encapsidation. The region spanning aa 114 to 163 was dispensable for viral RNA replication, but deletion of this region remarkably reduced encapsidation efficiency.
The L proteins of aphtho- and cardioviruses have been reported not to be essential for viral RNA replication. L deletion mutants of FMDV (29), TMEV (1, 6, 24, 40), and mengovirus (43) can replicate in BHK cells. An encephalomyocarditis virus mutant, in which a large portion of the L coding region is deleted, can also replicate in HeLa cells, albeit with lower efficiency compared to the wild type (11).
The L deletion mutant of the GDVII strain of TMEV cannot form plaques on mouse L929 cells. The mutant exhibits no severe defects in RNA replication or protein synthesis. It has been shown that the major cause of the inability of this mutant to form plaques on L929 cells is a defect in virion assembly (1). In L929 cells infected with this mutant, early viral assembly intermediates, like protomers and pentamers, were detected, albeit at reduced levels compared to the wild type, but mature virions and empty capsids were undetectable. Thus, the L protein of the GDVII strain is required for virion assembly at postpentameric stages in L929 cells (1). On the other hand, the present study showed that Δ114-163 has a defect in the formation of mature virions but not in the formation of empty capsids, indicating that the Aichi virus L protein is required for the encapsidation of viral RNA. That is, the L proteins of Aichi virus and GDVII must function at different stages during virion assembly.
In vitro translation-cleavage assays indicated that cleavage of the L-VP0 junction by 3C proteinase occurs at the Gln170-Gly171 pair (Fig. 1). In this experiment, we could detect a cleavage product corresponding to VP0 but failed to detect one corresponding to the L protein (Fig. 1C and D). In contrast, on immunoblot analysis with the anti-L antiserum, we detected a 17-kDa protein that accumulated in RNA-transfected cells with the passage of time (Fig. 3B). This protein detected by the anti-L antiserum migrated slightly faster than expected from the amino acid sequence of the L protein, 19.4 kDa. The L coding sequence contains no in-frame AUG codon other than the initiation codon. This suggests that the L protein is either partially degraded or posttranslationally modified in infected cells. It may be possible that some modification is essential for stability of the L protein and that instability of the L protein synthesized in vitro is due to no or little modification. In the in vitro translation analysis (Fig. 4B), the pLfs plasmid produced a unique protein (near the bottom of lane 13). This plasmid has frameshift mutations in the L coding region (Fig. 4A). This unique protein is likely to be a protein that is encoded by a reading frame different from that coding for the L protein. This protein, having an amino acid sequence different from that of L, would be stable in vitro. It is interesting to investigate what modifications, if any, are required for the stability of the L protein in vivo.
Further studies are required for understanding the mechanism by which the Aichi virus L protein is involved in viral RNA replication and encapsidation. In picornaviruses, viral RNA replication and encapsidation are coupled (26, 38). Picornavirus infection induces rearrangement of intracellular membranes into vesicles, which are associated with nonstructural proteins and viral RNAs (5, 38), and these membrane vesicles, called the replication complex, are the site of picornavirus RNA replication (4). In the encapsidation process, it is thought that newly synthesized positive-strand RNAs, which accumulate around the replication complex, are associated with capsid precursors attached to the surface of the replication complex (26, 28, 38). It has been reported that the poliovirus 2C protein is involved in viral RNA replication (19) and encapsidation (39). The 2C protein migrates to the rough endoplasmic reticulum to induce formation of the replication complex (37). In addition, the poliovirus 2C protein binds to RNA (2, 5, 32), and 2C is proposed to be responsible for the attachment of the viral RNA to the membrane vesicles (2, 5). Membrane binding domains (12, 37) and RNA binding domains (32) have been mapped within poliovirus 2C. We searched the amino acid sequence of the Aichi virus L protein for a transmembrane domain using the SOSUI program (16), but the program did not identify one. Also, the L sequence does not contain the endoplasmic reticulum retention motif, KDEL (25). Recently, it was found that a stem-loop structure formed at the 5′ end of the Aichi virus genome is involved in RNA replication and encapsidation (34, 35). Analyses of the localization of the L protein in infected cells, and its interaction with viral and host proteins, and viral RNA will help us understand the mechanism of Aichi virus replication.
In conclusion, our data indicate that the Aichi virus L protein is involved in viral RNA replication and encapsidation during virus replication. Such functions of the L protein are unique to Aichi virus among picornaviruses.
Acknowledgments
This work was supported in part by a grant from the Human Science Research Foundation of Japan and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Badshah, C., M. A. Calenoff, and K. Rundell. 2000. The leader polypeptide of Theiler's murine encephalomyelitis virus is required for the assembly of virions in mouse L cells. J. Virol. 74:875-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee, R., A. Echeverri, and A. Dasgupta. 1997. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 71:9570-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belsham, G. J. 1992. Dual initiation sites of protein synthesis on FMDV RNA are selected following internal entry and scanning of ribosomes in vivo. EMBO J. 11:1105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 5.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calenoff, M. A., C. S. Badshah, M. C. Dal Canto, H. L. Lipton, and M. K. Rundell. 1995. The leader polypeptide of Theiler's virus is essential for neurovirulence but not for virus growth in BHK cells. J. Virol. 69:5544-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinsangaram, J., M. Koster, and M. J. Grubman. 2001. Inhibition of L-deleted foot-and-mouth disease virus replication by alpha/beta interferon involves double-stranded RNA-dependent protein kinase. J. Virol. 75:5498-5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinsangaram, J., M. E. Piccone, and M. J. Grubman. 1999. Ability of foot-and-mouth disease virus to form plaques in cell culture is associated with suppression of alpha/beta interferon. J. Virol. 73:9891-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devaney, M. A., V. N. Vakharia, R. E. Lloyd, E. Ehrenfeld, and M. J. Grubman. 1988. Leader protein of foot-and-mouth disease virus is required for cleavage of the p220 component of the cap-binding protein complex. J. Virol. 62:4407-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doherty, M., D. Todd, N. McFerran, and E. M. Hoey. 1999. Sequence analysis of a porcine enterovirus serotype 1 isolate: relationships with other picornaviruses. J. Gen. Virol. 80:1929-1941. [DOI] [PubMed] [Google Scholar]
- 11.Dvorak, C. M. T., D. J. Hall, M. Hill, M. Riddle, A. Pranter, J. Dillman, M. Deibel, and A. C. Palmenberg. 2001. Leader protein of encephalomyocarditis virus binds zinc, is phosphorylated during viral infection, and affects the efficiency of genome translation. Virology 290:261-271. [DOI] [PubMed] [Google Scholar]
- 12.Echeverri, A. C., and A. Dasgupta. 1995. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208:540-553. [DOI] [PubMed] [Google Scholar]
- 13.Gerber, K., E. Wimmer, and A. V. Paul. 2001. Biochemical and genetic studies of the initiation of human rhinovirus 2 RNA replication: identification of a cis-replicating element in the coding sequence of 2Apro. J. Virol. 75:10979-10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinton, T. M., N. Ross-Smith, S. Warner, G. J. Belsham, and B. S. Crabb. 2002. Conservation of L and 3C proteinase activities across distantly related aphthoviruses. J. Gen. Virol. 83:3111-3121. [DOI] [PubMed] [Google Scholar]
- 16.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman, M. A., and A. C. Palmenberg. 1996. Revertant analysis of J-K mutations in the encephalomyocarditis virus internal ribosomal entry site detects an altered leader protein. J. Virol. 70:6425-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krumbholz, A., M. Dauber, A. Henke, E. Birch-Hirschfeld, N. J. Knowles, A. Stelzner, and R. Zell. 2002. Sequencing of porcine enterovirus groups II and III reveals unique features of both virus groups. J. Virol. 76:5813-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, J.-P., and D. Baltimore. 1988. Isolation of poliovirus 2C mutants defective in viral RNA synthesis. J. Virol. 62:4016-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobert, P.-E., N. Escriou, J. Ruelle, and T. Michiels. 1999. A coding RNA sequence acts as a replication signal in cardiovirus. Proc. Natl. Acad. Sci. USA 96:11560-11565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mason, P. W., S. V. Bezborodova, and T. M. Henry. 2002. Identification and characterization of a cis-acting replication element (cre) adjacent to the internal ribosome entry site of foot-and-mouth disease virus. J. Virol. 76:9686-9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKnight, K. L., and S. M. Lemon. 1998. The rhinovirus type 14 genome contains an internally located RNA structure that is required for viral replication. RNA 4:1569-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Medina, M., E. Domingo, J. K. Brangwyn, and G. J. Belsham. 1993. The two species of the foot-and-mouth disease virus leader protein, expressed individually, exhibit the same activities. Virology 194:355-359. [DOI] [PubMed] [Google Scholar]
- 24.Michiels, T., V. Dejong, R. Rodrigus, and C. Shaw-Jackson. 1997. Protein 2A is not required for Theiler's virus replication. J. Virol. 71:9549-9556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munro, S., and H. R. Pelham. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48:899-907. [DOI] [PubMed] [Google Scholar]
- 26.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parks, G. D., G. M. Duke, and A. C. Palmenberg. 1986. Encephalomyelitis virus 3C protease: efficient cell-free expressions from clones which link 5′ noncoding sequences to the P3 region. J. Virol. 60:376-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfister, T., L. Pasamontes, M. Troxler, D. Egger, and K. Bienz. 1992. Immunocytochemical localization of capped-related particles in subcellular fractions of poliovirus-infected cells. Virology 188:676-684. [DOI] [PubMed] [Google Scholar]
- 29.Piccone, M. E., E. Rieder, P. W. Mason, and M. J. Grubman. 1995. The foot-and-mouth disease virus leader proteinase gene is not required for viral replication. J. Virol. 69:5376-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Piccone, M. E., M. Zellner, T. F. Kumosinski, P. W. Mason, and M. J. Grubman. 1995. Identification of the active-site residues of the L proteinase of foot-and-mouth disease virus. J. Virol. 69:4950-4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pringle, C. R. 1999. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch. Virol. 144:2065-2070. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez, P. L., and L. Carrasco. 1995. Poliovirus protein 2C contains two regions involved in RNA binding activity. J. Biol. Chem. 270:10105-10112. [DOI] [PubMed] [Google Scholar]
- 33.Roos, R. P., W.-P. Kong, and B. L. Semler. 1989. Polyprotein processing of Theiler's murine encephalomyelitis virus. J. Virol. 63:5344-5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sasaki, J., Y. Kusuhara, Y. Maeno, N. Kobayashi, T. Yamashita, K. Sakae, N. Takeda, and K. Taniguchi. 2001. Construction of an infectious cDNA clone of Aichi virus (a new member of the family Picornaviridae) and mutational analysis of a stem-loop structure at the 5′ end of the genome. J. Virol. 75:8021-8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sasaki, J., and K. Taniguchi. 2003. The 5′-end sequence of Aichi virus, a picornavirus, contains an element critical for viral RNA encapsidation. J. Virol. 77:3542-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strebel, K., and E. Beck. 1986. A second protease of foot-and-mouth disease virus. J. Virol. 58:893-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Brinz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 71:8962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Troxler, M., D. Egger, T. Pfister, and K. Bienz. 1992. Intracellular localization of poliovirus RNA by in situ hybridization at the ultrastructural level using single-stranded riboprobes. Virology 191:687-697. [DOI] [PubMed] [Google Scholar]
- 39.Vance, L. M., N. Moscufo, M. Chow, and B. A. Heinz. 1997. Poliovirus 2C region functions during encapsidation of viral RNA. J. Virol. 71:8759-8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Pesch, V., O. van Eyll, and T. Michiels. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 75:7811-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamashita, T., S. Kobayashi, K. Sakae, S. Nakata, S. Chiba, Y. Ishihara, and S. Isomura. 1991. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J. Infect. Dis. 164:954-957. [DOI] [PubMed] [Google Scholar]
- 42.Yamashita, T., K. Sakae, H. Tsuzuki, Y. Suzuki, N. Ishikawa, N. Takeda, T. Miyamura, and S. Yamazaki. 1998. Complete nucleotide sequence and genetic organization of Aichi virus, a distinct member of the Picornaviridae associated with acute gastroenteritis in humans. J. Virol. 72:8408-8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zoll, J., J. M. D. Galama, F. J. M. van Kuppeveld, and W. J. G. Melchers. 1996. Mengovirus leader is involved in the inhibition of host cell protein synthesis. J. Virol. 70:4948-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zoll, J., W. J. G. Melchers, J. M. D. Galama, and F. J. M. van Kuppeveld. 2002. The mengovirus leader protein suppresses alpha/beta interferon production by inhibition of the iron/ferritin-mediated activation of NF-κB. J. Virol. 76:9664-9672. [DOI] [PMC free article] [PubMed] [Google Scholar]