Abstract
The conversion of [14C]benzoyl-coenzyme A (CoA) to nonaromatic products in the denitrifying β-proteobacterium Azoarcus evansii grown anaerobically on benzoate was investigated. With cell extracts and 2-oxoglutarate as the electron donor, benzoyl-CoA reduction occurred at a rate of 10 to 15 nmol min−1 mg−1. 2-Oxoglutarate could be replaced by dithionite (200% rate) and by NADPH (∼10% rate); in contrast NADH did not serve as an electron donor. Anaerobic growth on aromatic compounds induced 2-oxoglutarate:acceptor oxidoreductase (KGOR), which specifically reduced NADP+, and NADPH:acceptor oxidoreductase. KGOR was purified by a 76-fold enrichment. The enzyme had a molecular mass of 290 ± 20 kDa and was composed of three subunits of 63 (γ), 62 (α), and 37 (β) kDa in a 1:1:1 ratio, suggesting an (αβγ)2 composition. The native enzyme contained Fe (24 mol/mol of enzyme), S (23 mol/mol), flavin adenine dinucleotide (FAD; 1.4 mol/mol), and thiamine diphosphate (0.95 mol/mol). KGOR from A. evansii was highly specific for 2-oxoglutarate as the electron donor and accepted both NADP+ and oxidized viologens as electron acceptors; in contrast NAD+ was not reduced. These results suggest that benzoyl-CoA reduction is coupled to the complete oxidation of the intermediate acetyl-CoA in the tricarboxylic acid cycle. Electrons generated by KGOR can be transferred to both oxidized ferredoxin and NADP+, depending on the cellular needs. N-terminal amino acid sequence analysis revealed that the open reading frames for the three subunits of KGOR are similar to three adjacently located open reading frames in Bradyrhizobium japonicum. We suggest that these genes code for a very similar three-subunit KGOR, which may play a role in nitrogen fixation. The α-subunit is supposed to harbor one FAD molecule, two [4Fe-4S] clusters, and the NADPH binding site; the β-subunit is supposed to harbor one thiamine diphosphate molecule and one further [4Fe-4S] cluster; and the γ-subunit is supposed to harbor the CoA binding site. This is the first study of an NADP+-specific KGOR. A similar NADP+-specific pyruvate oxidoreductase, which contains all domains in one large subunit, has been reported for the mitochondrion of the protist Euglena gracilis and the apicomplexan Cryptosporidium parvum.
In the last decade numerous facultative and obligate anaerobic bacteria which under anaerobic conditions are able to use low-molecular-weight aromatic compounds as their sole sources of cell carbon and energy have been identified. These bacteria metabolize aromatic compounds to only a few common intermediates, among which benzoyl-coenzyme A (CoA) plays a central role (reviewed in references 9, 20, and 22). In facultative anaerobes, such as denitrifying or photosynthetic bacteria, benzoyl-CoA becomes dearomatized in a reductive process which requires electrons at an extremely low redox potential. The analogous reaction in organic synthesis (known as Birch reduction) requires solvated electrons, the most potent reductant (9). The enzymatic reaction is catalyzed by benzoyl-CoA reductase (BCR) (5). So far, BCR activity has solely been demonstrated in cell extracts of the denitrifying Thauera aromatica and the phototrophic Rhodopseudomonas palustris (31, 32). All biochemical information on BCR derives from T. aromatica (5-7, 9).
BCR catalyzes the ATP-driven two-electron reduction of the aromatic ring, yielding cyclohexa-1,5-diene-1-carbonyl-CoA (10); two molecules of ATP are hydrolyzed to ADP to drive the two-electron transfer reaction (6) (see Fig. 4 and Discussion). The 163-kDa enzyme consists of four different subunits with molecular masses of 49, 48, 44, and 30 kDa, resulting in an αβγδ composition (5). Its only recognized cofactors are three cysteine-ligated [4Fe-4S]+/2+ clusters with redox potentials more negative than −500 mV. In recent studies growing evidence that, in analogy to the chemical Birch reaction, enzymatic ring reduction proceeds via radical intermediates has been provided (40; reviewed in reference 9). BCR overcomes energetic limitations by using a low-potential electron donor (ferredoxin) (7, 8), by coupling the electron transfer to a stoichiometric ATP hydrolysis (two ATP molecules for each two electrons transferred) (5, 6), and by an appropriate binding of the substrate (40). The process of ATP-driven electron transfer in BCR can be considered analogous to the well-studied enzymatic dinitrogen reduction (24).
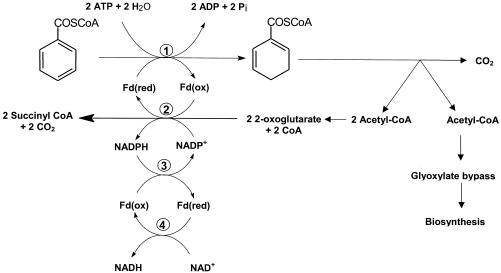
FIG. 4.
Proposed electron transfer reactions involved in 2-oxoglutarate oxidation and enzymatic ring reduction. In this scheme ferredoxin (Fd) is considered a one-electron carrier. Note that from one molecule of benzoyl-CoA three molecules of acetyl-CoA and one molecule of CO2 are formed. Approximately one of every three acetyl-CoA molecules is used for biosynthesis via the glyoxylate cycle, in which isocitrate is cleaved by isocitrate lyase (not shown). The residual two acetyl-CoA molecules are channeled into the TCA cycle, in which two isocitrate molecules are oxidized to 2-oxoglutarate molecules by isocitrate dehydrogenase. The surplus of electrons generated by KGOR are channeled to biosynthetic processes via NADPH. Alternatively, surplus NADPH can be used to reduce ferredoxin by a NADPH:ferredoxin oxidoreductase which equilibrates the NADP+/NADPH and oxidized/reduced ferredoxin pool. Reduced ferredoxin can transfer electrons also to NAD+. Enzymes: 1, BCR; 2, 2-oxoglutarate:acceptor oxidoreductase; 3, NADPH:ferredoxin oxidoreductase; 4, ferredoxin:NAD+ oxidoreductase.
In T. aromatica, a ferredoxin of the Chromatium vinosum type (33) containing two [4Fe-4S]+/2+ clusters serves as the electron donor for BCR in vivo (7, 8, 12). The ferredoxin-reducing enzyme was identified as 2-oxoglutarate:ferredoxin oxidoreductase (KGOR), which replaces the 2-oxoglutarate dehydrogenase complex of the tricarboxylic acid (TCA) cycle during anaerobic growth on aromatic compounds (16). The 200 ± 20-kDa enzyme has an (αβ)2 composition and contains two thiamine diphosphate (TPP) molecules and two [4Fe-4S]+/2+ clusters per dimer. It belongs to the Halobacterium type of 2-oxoacid oxidoreductases, which lack a ferredoxin-like module with two [4Fe-4S]+/2+ clusters (28-30, 44, 56).
In the phototrophic R. palustris and the two denitrifying bacteria T. aromatica and Azoarcus evansii clusters of genes coding for enzymes involved in anaerobic benzoate metabolism (bad gene clusters [benzoic acid degradation]) have been identified (12, 17, 20). They comprise the four structural genes of BCR, genes coding for enzymes of a modified β-oxidation (which are involved in oxidation and ring fission of the dearomatized intermediate), and the gene coding for the electron donor ferredoxin. In T. aromatica additionally the two structural genes encoding KGOR were found next to the bad gene cluster (16). The amino acid sequences of BCR and ferredoxin in T. aromatica and R. palustris are highly similar (64 to 76% identity). In contrast, the corresponding sequences in A. evansii show much lower similarities to those of BCR and ferredoxin of T. aromatica (e.g., 24 to 28% identity for the α- and δ-subunits).
Previously, an unusual NADP+-dependent 2-oxoglutarate dehydrogenase activity was observed in A. evansii cell extracts; this activity was induced during anaerobic growth on aromatic substrates (36). This suggested that NADPH rather than ferredoxin might be involved in transferring electrons to BCR. In addition, a benzoate-CoA ligase was purified and characterized (1); however, BCR activity could not be measured. The aim of this work was to demonstrate the reductive dearomatization process in A. evansii. A particular focus was on the electron transfer system, which stoichiometrically generates and transfers electrons to the aromatic ring.
We identified and characterized KGOR as electron-generating enzyme for enzymatic ring reduction. In contrast to the enzyme from T. aromatica (16) and most other 2-oxoacid:acceptor oxidoreductases (4, 13, 25, 28, 29, 30, 38, 44, 46, 49-51, 53, 54, 56), KGOR accepted both NADP+ and one-electron carriers as a substrate. This is the first study of an NADP+-specific KGOR. A similar NADP+-specific pyruvate oxidoreductase, which, however, contains all domains in one large subunit, has been reported for the mitochondrion of the protist Euglena gracilis and the apicomplexan Cryptosporidium parvum (26, 42, 47).
MATERIALS AND METHODS
Growth of bacterial cells.
A. evansii KB 740 (2) (DSM 6869; Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany), formerly designated Pseudomonas sp. strain KB740, was grown anoxically at 30°C in mineral salt media (1, 2) in a 200-liter fermentor. The substrates were continuously fed from a concentrated stock solution, pH 7.4, containing 0.5 M aromatic acid and 1.8 M KNO3 (molar ratio of benzoate to nitrate of 1:3.6), which served as the sole source of energy and cell carbon (2). The pumping rate was electronically controlled and increased exponentially. The pumping rate p (in milliliters per minute) is given by the equation p = [ln 2 × A0RV × 1,000 × et(ln 2/td)]/tdYc, where A0 is the optical density of the culture at the beginning of the feeding; R is the content of cell dry mass per liter of culture at an optical density (OD) of 1 (approximately 0.38 g liter−1), V is the culture volume (200 liters), td is the chosen preset generation time (in minutes; normally 480 min), t is the time (in minutes), Y is the molar growth yield (approximately 50 g of dry cell mass formed per mol of benzoate consumed), and c is the concentration (moles per liter) of benzoate in the stock solution (0.5 M). Cells were harvested at an OD at 578 nm (OD578) of ∼4 by continuous centrifugation. When cells were grown on acetate-nitrate, the molar ratio of substrate supply was 1:1. Aerobic cultivation on benzoate (5 mM) or acetate (20 mM) with NH4Cl (10 mM) as the nitrogen source used the same basal medium but without nitrate.
Cell extract.
For the preparation of cell extracts 80 g of cells (wet mass) of A. evansii grown on benzoate and nitrate was anaerobically suspended in 120 ml of 100 mM Tris-HCl (pH 7.8)-20% (wt/vol) glycerol-2 mM dithioerythritol (DTE)-1 to 2 mg of DNase I. French press treatment (137 MPa) and ultracentrifugation (100,000 × g) were performed under anaerobic conditions. Small-scale preparation of cell extract was performed accordingly.
Protein purification.
For large-scale protein purification all steps up to the affinity chromatography on Reactive Red agarose were performed twice. Anaerobic buffers contained 2 mM DTE to minimize loss of enzyme activity. DTE was omitted for the steps involving Reactive Red agarose and gel filtration chromatography; instead these purification steps were performed in an anaerobic glove box under a N2-H2 (95%-5% [vol/vol]) atmosphere.
(i) PEG 6000 precipitation.
The cell extract obtained after ultracentrifugation (144 ml) was precipitated by titration with a polyethylene glycol (PEG) 6000 stock solution (25% by volume in H2O) to a final concentration of 6% (by volume). After a 20-min stirring on ice and centrifugation (14,000 × g), KGOR activity was found in the supernatant, which was again precipitated with the same PEG 6000 stock solution to a final concentration of 11% (by volume) and treated as described above.
(ii) DEAE-Sepharose chromatography.
For the following chromatographic steps all buffers used were made anaerobic by flushing with nitrogen gas before use (100%, 2 × 104 Pa, 10 min). All columns and protein fractions were cooled to 6 to 10°C during the purification. For the first chromatographic step the protein pellet obtained after PEG 6000 precipitation was resuspended in 40 ml of 10 mM Tris-HCl (pH 7.8)-20% (wt/vol) glycerol-2 mM DTE (buffer A). The protein solution was applied to a DEAE-Sepharose column (Amersham Biosciences, Freiburg, Germany; column volume, 280 ml; column diameter, 5 cm; flow rate, 6 ml min−1), which had been equilibrated with buffer A. The column was washed with 1.6 bed volumes of buffer A plus 100 mM KCl. The washing step was followed by a gradient from buffer A plus 100 mM KCl to buffer A plus 300 mM KCl in 8 column volumes. KGOR activity eluted between 165 and 225 mM KCl in a volume of 580 ml.
(iii) Q-Sepharose chromatography.
The active DEAE fractions were combined and diluted 1:1.4 with buffer A to a final concentration of 140 mM KCl. This fraction was applied on a High Load Q-Sepharose column (Amersham Biosciences; column volume, 80 ml; column diameter, 2.6 cm; flow rate, 3.4 ml min−1), which had been equilibrated with 20 mM morpholinopropanesulfonic acid-KOH buffer (pH 7)-20% (wt/vol) glycerol-2 mM DTE (buffer B). The column was washed with 2.5 bed volumes of buffer B containing 100 mM KCl, followed by 560 ml of a linear KCl gradient from 100 to 240 mM in buffer B. The activity eluted between 160 and 240 mM KCl. The fractions containing KGOR activity were pooled and gassed with N2 gas (see above).
(iv) Reactive Red agarose chromatography.
Reactive Red agarose chromatography (Sigma, Taufkirchen, Germany; column volume, 220 ml; column diameter, 5 cm; flow rate, 7 ml min−1) was performed in an anaerobic glove box. The column was equilibrated with anoxic 10 mM Tris-HCl (pH 7.8) containing 10% (wt/vol) glycerol (buffer C). After the protein pool containing KGOR activity was applied, the column was washed with 600 ml of buffer C plus 190 mM KCl. KGOR activity was eluted in a single step with 600 mM KCl in buffer C in a volume of 255 ml.
(v) Concentration steps.
To avoid loss of activity during concentration, the high KCl concentration had to be reduced. The pooled fractions were first concentrated by ultrafiltration with a 30-kDa-cutoff membrane (Amicon, Beverly, Mass.) to a volume of approximately 30 ml. After dilution with buffer C to a volume of 210 ml (KCl concentration of less than 100 mM) the enzyme solution was applied to a second DEAE-Sepharose column (volume, 35 ml; diameter, 5 cm; flow rate, 5 ml min−1), which had been equilibrated with buffer C. The KGOR activity was eluted with buffer C plus 220 mM KCl in a volume of 77 ml and was again concentrated by ultrafiltration to 6 ml.
(vi) Gel filtration.
Gel filtration chromatography on a Superdex 200 column (Amersham Biosciences; column volume, 300 ml; column diameter, 2.6 cm; flow rate, 1.5 ml min−1) was run twice with buffer C containing 100 mM KCl (3 ml was applied per run). The fractions containing KGOR activity eluted between 127 and 156 ml; the fractions of both runs were combined and concentrated by ultrafiltration to a volume of 4.8 ml (1 mg/ml). In this form KGOR was stored at −80°C for several weeks without significant loss of activity.
Oxidoreductase enzyme assays. (i) KGOR activity assay.
KGOR activity was routinely determined in a continuous spectrophotometric assay following the enzyme-, substrate-, and time-dependent reduction of NADP at a λ of 365 nm at 37°C. The typical assay mixture (total volume, 500 μl) contained 100 mM Tris-HCl (pH 7.8), 5 mM MgCl2, 0.5 mM TPP, 0.5 mM CoA, 0.5 mM NADP+, 2 mM DTE, 5 to 50 μl of protein fractions containing KGOR activity, and 5 mM 2-oxoglutarate. The test was routinely started by adding 2-oxoglutarate. For the determination of the Km values of KGOR for 2-oxoglutarate, NADPH, and CoA, all substrates except the one to be tested were added in nearly saturating concentrations (as indicated above); for Km determination the initial rates were plotted against the substrate concentration by using the Lineweaver-Burk transformation. For testing the substrate specificity, 2-oxoacids were added at 5 mM. To test the electron acceptor specificity, benzyl viologen (1 mM) and methyl viologen (1 mM) were added to anaerobic gas-tight glass cuvettes (gas phase: 95% N2-5% H2 by volume) which contained in addition 0.02 mM dithionite, giving an OD600 of 0.2 to 0.4; the assay was monitored at a λ of 600 nm. The following molar absorption coefficients (ɛ) were used: NADH, ɛ365 = 3.4 mM−1 cm−1; NADPH, ɛ365 = 3.4 mM−1 cm−1; benzyl viologen, ɛ600 = 10 mM−1 cm−1 (18); methyl viologen, ɛ600 = 13 mM−1 cm−1 (18). The isotope exchange reaction between [14C]CO2 and the carboxyl of 2-oxoglutarate (45) was measured as described previously (16).
(ii) NADPH:acceptor oxidoreductase assay.
For measuring the NADPH:acceptor oxidoreductase activities the reduction of the artificial electron acceptor benzyl viologen was monitored in a continuous spectrophotometric assay at 600 nm and 37°C. The test was performed under anaerobic conditions in gas-tight glass cuvettes (gas phase: 95% N2-5% H2 by volume) that contained 100 mM Tris-HCl (pH 7.8)-1 mM benzyl viologen, reduced by 0.02 mM dithionite to an OD600 of 0.2 to 0.4. The test was started by adding NADPH or NADH to a final concentration of 1 mM.
BCR assay.
BCR activity was tested in a radioactive assay as described previously (5, 32); each step was performed under strictly anaerobic conditions. The test was carried out in gas-tight glass tubes at 30°C. The standard assay mixture (0.35 ml) contained 100 mM morpholinopropanesulfonic acid-KOH buffer (pH 7.2), 10 mM MgCl2, 0.5 mM CoA, 7 mM ATP, 10 mM phosphoenolpyruvate, 12.3 U of pyruvate kinase, 200 μM [ring-14C]benzoate (4,810 kBq/μmol; Amersham Biosciences), and 70 μl of cell extract with a protein concentration of 25 mg ml−1. The activity of BCR was tested in the presence of the following electron donors/control compounds: 2-oxoglutarate (5 mM), NADPH (1 mM), NADP+ (1 mM), and a combination of 2-oxoglutarate (5 mM) and NADP+ (1 mM). For the formation of benzoyl-CoA by benzoate-CoA ligase, the mixture was preincubated for 3 min at 30°C with 10 μl of a partially purified fraction containing benzoate-CoA ligase activity of T. aromatica (6 mg ml−1, 0.3 μmol min−1 mg−1). Reducing agents were removed from the cell extract of A. evansii (for preparation see above) by passing the extract over a desalting column (PD-10, prepacked with 10 ml of Sephadex G-25 M; Amersham Biosciences) which had been equilibrated with 10 mM Tris-HCl (pH 7.8). The fractions with the highest protein concentration (25 mg ml−1) were combined, and 70 μl was used to start the test. Samples (50 μl) were taken at 0, 5, 10, 20, 30, and 40 min, stopped with 5 μl of 4 M KOH, and hydrolyzed by heating at 80°C for 10 min. After acidification to pH 2 by addition of 2 μl of 50% H2SO4 and centrifugation, 5 μl of each sample was analyzed by thin-layer chromatography (TLC) on silica gel (Kieselgel 60, F254; Merck, Darmstadt, Germany; solvent system: 25% 1-butanol-75% diisopropyl ether, by volume). Radioactive bands were visualized by autoradiography with a phosphorimager plate (Fuji Photo Film, Kanagawa, Japan). Semiquantitative analysis was carried out using the Quantity One software (Bio-Rad, Münich, Germany).
UV/visible absorption.
For recording the UV/visible spectrum of KGOR, a gel filtration fraction of the highest purity (0.5 mg/ml) which did not contain any reductant was used. Spectra were recorded anaerobically in gas-tight quartz cuvettes. Reduction was carried out by addition of 10 μl of an anaerobically prepared 2 mM sodium dithionite stock solution (final concentration of dithionite in the cuvette was ∼50 μM).
Analytical methods.
The native molecular mass of KGOR was determined by gel filtration using the Superdex 200 column. Ovalbumin (45 kDa), bovine serum albumin (67 kDa), aldolase (158 kDa), catalase (232 kDa), ferritin (450 kDa), and thyroglobulin (669 kDa) were used as molecular mass standard proteins. For N-terminal amino acid sequencing the purified enzyme was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane. The transblotted proteins were detected by Coomassie staining and cut off. N-terminal amino acid sequences were obtained by gas/liquid phase sequencing with an Applied Biosystems 473A sequencer, as describer earlier (23). Sequencing was performed by H. Schägger (Universität Frankfurt, Frankfurt, Germany). Protein was determined by the method of Bradford (11) using bovine serum albumin as the standard. SDS-PAGE analysis (8 and 10% polyacrylamide) was performed as described by Laemmli (34), and visualization of the proteins was by Coomassie staining (55). The following mass standards were used: phosphorylase b (97 kDa), bovine serum albumin (67 kDa), ovalbumin (45 kDa), l-lactic acid dehydrogenase (34 kDa), carbonic acid anhydrase (29 kDa), and lysozyme (14 kDa). Iron was determined as described by Lovenberg et al. (37) with ferrous ammonium sulfate as the standard. The acid-labile sulfur content of a DTE-free sample was determined photometrically at 670 nm, with Na2S · 9 H2O as the standard (3). For determination of the flavin cofactor an aliquot of purified KGOR was applied to a desalting column (PD-10, prepacked with 10 ml of Sephadex G-25 M; Amersham Biosciences) which had been equilibrated with 10 mM Tris-HCl (pH 7.8). All steps of the extraction of the flavin cofactor were carried at 4°C in the dark. The enzyme was denatured by adding perchloric acid to a final pH of 3.5. After an incubation for 1 h at 4°C, precipitated protein was removed by centrifugation (10,000 × g, 10 min, 4°C). The pellet was washed twice with water, after which 2 M potassium carbonate was added to the combined supernatants, giving a final pH of 6. After the combined supernatants were freeze-dried and dissolved in water, the flavin cofactor was identified by high-pressure liquid chromatography using an RP-C18 column (125 by 4 mm; flow rate, 1 ml min−1; Merck, Darmstadt, Germany) and 9% acetonitrile in 20 mM ammonium acetate-HCl (pH 6) as the solvent. Detection was at 375 nm. The retention times of the standards flavin mononucleotide, flavin adenine dinucleotide (FAD), and riboflavin were 6.9, 5.5, and 17.4 min, respectively. The amount of extracted flavin cofactor was determined spectrophotometrically (ɛ450 = 11.3 mM−1 cm−1). The determination of thiamine derivatives was as described by Penttinen (43) and proceeded via the oxidation of TPP to thiochrome, which was quantitatively determined by fluorescence spectroscopy. The excitation wavelength was 365 nm, and the emission was recorded at 430 nm.
Syntheses.
Benzoyl-CoA was synthesized as reported previously (48).
RESULTS
BCR activity in extracts of cells grown anaerobically on benzoate and nitrate.
BCR activity in extracts of cells grown anaerobically on benzoate and nitrate was tested. For this purpose an anaerobic radioactive assay which was previously developed for testing BCR in extracts from T. aromatica and R. palustris was used (32). The assay mixture contained the substrates for benzoate-CoA ligase (MgCl2, ATP, CoA, and [ring-14C]benzoate), which generates the substrate benzoyl-CoA. For testing the reductive dearomatization of the formed [14C]benzoyl-CoA, dithionite, NADPH, and 2-oxoglutarate were tested as electron donors. The last compound might serve as electron donor by means of the combined action of KGOR and ferredoxin. Residual [14C]benzoyl-CoA and derived 14C-labeled CoA ester products were hydrolyzed under alkaline conditions, and the released carboxylic acids were analyzed by TLC and semiquantitative autoradiography (Fig. 1). Note that in T. aromatica BCR reduces benzoyl-CoA to cyclic diene and monoene products; these early products migrate faster than benzoate. The subsequent formation of late products, which migrate slower than benzoate, is catalyzed by a hydratase (hydrating a double bond), a dehydrogenase (oxidizing a hydroxyl group), and a hydrolase (cleaving a C-C bond and thus opening the ring).
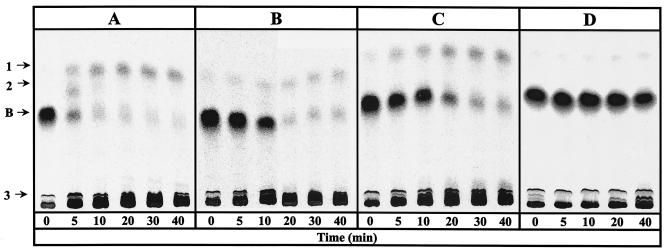
FIG. 1.
Conversion of [ring-14C]benzoate to nonaromatic products by cell extracts of A. evansii. The anaerobic assay mixture (0.35 ml) contained CoA, MgATP, phosphoenolpyruvate, pyruvate kinase, and [ring-14C]benzoate. Benzoyl-CoA was formed enzymatically by preincubation with partially purified benzoate-CoA ligase of T. aromatica. Time zero indicates the end of the preincubation (3 min) and the addition of reductant-free cell extract of A. evansii. Products were separated after hydrolysis of the thiol esters to free acids by TLC. Conversion of benzoyl-CoA in the presence of 5 mM 2-oxoglutarate (A), 5 mM 2-oxoglutarate-1 mM NADP+ (B), 1 mM NADPH (C), and 1 mM NADP+ (D) is shown. Radioactive spots are labeled 1, 2, 3, and B (for residual benzoate).
Figure 1A shows the time- and protein-dependent consumption of benzoyl-CoA and formation of products with A. evansii cell extract and 5 mM 2-oxoglutarate as the electron donor. After 5 min most of the formed [ring-14C]benzoyl-CoA (0.2 mM) was converted to two less-polar products (products 1 and 2) and to more-polar products (summarized as product 3). The pattern of products formed from [ring-14C]benzoyl-CoA, as analyzed by TLC and autoradiography, was similar to the one observed with T. aromatica. Product 2, which was formed early in the course of the reaction, most likely is a cyclic diene product which is formed in a two-electron reduction of benzoyl-CoA. The specific activity of benzoyl-CoA reduction was estimated to be 10 to 15 nmol min−1 mg of protein−1 with 2-oxoglutarate. When 2-oxoglutarate was replaced by 1 mM sodium dithionite, the reduction rate increased to 20 to 30 nmol min−1 mg−1 (not shown). Addition of 1 mM NADP+ to the reaction mixture containing 5 mM 2-oxoglutarate as the reductant slowed down the rate by a factor of 5 to 7 (Fig. 1B). Replacement of 2-oxoglutarate and NADP+ by NADPH (1 mM) resulted in a similarly lower rate (Fig. 1C). Virtually no conversion was obtained when NADPH was replaced by NADP+ in the absence of 2-oxoglutarate (Fig. 1D).
The results suggest that electrons can be transferred from 2-oxoglutarate to either benzoyl-CoA or to NADP+. In the latter case the formed NADPH serves as a poor electron donor for BCR; electron transfer from NADPH most probably proceeds indirectly by means of a combined action of NADPH:ferredoxin oxidoreductase and ferredoxin, which are present in extracts (see below).
Induction of 2-oxoglutarate:acceptor oxidoreductase activity under different growth conditions.
Earlier studies showed that NADP+ is the electron acceptor of a 2-oxoglutarate:acceptor oxidoreductase; therefore, we monitored the 2-oxoglutarate- (5 mM) and CoA (0.5 mM)-dependent reduction of NADP+ (0.5 mM) by cell extracts. The specific activity was 250 nmol of 2-oxoglutarate oxidized min−1 mg of protein−1 in cells grown anaerobically on benzoate and nitrate. Extracts catalyzed only a very slow isotope exchange reaction between [14C]CO2 and the carboxyl group of 2-oxoglutarate (≤5 nmol min−1 mg of protein−1). All KGOR activity was found in the water-soluble protein fraction (supernatant ultracentrifuged at 100,000 × g). KGOR was not detectable in cells grown anaerobically on 20 mM acetate and nitrate as the sole energy source. In cells grown aerobically on benzoate the activity was 10% of the activity in anaerobically grown cells. These cells contained, instead of KGOR, a NAD+-dependent 2-oxoglutarate dehydrogenase (120 nmol min−1 mg−1). Thus, KGOR activity was induced in A. evansii during anaerobic growth on an aromatic substrate.
Purification of 2-oxoglutarate:acceptor oxidoreductase.
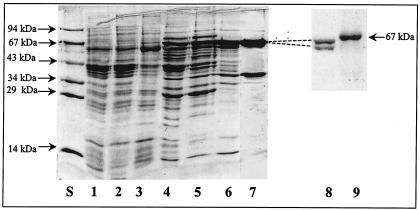
KGOR was purified from extracts of A. evansii grown anaerobically on benzoate and nitrate with a 76-fold enrichment and a yield of 3 to 4% (Table 1). Purification was performed by PEG 6000 precipitation and chromatography on DEAE-Sepharose, hydroxyapatite, Reactive Red agarose, DEAE-Sepharose again, and a Superdex 200 molecular sieve. Purified KGOR was brownish and exhibited a weak sensitivity to oxygen; approximately 60% of the activity was recovered after oxic storage overnight at 4°C compared to the anoxically stored enzyme. Therefore, buffers for all purification steps were made anaerobic and contained 2 mM DTE except for the buffers for the Reactive Red agarose column and the gel filtration; these purification steps were performed in an anaerobic glove box in the absence of DTE. Generally, 10 to 20% glycerol was added to KGOR solutions and buffers, as these additions had a stabilizing effect on KGOR. Approximately 4.8 mg of purified KGOR was obtained from 160 g of cells (wet mass); taking into account the yield during the purification, KGOR makes up to 1.3% of the water-soluble protein fraction in A. evansii cell extracts. Figure 2 shows a representative SDS-PAGE gel of the KGOR activity-containing protein fractions obtained after each purification step. After the last purification step (gel filtration) two protein bands (∼63 and 37.5 kDa) with a 2:1 stoichiometry (after normalization for the different molecular masses) were obtained. The protein band at the molecular mass of ∼63 kDa could be separated into two equally staining bands of 63 and 62 kDa by SDS-8% PAGE (Fig. 2, lane 8), which was run until the molecular mass standard of ovalbumin (45 kDa) nearly reached the front.
TABLE 1.
Protocol for purification of KGOR from A. evansiia
| Purification step | Protein (mg) | Total activity (U) | Sp act (U mg−1) | Enrichment n-(fold) | Recovery (%) |
|---|---|---|---|---|---|
| Cell extract | 11,500 | 2,900 | 0.25 | 1 | 100 |
| PEG 6% supernatant | 11,500 | 2,300 | 0.2 | 0.8 | 79 |
| PEG 11% pellet | 4,100 | 2,370 | 0.6 | 2.2 | 81 |
| DEAE-Sepharose | 420 | 2,040 | 4.9 | 19.3 | 70 |
| Q-Sepharose | 210 | 1,600 | 7.1 | 28.1 | 52 |
| Reactive Red agarose | 63 | 940 | 14.8 | 58.5 | 32 |
| Superdex 200 gel filtration | 4.8 | 92 | 19.3 | 76.2 | 3.2 |
KGOR was purified from 160 g of cells of A. evansii grown anaerobically on benzoate under denitrifying conditions. One unit corresponds to 1 μmol of NADPH formed per min. Note that protein determination in cell extracts of A. evansii is within an error range of ±10%.
FIG. 2.
SDS-PAGE analysis (10% acrylamide) of KGOR-containing fractions obtained during purification of KGOR. Lanes: S, molecular mass standard; 1, supernatant (100,000 × g centrifugation) cell extract; 2, PEG-precipitated 6% supernatant; 3, PEG-precipitated 11% pellet; 4, DEAE-Sepharose fraction; 5, Q-Sepharose fraction; 6, Reactive Red agarose fraction; 7, gel filtration fraction; 8, gel filtration fraction (1 μg) separated by a SDS-8% PAGE, which was run until the molecular mass standard of ovalbumin nearly reached the front; 9, 67-kDa marker bovine serum albumin. For lanes S and 1 to 7, 8 to 25 μg of protein was applied.
Molecular and UV/visible spectroscopic properties of KGOR.
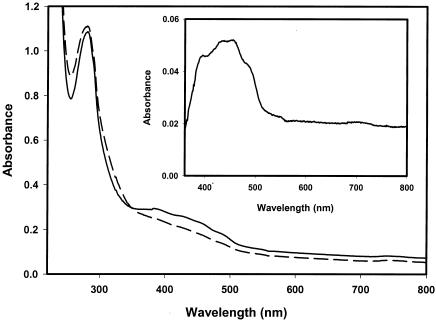
Table 2 summarizes the properties of KGOR. The molecular mass of KGOR was estimated by Superdex 200 chromatography to be approximately 290 ± 20 kDa (not shown). SDS-PAGE analysis showed that purified KGOR consisted of three subunits of 63 (γ), 62(α), and 37.5 kDa (β), which were present in equal amounts, suggesting an (αβγ)2 composition (Fig. 2); the calculated molecular mass was 324 kDa. One mole of enzyme contained 24 mol of nonheme iron and 23 mol of acid-labile sulfur. The other cofactors determined were 1.4 mol of FAD and 0.95 mol of TPP per mol of protein. With the proposed dimeric composition of the enzyme, this suggests that some of the cofactors were lost during the purification procedure. Incubation of KGOR with TPP or FAD (1 mM each) for 1 h at 30°C did not result in an increase of enzyme activity. The UV/visible spectrum of isolated KGOR in the absence of a reducing agent exhibited absorption maxima at 390 and 455 nm, with a shoulder at 490 nm (Fig. 3). The spectrum is typical for a flavin- and iron-sulfur-containing protein. The estimated molar absorption coefficients were as follows: ɛ280 = 387,000 M−1 cm−1, ɛ382 = 104,000 M−1 cm−1, and ɛ420 = 94,000 M−1 cm−1. Reduction of the enzyme by dithionite resulted in a loss of absorption between 350 and 700 nm (Fig. 3, inset).
TABLE 2.
General properties of KGOR from A. evansii
| Property | Value |
|---|---|
| Reaction catalyzed | 2-Oxoglutarate + CoA + NADP+→succinyl-CoA + CO2 + NADPH + H+ |
| Electron donor | 2-Oxoglutarate |
| Electron acceptors | NADP+ (100%), benzyl viologen (60%), methyl viologen (40%) |
| Apparent Kms | 2-Oxoglutarate, 1.2 mM; CoA, 32 μM; NADP, 16 μM |
| Sp act | 19.3 μmol of 2-oxoglutarate min−1 mg−1 |
| Turnover no. | 93 s−1 |
| pH optimum | 7.8 (determined in Tris-HCl buffer) |
| Molecular mass | 290 kDa (determination by gel filtration) |
| Subunits | 63 (γ), 62 (α), and 37 (β) kDa (determined by SDS-PAGE) |
| Suggested composition | (αβγ)2 |
| Iron | 24 mol/mol |
| Acid-labile sulfur | 23 mol/mol |
| Cofactors | FAD, 1.4 mol/mol; TPP, 0.95 mol/mol |
| Absorption coefficients | ɛ280 = 385,000 M−1 cm−1, ɛ450 = 85,000 M−1 cm−1 |
| Stabilizer | Glycerol (20%) |
FIG. 3.
UV/visible spectra of KGOR (1.7 μM). Solid line, oxidized enzyme as isolated in 100 mM Tris-HCl (pH 7.8)-100 mM KCl; dashed line, enzyme after reduction with dithionite. The inset shows a part of the difference spectrum of oxidized KGOR minus reduced KGOR.
N-terminal amino acid sequences of the subunits of KGOR.
The N-terminal amino acid sequences of the three subunits were determined by an automated Edman degradation (Table 3). The three sequences showed significant similarities to the deduced N termini encoded by three adjacently located open reading frames in the Bradyrhizobium japonicum genome; no significant similarities to other proteins were found. The products of the three open reading frames were preliminarily identified as the small β-subunit of a glutamate synthase (82% identity) and two subunits of a 2-oxoacid oxidoreductase. Throughout this work the proteins deduced from these open reading frames in B. japonicum are referred to as BjOORa (annotated as the glutamate synthase β-subunit) and BjOORb and -c (annotated as the two subunits of a 2-oxoacid oxidoreductase). The deduced molecular masses of the three putative proteins from B. japonicum fitted well to those of the corresponding subunits of KGOR from A. evansii (Table 3). The impact of these similarities on the proposed subunit/cofactor architecture of KGOR is described in Discussion.
TABLE 3.
Alignment of the N-terminal amino acid sequences of the three subunits of KGOR from A. evansii to subunits from B. japonicum
| Organism | N-terminal sequence | Subunit | Molecu- lar mass (kDa) | Source or accession no. |
|---|---|---|---|---|
| A. evansii | 1MKPTDISNPDYLHKVVD17 | KGOR α | 62a | This work |
| B. japonicum | 1MKPTDIAAPDYFHKVVD17 | BjOORa | 70b | BAC52007 |
| A. evansii | 1GYLAKPxLLA10 | KGOR β | 37a | This work |
| B. japonicum | 1MTYIAKPKFHH11 | BjOORb | 37b | BAC52009 |
| A. evansii | 01PAIQHTNDFVIKFANV16 | KGOR γ | 63a | This work |
| B. japonicum | 32ISSVNDFVVRFANV45 | BjOORc | 66b | BAC52008 |
Calculated from the deduced amino acid composition.
Determined by SDS-PAGE analysis.
Catalytic properties of KGOR.
The maximal specific activity of KGOR was 19.3 μmol min−1 mg−1 (37°C), resulting in a turnover number of 93 s−1 assuming a molecular mass of 290 kDa. The pH optimum determined in Tris-HCl buffer was 7.8. To test the substrate specificity of purified KGOR, two other 2-oxoacids (5 mM each) and three more electron acceptors were tested. KGOR was specific for 2-oxoglutarate, and virtually no NADP+ reduction was obtained with oxaloacetate and pyruvate. The enzyme used NADP+ most efficiently as an electron acceptor. Reduction of oxidized benzyl viologen (1 mM) by 2-oxoglutarate was 60% ± 10% of the maximal rate, and reduction of oxidized methyl viologen (1 mM) was 40% ± 10% of the maximal rate; variations were due to different enzyme batches. NAD+ (0.5 mM) did not serve as an electron acceptor for KGOR. When both NADP+ (0.5 mM) and oxidized benzyl viologen (1 mM) were simultaneously added to the assay, electrons were solely transferred from 2-oxoglutarate to benzyl viologen, not to NADP+. This indicates that small artificial one-electron carriers such as oxidized viologen dyes trap electrons before they are transferred to NADP+. The apparent Km values were 1.2 mM for 2-oxoglutarate, 32 μM for CoA, and 16 μM for NADP+ (Table 2). The purified enzyme did not catalyze a significant isotope exchange reaction between [14C]CO2 and the carboxyl group of 2-oxoglutarate. Omission of TPP, which was routinely added to the assay mixture, had no effect on KGOR activity.
NADPH:acceptor oxidoreductase partial activity of KGOR.
The results obtained so far indicated that KGOR from A. evansii can use both NADP+ (a hydride-accepting molecule) and one-electron acceptors. In a further experiment we tested purified KGOR for NADPH:acceptor oxidoreductase partial activity using benzyl viologen as the artificial electron acceptor. KGOR catalyzed electron transfer from NADPH (0.5 mM) to oxidized benzyl viologen (1 mM) at an initial rate of 7 μmol min−1 mg−1, corresponding to 37% of the 2-oxoglutarate:NADP+ oxidoreductase activity (Table 4). In contrast to the overall KGOR activity, this partial activity leveled off when only 20 to 40 μM benzyl viologen was reduced. As expected, NADH could not substitute for NADPH in this assay. Notably, KGOR did not catalyze the reverse reaction, the transfer of electrons from reduced benzyl viologen to NADP+ or NAD+.
TABLE 4.
Oxidoreductase activities in cell extracts and catalyzed by purified KGOR from A. evansii
| Cell extract or enzyme | Donor→acceptor | Activity (U mg−1)a |
|---|---|---|
| Anaerobic benzoate | NADPH→benzyl viologenox | 4.9 |
| NADH→benzyl viologenox | 0.3 | |
| Aerobic benzoate | NADPH→benzyl viologenox | 0.2 |
| NADH→benzyl viologenox | 2.7 | |
| Purified KGOR | NADPH→benzyl viologenox | 7.1 |
| NADH→benzyl viologenox | <0.1 | |
| Benzyl viologenred→NADP+ | <0.1 | |
| Benzyl viologenred→NAD+ | <0.1 |
One unit corresponds to 1 μmol of NAD(P)H oxidized or NAD(P)+ reduced min−1.
NADPH:acceptor oxidoreductase activities in extracts of cells grown under different conditions.
To test whether KGOR is the only enzyme that catalyzes electron transfer from NADPH to benzyl viologen, we measured NADPH:acceptor (benzyl viologen) oxidoreductase activity in cell extracts of A. evansii grown anaerobically on benzoate (Table 4). A very high NADPH:acceptor (benzyl viologen) oxidoreductase activity was observed (4.9 μmol min−1 mg−1). With NADH as the electron donor the activity was only 7% of that with NADPH. Taking into account the relative amount of KGOR in the soluble cell fraction (1.3%) and the specific NADPH:acceptor partial activity of purified KGOR (7 μmol min−1 mg−1), the contribution of KGOR to the overall NADPH:acceptor oxidoreductase activity in cell extracts (4.9 μmol min−1 mg−1) is estimated to be approximately 0.1 μmol min−1 mg−1 (2%). Moreover, in contrast to NADPH oxidation associated with purified KGOR, the activity measured in cell extracts slowly leveled off at much higher concentrations of benzyl viologen (>150 μM). Clearly, the very high NADPH oxidizing activity in cell extracts results from a true NADPH:acceptor oxidoreductase.
The NADPH:acceptor oxidoreductase was strictly regulated. In cells grown aerobically on benzoate the specificity of the NADPH:acceptor oxidoreductase activities was inverse: NADPH:acceptor oxidoreductase activity dropped 20-fold. Instead, a high NADH:acceptor (benzyl viologen) oxidoreductase activity, which was more than 10-times higher than NADPH:acceptor (benzyl viologen) oxidoreductase activity, was observed. Hence, anaerobic growth on benzoate induced not only KGOR but also a highly active NADPH:acceptor oxidoreductase. In contrast, aerobic growth induced a normal 2-oxoglutarate dehydrogenase complex (NAD+ specific) and a NADH:acceptor oxidoreductase.
DISCUSSION
Role of KGOR in enzymatic reduction of benzoyl-CoA in A. evansii.
So far, enzymatic ring reduction has been demonstrated and studied in some detail solely for T. aromatica (5, 10, 31, 32) and R. palustris (19, 31). Under anaerobic growth on aromatic compounds these two bacteria produce highly similar BCRs. In contrast, the enzyme from A. evansii shows marked differences in the amino acid sequences of the putative α- and δ-subunits of BCR harboring the ATP binding sites (20) (accession no. CAD21628 and CAD21629). This difference is also reflected in the immunological properties of the proteins (39). With 2-oxoglutarate as the electron donor, the rates of BCR activity in cell extracts of an Azoarcus species were reasonable; hence, 2-oxoglutarate oxidation by KGOR drives benzoyl-CoA reduction. TLC analysis of the labeled products formed from [14C]benzoyl-CoA (after alkaline hydrolysis) revealed a product pattern similar to the one reported for T. aromatica and R. palustris (31), despite some differences in the enzyme properties in the three Proteobacteria (see below) (reviewed in reference 20).
The rate of benzoyl-CoA reduction with 2-oxoglutarate is comparable to the one reported for T. aromatica extract with Ti(III) as the electron donor, corresponding to 15 to 30% of the assumed in vivo rate (5). With dithionite as the electron donor the rate was even higher (30 to 50% of the in vivo rate), suggesting that in cell extracts the electron transfer chain from 2-oxoglutarate to BCR was rate-limiting. In contrast to results for T. aromatica and R. palustris, NADPH, but not NADH, also served as an electron donor for benzoyl-CoA reduction in A. evansii cell extracts, albeit at only 15 to 20% of the rate with 2-oxoglutarate. Based on these results we propose an electron transfer chain from 2-oxoglutarate to benzoyl-CoA in A. evansii in which KGOR replaces the NAD+-dependent 2-oxoglutarate dehydrogenase complex of the TCA cycle (Fig. 4). Oxidation of 2-oxoglutarate by KGOR ultimately results in the reduction of ferredoxin, which is likely to serve as an immediate electron donor for BCR. This proposal is supported by several arguments. (i) A gene coding for a ferredoxin is located adjacent to the structural genes of BCR in the operon of A. evansii (20) (accession no. CAD21632), suggesting coexpression, as in the case of T. aromatica (7, 12). (ii) Benzoyl-CoA reduction was five to seven times faster with 2-oxoglutarate than with NADPH. (iii) Addition of NADP+ to 2-oxoglutarate slowed down the rate of benzoyl-CoA reduction in cell extracts, most likely because NADP+ competes for electrons by serving as an alternative electron acceptor (instead of ferredoxin; see below).
Additional NADP+-reducing activity of KGOR.
In the aromatic metabolism of facultatively anaerobic bacteria, one molecule of benzoyl-CoA is reduced by two electrons prior to oxidation to one molecule of CO2 and three molecules of acetyl-CoA. Most of the acetyl-CoA molecules are channeled into the TCA cycle, yielding 2-oxoglutarate (Fig. 4). From the growth yield (50 g of dry cell mass formed per mol of benzoate consumed) one can estimate that approximately two-thirds of the acetyl-CoA molecules (i.e., two of every three molecules) are oxidized. Only approximately one-third (i.e., one of every three molecules) are used for biosynthesis via the glyoxylate bypass. Hence, oxidative decarboxylation of two molecules of 2-oxoglutarate generates four electrons per benzoyl-CoA molecule metabolized.
Obviously, KGOR differs from 2-oxoacid:acceptor oxidoreductases by an additional module. This module confers the capability to transfer electrons to NADP+ at rates similar to those at which electrons are transferred to viologen dyes. This may allow both oxidized one-electron acceptors (e.g., benzyl viologen and ferredoxin) and NADP+ to serve as electron acceptors at the same time. However, the presence of NADP+ did not prevent or slow down benzyl viologen reduction, and electrons were not transferred from reduced benzyl viologen to NADP+. This may indicate that only an electron overflow would result in NADP+ reduction. Hence, the NADP+-reducing module of KGOR may function as a valve for surplus electrons.
Role of NADPH:acceptor (ferredoxin) oxidoreductase and metabolic electron flow.
The above interpretation is based on the assumption that viologen dyes and ferredoxin behave similarly as electron acceptors for KGOR. This assumption needs to be tested with purified ferredoxin from A. evansii. In any case, electrons may be transferred back from NADPH to ferredoxin by an active NADPH:acceptor (benzyl viologen or ferredoxin) oxidoreductase which is induced under anaerobic growth on aromatic compounds (Fig. 4). Thus, the mechanism by which low-potential electrons generated by KGOR are transferred to BCR probably depends on the cellular reduction charge of the two redox couples reduced/oxidized ferredoxin and NADPH/NADP+. The combined action of KGOR and NADPH:acceptor oxidoreductase guarantees the rapid setting of the equilibria between these redox couples. However, as only two electrons are required for the initial benzoyl-CoA reduction step, reduced ferredoxin and/or NADPH would accumulate and block the TCA cycle since no other ferredoxin-oxidizing reaction in the anaerobic catabolism of benzoate in A. evansii is known. In this case, a surplus of electrons may be channeled to NAD+ by the lower NADH:acceptor oxidoreductase activity present in extracts.
In line with the proposed role and regulation of NADPH:acceptor (ferredoxin) oxidoreductase, an open reading frame coding for a 91-kDa protein is located in the benzoate degradation gene cluster of A. evansii. This protein, identified as a putative dehydrogenase (accession no. AJ 428529), has high similarities to NADPH:acceptor oxidoreductases and to the 62-kDa α-subunit of KGOR from A. evansii, suggesting that the presence of FAD and [4Fe-4S] cluster binding sites. We propose that this 91-kDa protein represents the induced NADPH:acceptor oxidoreductase.
When grown on benzoate and nitrate, A. evansii contains an NADP+-specific isocitrate dehydrogenase activity (430 nmol min−1 mg−1) (our unpublished data). The stoichiometric formation of NADPH by isocitrate dehydrogenase normally fulfills the role of supplying NADPH for biosynthetic reduction processes. The maintenance of the electron transfer balance between ferredoxin, NADP+, and NAD+ during anaerobic growth on aromatic compounds is solved in a different, less complex way in T. aromatica (16) (compare with Fig. 4). There, KGOR reduces ferredoxin only; NADP+ is not accepted. The surplus of electrons from reduced ferredoxin is shuttled to NAD+ by a ferredoxin:NAD+ oxidoreductase, which is induced under anaerobic growth on aromatic compounds.
Comparison of KGOR from A. evansii with other enzymes and subunit/cofactor architecture of KGOR βγ-core enzyme.
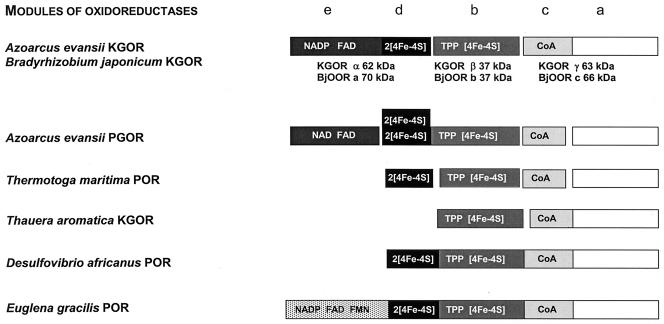
2-Oxoacid:acceptor oxidoreductases are grouped into a single protein family (30, 56) whose members are composed of four conserved modules, termed a to d, with molecular masses between 10 and 50 kDa (Fig. 5). The modules are either located on individual subunits (e.g., in several archaeal oxidoreductases) (4, 27, 28, 30, 38, 44, 49, 54, 56) or fused together on single polypeptides (e.g., in several eubacterial oxidoreductases) (14, 15, 53). The b module usually binds a [4Fe-4S]+/2+ cluster and one TPP molecule (21, 35, 41), the c module harbors the CoA binding site, and the ferredoxin-like d module coordinates two additional [4Fe-4S]+/2+ clusters. The a module has not been ascribed a distinct function yet. Comparison of the N-terminal amino acid sequences of the three subunits of KGOR from A. evansii with those of other proteins revealed that the 37- (β-) and 63-kDa (γ-) subunits are highly similar to the products of two open reading frames in the B. japonicum genome (Table 3). These products, termed BjOORb (37 kDa) and BjOORc (66 kDa), were identified as two subunits of a 2-oxoacid:ferredoxin oxidoreductase with similar sizes (Table 3). To relate the two subunits of KGOR from A. evansii to the typical modules of 2-oxoacid oxidoreductases, we analyzed BjOORb and BjOORc for typical cofactor binding motifs.
FIG. 5.
Comparison of the subunits and modular compositions of 2-oxoacid oxidoreductases. The assignment of the modules and cofactors to the individual subunits of 2-oxoglutarate:NADP+ oxidoreductase from A. evansii was based on conserved cofactor binding motifs encoded by the three open reading frames of B. japonicum (for details see text). The typical four modules found in 2-oxoacid oxidoreductases are termed a to d (30); the additional e module harbors an NADP and a flavin nucleotide binding site. Homologous modules are shown in the same grey scale. Abbreviations: POR, pyruvate:acceptor (ferredoxin or NADP+) oxidoreductase; PGOR, phenylglyoxylate:NAD+ oxidoreductase.
BjOORb (corresponding to the β-subunit of KGOR) contained the two conserved TPP binding motifs 108G(V)SGDG113 and 146TKGQ(F)SA152 (the amino acids in parentheses are not part of the conserved motif). In addition, two of the four conserved cysteines, which coordinate a [4Fe-4S] cluster and which are found in the motifs 66IGCSS70 and 216SPCIA220, were identified in the same subunit. However, the two other cysteines involved in cluster binding, typically identified in an N-terminal WCPGCG motif, are missing and might be replaced by two of the other four cysteines of BjOORb. The β-subunit of KGOR from A. evansii and BjOORb are assigned to the TPP- and [4Fe-4S] cluster-binding b module (Fig. 5). This is further supported by the low molecular mass of BjOORb; this mass is in the typical range for b modules, which excludes the possibility that an additional module is fused to it (Table 3). As a consequence, the 66-kDa BjOORc and the corresponding 63-kDa γ-subunit of KGOR from A. evansii should be a fusion of the a and c modules of 2-oxoacid oxidoreductases. In addition, a glycine-rich CoA binding site (47GSGSAS52) is present in the more-N-terminal region of BjOORc.
In summary, the 63- (γ-) and 37-kDa (β-) subunits of KGOR together form a complete 2-oxoacid oxidoreductase which contains three typical modules, a, b, and c, on two polypeptides (Fig. 5). This architecture is characteristic of the Halobacterium type of 2-oxoacid:ferredoxin oxidoreductases lacking the ferredoxin-like module (28). Note that KGOR from T. aromatica is also a Halobacterium type 2 oxoacid:ferredoxin oxidoreductase (16) (Fig. 5). This core enzyme is probably responsible for the reduction of benzyl viologen. The electrons may come from the γ-subunit harboring TPP and a [4Fe-4S] cluster of probably rather low redox potential (e.g., E′° of the corresponding cluster in T. aromatica KGOR is estimated to be more negative than −500 mV). This may explain why the enzyme did not catalyze the flow of electrons back from reduced benzyl viologen to the enzyme and from there to NADP+.
New interpretation of three B. japonicum genes and cofactor architecture of the KGOR α-subunit.
The additional α-subunit of KGOR (62 kDa) from A. evansii is similar to the 70-kDa BjOORa, whose open reading frame is adjacent to those of BjOORb and BjOORc in the B. japonicum genome (Table 3; Fig. 5). This open reading frame is highly similar to those of a number of β-subunits of glutamate synthases from other organisms and has been annotated as such. The proteins usually harbor one FAD molecule, one NADP+ binding site, and most likely two [4Fe-4S]+/2+ clusters (Fig. 5). The two clusters are usually located in a ferredoxin-like module with two typical CXXCXXC motifs at the C terminus of the small β-subunit of glutamate synthases; such motifs are also present in BjOORa. It has been suggested that the small subunit of glutamate synthase represents a prototype of a novel class of oxidoreductases containing FAD and [4Fe-4S] centers which catalyze electron transfer between two-electron and one-electron redox carriers (52). The absence of the gene for the large α-subunit of glutamate synthase in the vicinity of the BjOORa to -c genes supports the idea that BjOORa does not represent the small subunit of a glutamate synthase but rather an additional subunit of the 2-oxoacid oxidoreductase.
Using the BjOORb and BjOORc open reading frames, coding for the putative two subunits of a 2-oxoacid oxidoreductase, we searched the B. japonicum genome for genes encoding other putative 2-oxoacid oxidoreductases but did not find any. In contrast, another gene like the BjOORa gene was found next to the gene for a putative large α-subunit of glutamate synthase (accession no. NP_774383 and NP_774384), indicating that these two genes code for the true glutamate synthase in B. japonicum. This finding corroborates our conclusion concerning the role of BjOORa.
The KGOR model in A. evansii predicts a total of six [4Fe-4S]centers, two FAD molecules, and two TPP molecules per native (αβγ)2 enzyme, a prediction which is close to the experimentally determined values (24 mol of Fe/mol, 23 mol of acid-labile sulfur/mol, 1.4 mol of FAD/mol, and 0.95 mol of TPP/mol). The presence of FAD in the α-subunit may explain why the enzyme did not catalyze a significant exchange of 14CO2 with the carboxyl group of 2-oxoglutarate. The flavin simply acts as an electron sink, rendering the oxidative decarboxylation of 2-oxoglutarate virtually irreversible.
Possible role of KGOR in N2 fixation.
The enzyme generating low-potential electrons for dinitrogen fixation by nitrogenase in the symbiotic B. japonicum is unknown. A 2-oxoglutarate-oxidizing enzyme is crucial for the maintenance of the TCA cycle in B. japonicum. Note that, in symbiotic associations, dicarboxylic acids are provided by the plant to the bacterial symbiotic partner, which uses them as the main sources of cell carbon and energy. It has been reported that mutants deficient in the conventional 2-oxoglutarate dehydrogenase complex are still able to grow on dicarboxylic acids such as malate or succinate (19a). To explain the phenotype of the 2-oxoglutarate dehydrogenase-deficient mutant, a reaction that circumvents this block was assumed: 2-oxoglutarate decarboxylase forms succinate semialdehyde, which subsequently is oxidized to succinate; however, no evidence for the operation of this shuttle has been provided yet. We propose that, alternatively, a three-component KGOR (BjOORa to -c) could represent the missing link between the TCA cycle and dinitrogen fixation.
Other 2-oxoacid oxidoreductases in A. evansii.
KGOR represents the second unusual 2-oxoacid:acceptor oxidoreductase in the anaerobic aromatic metabolism of A. evansii. A phenylglyoxylate:NAD+ oxidoreductase has been identified and characterized; it plays a role in the anaerobic phenylalanine metabolism and catalyzes the oxidative decarboxylation of phenylglyoxylate to benzoyl-CoA (23). This enzyme is different from the pyruvate dehydrogenase complex; its module composition is similar to that of KGOR, but it is composed of five subunits (Fig. 5). Phenylglyoxylate:NAD+ oxidoreductase contains all four modules of 2-oxoacid:acceptor oxidoreductases on four separate subunits and, in addition, a fifth subunit harboring one FAD molecule and a NAD+ binding site (Fig. 5).
Similarity of KGOR to pyruvate:NADP+ oxidoreductase from the mitochondrion of E. gracilis and from the apicomplexan C. parvum.
Most eukaryotes perform the oxidative decarboxylation of pyruvate in mitochondria using a NAD+-dependent pyruvate dehydrogenase complex. Eukaryotes that lack mitochondria also lack pyruvate dehydrogenase, using instead the oxygen-sensitive enzyme pyruvate:ferredoxin oxidoreductase, which is localized either in the cytosol or in hydrogenosomes. The facultatively anaerobic mitochondria of the photosynthetic protist E. gracilis constitute an exception in that these mitochondria oxidize pyruvate with the oxygen-sensitive enzyme pyruvate:NADP+ oxidoreductase (26, 42, 47). The same holds true for the apicomplexan C. parvum (47). These findings have been interpreted in terms of eukaryotic pyruvate:ferredoxin oxidoreductase domains being biochemical relics inherited from a facultatively anaerobic, eubacterial ancestor of mitochondria and hydrogenosomes (47).
Our work showed that similar oxidoreductases acting on phenylglyoxylate or 2-oxoglutarate exist in facultatively anaerobic members of the β-proteobacteria (an Azoarcus sp.) and probably also in α-proteobacteria (a Bradyrhizobium sp.). All these NADP+-accepting oxidoreductases are similarly built up from domains of typical 2-oxoacid:ferredoxin oxidoreductases and an additional electron-transferring subunit that mediates the transfer of electrons via a flavin module to NADP+ (Fig. 5). These enzymes mainly differ in the distribution of the individual modules on different subunits, whose numbers range from five (PGOR from A. evansii) to one (POR from several members of the Eubacteria). In the absence of NADP+, the oxidoreductases may even be able to transfer electrons to ferredoxin since the enzymes accept viologen dyes as electron acceptors. This possibility needs to be tested.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft and by the Fonds der Chemische Industrie.
We thank H. Schägger, Frankfurt, for N-terminal amino acid sequencing.
REFERENCES
- 1.Altenschmidt, U., B. Oswald, and G. Fuchs. 1991. Purification and characterization of benzoate-coenzyme A ligase and 2-aminobenzoate-coenzyme A ligases from a denitrifying Pseudomonas sp. J. Bacteriol. 173:5494-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anders, H. J., A. Kaetzke, P. Kämpfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position of aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera, as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the beta subclass of the Proteobacteria. Int. J. Syst. Bacteriol. 45:327-333. [DOI] [PubMed] [Google Scholar]
- 3.Beinert, H. 1983. Semi-micro methods for analysis of labile sulfide plus sulfane in usually stable iron-sulfur proteins. Anal. Biochem. 131:373-378. [DOI] [PubMed] [Google Scholar]
- 4.Bock, A. K., J. Kunow, J. Glasemacher, and P. Schönheit. 1996. Catalytic properties, molecular composition and sequence alignments of pyruvate:ferredoxin oxidoreductase from the methanogenic archaeon Methanosarcina barkeri (strain Fusaro). Eur. J. Biochem. 237:35-44. [DOI] [PubMed] [Google Scholar]
- 5.Boll, M., and G. Fuchs. 1995. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. ATP dependence of the reaction, purification and some properties of the enzyme from Thauera aromatica strain K172. Eur. J. Biochem. 234:921-933. [DOI] [PubMed] [Google Scholar]
- 6.Boll, M., S. J. P. Albracht, and G. Fuchs. 1997. Benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. A study of adenosinephosphate activity, ATP stoichiometry of the reaction and EPR properties of the enzyme. Eur. J. Biochem. 244:840-851. [DOI] [PubMed] [Google Scholar]
- 7.Boll, M., and G. Fuchs. 1998. Identification and characterization of the natural electron donor ferredoxin and of FAD as a possible prosthetic group of benzoyl-CoA reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. Eur. J. Biochem. 251:946-954. [DOI] [PubMed] [Google Scholar]
- 8.Boll, M., G. Fuchs, G. Tilley, F. A. Armstrong, and D. J. Lowe. 2000. Unusual spectroscopic and electrochemical properties of the 2[4Fe-4S] ferredoxin of Thauera aromatica. Biochemistry 39:4929-4938. [DOI] [PubMed] [Google Scholar]
- 9.Boll, M., G. Fuchs, and J. Heider. 2002. Anaerobic oxidation of aromatic compounds and hydrocarbons. Curr. Opin. Biol. Chem. 6:604-611. [DOI] [PubMed] [Google Scholar]
- 10.Boll, M., D. Laempe, W. Eisenreich, A. Bacher, T. Mittelberger, J. Heinze, and G. Fuchs. 2000. Non-aromatic products formed from anoxic conversion of benzoyl-CoA with benzoyl-CoA reductase and cyclohexa-1,5-diene-1-carbonyl-CoA hydratase. J. Biol. Chem. 275:21889-21895. [DOI] [PubMed] [Google Scholar]
- 11.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Breese, K., M. Boll, J. Alt-Mörbe, H. Schägger, and G. Fuchs. 1998. Genes coding for the benzoyl-CoA pathway of anaerobic aromatic metabolism in the bacterium T. aromatica. Eur. J. Biochem. 256:148-154. [DOI] [PubMed] [Google Scholar]
- 13.Brostedt, E., and S. Nordlund. 1991. Purification and partial characterization of pyruvate oxidoreductase from the photosynthetic bacterium Rhodospirillum rubrum grown under nitrogen-fixing conditions. Biochem. J. 279:155-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chabriere, E., M. H. Charon, A. Volbeda, L. Pieulle, E. C. Hatchikian, and J. C. Fontecilla-Camps. 1999. Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate. Nat. Struct. Biol. 6:182-190. [DOI] [PubMed] [Google Scholar]
- 15.Charon, M.-H., A. Volbeda, E. Chabriere, L. Pieulle, and J. C. Fontecilla-Camps. 1999. Structure and electron transfer mechanism of pyruvate:ferredoxin oxidoreductase. Curr. Opin. Struct. Biol. 9:663-669. [DOI] [PubMed] [Google Scholar]
- 16.Dörner, E., and M. Boll. 2002. Properties of 2-oxoglutarate:ferredoxin oxidoreductase from Thauera aromatica and its role in enzymatic reduction of the aromatic ring. J. Bacteriol. 184:3975-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egland, P. G., D. A. Pelletier, M. Dispensa, J. Gibson, and C. S. Harwood. 1997. A cluster of bacterial genes for anaerobic benzene ring biodegradation. Proc. Natl. Acad. Sci. USA 94:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraisse, L., and H. Simon. 1988. Observations on the reduction of non-activated carboxylates by Clostridium formicoaceticum with carbon monoxide or formate and the influence of various viologens. Arch. Microbiol. 150:381-386. [Google Scholar]
- 19.Gibson, K. J., and J. Gibson. 1992. Potential early intermediates in anaerobic benzoate degradation by Rhodopseudomonas palustris. Appl. Environ. Microbiol. 58:696-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Green, L. S., Y. Li, D. W. Emerich, F. J. Bergersen, and D. A. Day. 2002. Catabolism of α-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: evidence for an alternative tricarboxylic acid cycle. J. Bacteriol. 182:2838-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harwood, C. S., G. Burchhardt, H. Herrmann, and G. Fuchs. 1999. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22:439-458. [Google Scholar]
- 21.Hawkins, C. F., A. Borges, and R. N. Perham. 1989. A common structural motif of thiamin pyrophosphate-binding enzymes. FEBS Lett. 255:77-82. [DOI] [PubMed] [Google Scholar]
- 22.Heider, J., and G. Fuchs. 1997. Anaerobic metabolism of aromatic compounds. Eur. J. Biochem. 243:577-596. [DOI] [PubMed] [Google Scholar]
- 23.Hirsch, W., and G. Fuchs. 1998. Phenylglyoxylate:NAD+ oxidoreductase (CoA benzoylating), a new enzyme of anaerobic phenylalanine metabolism in the denitrifying bacterium Azoarcus evansii. Eur. J. Biochem. 251:907-915. [DOI] [PubMed] [Google Scholar]
- 24.Howard, J. B., and D. C. Rees. 2000. Nitrogenase: standing at the crossroads. Curr. Opin. Chem. Biol. 4:559-566. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, N. J., C. L. Clayton, P. A. Chalk, and D. J. Kelly. 1998. Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180:1119-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inui, H., K. Ono, K. Miyatake, Y. Nakano, and S. Kitaoka. 1987. Purification and characterization of pyruvate:NADP+ oxidoreductase in Euglena gracilis. J. Biol. Chem. 262:9130-9135. [PubMed] [Google Scholar]
- 27.Iwasaki, T., T. Wakagi, and T. Oshima. 1995. Ferredoxin-dependent redox system of a thermoacidophilic archaeon, Sulfolobus sp. strain 7. Purification and characterization of a novel reduced ferredoxin-reoxidizing iron-sulfur flavoprotein. J. Biol. Chem. 270:17878-17883. [DOI] [PubMed] [Google Scholar]
- 28.Kerscher, L., and D. Oesterhelt. 1981. Purification and properties of two 2-oxoacid:ferredoxin oxidoreductases from Halobacterium halobium. Eur. J. Biochem. 116:587-594. [DOI] [PubMed] [Google Scholar]
- 29.Kerscher, L., and D. Oesterhelt. 1981. The catalytic mechanism of 2-oxoacid:ferredoxin oxidoreductases from Halobacterium halobium. One-electron transfer at two distinct steps of the catalytic cycle. Eur. J. Biochem. 116:595-600. [DOI] [PubMed] [Google Scholar]
- 30.Kletzin, A., and M. W. W. Adams. 1996. Molecular and phylogenetic characterization of pyruvate and 2-ketoisovalerate ferredoxin oxidoreductases from Pyrococcus furiosus and pyruvate ferredoxin oxidoreductase from Thermotoga maritima. J. Bacteriol. 178:248-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koch, J., W. Eisenreich, A. Bacher, and G. Fuchs. 1993. Products of enzymatic reduction of benzoyl-CoA, a key reaction in anaerobic aromatic metabolism. Eur. J. Biochem. 211:649-661. [DOI] [PubMed] [Google Scholar]
- 32.Koch, J., and G. Fuchs. 1992. Enzymatic reduction of benzoyl-CoA to alicyclic compounds, a key reaction in anaerobic aromatic metabolism. Eur. J. Biochem. 205:195-202. [DOI] [PubMed] [Google Scholar]
- 33.Kyritsis, P., O. M. Hatzfeld, T. A. Link, and J.-M. Moulis. 1998. The two [4Fe-4S] clusters in Chromatium vinosum ferredoxin have largely differing reduction potentials. J. Biol. Chem. 273:15404-15411. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 35.Lindqvist, Y., and G. Schneider. 1993. Thiamine pyrophosphate-dependent enzymes: transketolase, pyruvate oxidase and pyruvate decarboxylase. Curr. Opin. Struct. Biol. 3:896-901. [Google Scholar]
- 36.Lochmeyer, C., and G. Fuchs. 1990. NADP+ specific 2-oxoglutarate dehydrogenase in denitrifying Pseudomonas species. Arch. Microbiol. 153:226-229. [Google Scholar]
- 37.Lovenberg, W., B. B. Buchanan, and J. Rabinowitz. 1963. Studies on the chemical nature of ferredoxin. J. Biol. Chem. 238:3899-3913. [PubMed] [Google Scholar]
- 38.Mai, X., and M. W. Adams. 1996. Characterization of a fourth type of 2-keto acid-oxidizing enzyme from a hyperthermophilic archaeon: 2-ketoglutarate ferredoxin oxidoreductase from Thermococcus litoralis. J. Bacteriol. 178:5890-5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mechichi, T., E. Stackebrandt, N. Gad'on, and G. Fuchs. 2002. Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions, and description of Thauera phenylacetica sp. nov., Thauera aminoaromatica sp. nov., and Azoarcus buckelii sp. nov. Arch. Microbiol. 178:26-35. [DOI] [PubMed] [Google Scholar]
- 40.Möbitz, H., and M. Boll. 2002. A Birch-like mechanism in enzymatic benzoyl-CoA reduction—a kinetic study of substrate analogues combined with an ab initio model. Biochemistry 41:1752-1758. [DOI] [PubMed] [Google Scholar]
- 41.Muller, Y. A., Y. Lindqvist, W. Furey, G. E. Schulz, F. Jordan, and G. S. Schneider. 1993. A thiamine diphosphate binding fold revealed by comparison of the crystal structures of transketolase, pyruvate oxidase and pyruvate decarboxylase. Structure 1:95-103. [DOI] [PubMed] [Google Scholar]
- 42.Nakazawa, M., H. Inui, R. Yamaji, T. Yamamoto, S. Takenaka, M. Ueda, Y. Nakano, and K. Miyatake. 2000. The origin of pyruvate: NADP+ oxidoreductase in mitochondria of Euglena gracilis. FEBS Lett. 479:155-156. [DOI] [PubMed] [Google Scholar]
- 43.Penttinen, H. K. 1979. Fluorometric determination of thiamine and its mono-, di- and triphosphate esters. Methods Enzymol. 62:58-59. [DOI] [PubMed] [Google Scholar]
- 44.Plaga, W., F. Lottspeich, and D. Oesterhelt. 1992. Improved purification, crystallization and primary structure of pyruvate:ferredoxin oxidoreductase from Halobacterium halobium. Eur. J. Biochem. 205:391-397. [DOI] [PubMed] [Google Scholar]
- 45.Raeburn, S., and J. C. Rabinowitz. 1971. Pyruvate: ferredoxin oxidoreductase. I. The pyruvate-CO2 exchange reaction. Arch. Biochem. Biophys. 146:9-20. [DOI] [PubMed] [Google Scholar]
- 46.Raeburn, S., and J. C. Rabinowitz. 1971. Pyruvate: ferredoxin oxidoreductase. II. Characteristics of the forward and reverse reactions and properties of the enzyme. Arch. Biochem. Biophys. 146:21-33. [DOI] [PubMed] [Google Scholar]
- 47.Rotte, C., F. Stejskal, G. Zhu, J. S. Keithly, and W. Martin. 2001. Pyruvate:NADP+ oxidoreductase from the mitochondrion of Euglena gracilis and from the apicomplexan Cryptosporidium parvum: a biochemical relic linking pyruvate metabolism in mitochondriate and amitochondriate protists. Mol. Biol. Evol. 18:710-720. [DOI] [PubMed] [Google Scholar]
- 48.Schachter, D., and V. J. Taggart. 1953. Benzoyl coenzyme A and hippurate synthesis. J. Biol. Chem. 203:925-933. [PubMed]
- 49.Tersteegen, A., D. Linder, R. K. Thauer, and R. Hedderich. 1997. Structures and functions of four anabolic 2-oxoacid oxidoreductases in Methanobacterium thermoautotrophicum. Eur. J. Biochem. 244:862-868. [DOI] [PubMed] [Google Scholar]
- 50.Uyeda, K., and J. C. Rabinowitz. 1971. Pyruvate-ferredoxin oxidoreductase. 3. Purification and properties of the enzyme. J. Biol. Chem. 246:3111-3119. [PubMed] [Google Scholar]
- 51.Uyeda, K., and J. C. Rabinowitz. 1971. Pyruvate-ferredoxin oxidoreductase. IV. Studies on the reaction mechanism. J. Biol. Chem. 246:3120-3125. [PubMed] [Google Scholar]
- 52.Vanoni, M. A., and B. Curti. 1999. Glutamate synthase: a complex iron-sulfur flavoprotein. Cell. Mol. Life Sci. 55:617-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wahl, R. C., and W. H. Orme-Johnson. 1987. Clostridial pyruvate oxidoreductase and the pyruvate-oxidizing enzyme in Klebsiella pneumoniae are similar enzymes. J. Biol. Chem. 262:10489-10496. [PubMed] [Google Scholar]
- 54.Yun, N. R., H. Arai, M. Ishii, and Y. Igarashi. 2001. The genes for anabolic 2-oxoglutarate:ferredoxin oxidoreductase from Hydrogenobacter thermophilus TK-6. Biochem. Biophys. Res. Commun. 282:589-594. [DOI] [PubMed] [Google Scholar]
- 55.Zehr, B. D., T. J. Savin, and R. E. Hall. 1989. A one-step, low background Coomassie staining procedure for polyacrylamide gels. Anal. Biochem. 182:157-159. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, Q., T. Iwasaki, T. Wakagi, and T. Oshima. 1996. 2-Oxoacid:ferredoxin oxidoreductase from the thermoacidophilic archaeon, Sulfolobus sp. strain 7. J. Biochem. 120:587-599. [DOI] [PubMed] [Google Scholar]