Plant cell wall-degrading enzymes have become increasingly important, since the development of efficient biomass degradation methods and the conversion of sugars to valuable products such as butanol and amino acids and utilizable forms of energy such as ethanol and methane could lead to less dependence on imported petroleum as a fuel and chemical source. Plant biomass is an abundant renewable resource. Since cellulose and hemicellulose comprise about 40 to 50% of plant cell walls and are considered to be the largest components of the earth's biomass, efficient conversion of this material by engineered enzymes and/or microorganisms would be highly desirable. The rate-limiting step in biomass degradation is the conversion of the cellulose and hemicellulose polymers to sugars.
Basic research is essential to elucidate and improve the properties of enzymes that can degrade plant biomass, such as cellulose. Generally two types of cellulase systems occur. One type consists of independent extracellular cellulases that act synergistically to degrade cellulose, while the second type is represented by an enzyme complex called the “cellulosome,” which consists of a nonenzymatic scaffolding protein associated with various enzymatic subunits that act in concert to degrade cellulose and hemicellulose. This minireview will focus on describing the properties of cellulosomes from mesophilic bacteria and recent findings related to this interesting enzyme complex. The cellulosome from Clostridium thermocellum has been discussed in a number of recent reviews (2, 4, 5, 29). Reviews on cellulosomes (1, 3, 14, 15, 20, 57) and noncellulosomal cellulases (37, 48, 65) should be consulted for previous and complementary discussions of this subject.
CELLULOSOMES
What are cellulosomes? Cellulosomes are large extracellular enzyme complexes that are capable of degrading cellulose, hemicelluloses, and pectin and are produced by anaerobic bacteria such as Clostridium, Acetivibrio, Bacteroides, and Ruminococcus (Table 1). These microorganisms are found in various environmental niches, including soil, wood chip piles, sewage, and rumens. Cellulosomes actually function to degrade plant cell wall material and not only crystalline cellulose as its initial nomenclature suggested. Cellulosomes may be the largest extracellular enzyme complexes found in nature, since polycellulosomes have been reported to be as large as 100 MDa, although the individual cellulosomes range in mass from about 650,000 Da to 2.5 MDa.
TABLE 1.
Cellulosome-producing anaerobic bacteria
| Species | Optimal growth tempa | Source | Reference |
|---|---|---|---|
| Acetivibrio cellulolyticus | M | Sewage | 10 |
| Bacteroides cellulosolvens | M | Sewage | 11 |
| Butyrivibrio fibrisolvens | M | Rumen | 7 |
| Clostridium acetobutylicum | M | Soil | 66 |
| Clostridium cellulovorans | M | Wood fermenter | 61 |
| Clostridium cellobioparum | M | Rumen | 30 |
| Clostridium cellulolyticum | M | Compost | 49 |
| Clostridium josui | M | Compost | 25 |
| Clostridium papyrosolvens | M | Paper mill | 51 |
| Clostridium thermocellum | T | Sewage soil | 29 |
| Ruminococcus albus | M | Rumen | 48 |
| Ruminococcus flavefaciens | M | Rumen | 12 |
| Ruminococcus succinogenes | M | Rumen | 39 |
M, mesophilic; T, thermophilic (above 50°C).
Cellulosomes were first observed by Bayer and Lamed as large protuberances on the surface of C. thermocellum (2, 31), and the complex was found to consist of a nonenzymatic scaffolding protein to which were attached a number of enzymatic subunits (3). These observations on the complex structure were supported by the findings of Mayer et al. (38), who found by electron microscopy that cellulosomes had an ordered arrangement of subunits. There has been a relatively rapid expansion in identification of cellulosomes from various anaerobic cellulolytic bacteria (Table 1), and it is likely that cellulosomes will be identified in a large number of anaerobic bacteria. Cellulosomes are now thought to degrade not only crystalline cellulose, but also hemicelluloses, chitin, and even pectin, depending on the source of the cellulosomes. Interestingly, in one case, cellulosome-like structures were observed with Clostridium acetobutylicum, but they did not have any activity in degrading crystalline forms of cellulose (55). Recent studies have been devoted to characterizing and comparing the properties of the scaffolding protein and the cellulosomal enzymes from various bacterial sources. They have provided a glimpse into the diversity of cellulosomes and a partial rationale for their existence.
CELLULOSOME COMPONENTS
Scaffolding protein (scaffoldin).
What are the components of the cellulosome? The cellulosome consists of a scaffolding protein called CbpA (59, 60), CipA (22, 25, 56), or CipC (50) complexed with a number of cellulosomal enzymes. The scaffolding proteins, also called “scaffoldins” (1), are large nonenzymatic proteins that usually contain a number of cohesin domains (Coh) and cellulose binding domains (CBDs); however, hydrophilic domains, dockerin II domains, the enzyme coding domain, and a number of unidentified domains whose functions remain unknown have also been observed in some of the scaffoldins (Fig. 1).
FIG. 1.
Structural properties of cellulosomal scaffolding proteins (scaffoldins) from various bacterial species.
WHAT IS THE FUNCTION OF THE COHESINS?
The cohesins are always present in scaffoldins and represent the binding sites for the dockerin domains of cellulosomal enzymes. The cohesin-dockerin interaction plays a key role in the assembly of the cellulosome. The number of cohesins present in various scaffoldins varies, as Fig. 1 illustrates. There is a considerable degree of amino acid homology between cohesins from different species. However, the cohesins appear to be quite specific in their interactions with the dockerin domains that are present in all cognate cellulosomal enzymes. This was demonstrated when the cohesins present in CipA of C. thermocellum did not interact with the dockerins present in C. cellulolyticum enzymes and vice versa (49). This high specificity in cohesin-dockerin interaction has allowed the development of an interesting test system for synergy, which will be discussed later.
CBDs (CBMs)
The CBD (also called the carbohydrate binding module [CBM]) of scaffoldins binds the cellulosome tightly to the cellulose substrate (8). The CBD appears to bind the cellulosome to the crystalline form of cellulose more efficiently than to amorphous forms of cellulose (23). In addition, the CBD of the C. cellulovorans scaffoldin can bind to chitin, which has a backbone and structure similar to those of cellulose (23). In one case, Ruminococcus flavefaciens ScaB apparently does not contain a CBM (Fig. 1), although a CBM sequence may be found in an ancillary protein that works in conjunction with ScaB. The CBDs are variable and are divided into a number of families based on their amino acid sequences and occur not only in scaffoldins, but as domains of various cellulosomal and noncellulosomal cellulases (8). In the context of the CBMs illustrated in Fig. 1, the family 3 CBMs are capable of binding crystalline cellulose.
HLDs
The various scaffoldins also contain domains the functions of which have not been clearly identified or which are present somewhat uniquely in the scaffoldin. The presence of hydrophilic domains (HLDs) has been observed in a number of scaffoldins, and there are up to four of them in C. cellulovorans CbpA (59). It has been proposed that these HLDs or surface layer homology domains (SLHs) in C. cellulovorans play a role in binding the cellulosome to the cell surface (27, 62). This idea is based on data from the SLHs present in endoglucanase E (EngE) (62), which will be discussed later. During growth, there is a tight association between the cell and the cellulose substrate. This association is facilitated by the cellulosome, which binds to both the substrate and the cell surface. This close association presumably facilitates degradation of the substrate and ready assimilation of the sugars as they are produced by the cellulosomal enzymes.
OTHER DOMAINS OF SCAFFOLDIN
The other domains of scaffoldin include a different type of dockerin, called dockerin II, in the C. thermocellum scaffoldin CipA (26). The dockerin II binds to cohesin II domains found in proteins SdbA, ORF2p, and OlpB, which also contain SLHs. These three proteins can bind cellulosomes through the dockerin II-cohesin II interaction, and these complexes can then bind to the a cell surface binding complex that attaches the cellulosome to the surface of the C. thermocellum cell through their SLH domains (19, 32, 34-36). The dockerin II domain has not been observed in any of the scaffoldins of mesophilic cells. Thus, a different mechanism may exist for attaching the cellulosome to the surface of mesophilic bacteria. Thus, in this case, the binding of the cellulosome to the cell surface does not depend on SLH domains and illustrates the existence of another type of cellulosome-cell association mechanism.
In the case of R. flavefaciens, the scaffoldin-type ScaA contains three cohesin domains, a C-terminal dockerin, and a unique N-terminal X domain (12). ScaA does not contain an identifiable CBM and yet is able to bind to cellulose. Thus, it appears that either a novel CBM has not been identified for ScaA, or some other factor is involved in binding ScaA to the substrate. It is proposed that ScaA interacts with a surface-bound ScaB protein and that the cellulosome is bound to the cell surface by this interaction.
An unusual situation exists with Acetivibrio cellulolyticus, since its scaffoldin not only serves as a scaffolding protein for a number of enzymatic subunits but also contains a family 9 glycosyl hydrolase enzymatic domain that can function to degrade cellulose (10).
Thus, the properties of scaffoldins suggest that they not only serve as scaffolding proteins to bind cellulosomal enzymes, but some have evolved or combined multiple functions, including cell surface binding functions and even enzymatic activity. It is likely that as more scaffoldins are characterized, diverse structures and functions will be found.
CELLULOSOMAL ENZYMES
Cellulosomal enzymes contain minimally a catalytic domain and a 22-amino-acid-long duplicated sequence, the dockerin (1). The presence of a dockerin in an enzyme usually indicates that it is a cellulosomal enzyme, since the dockerin interacts with the cohesins of the scaffolding protein to form the enzyme complex. Several other domains besides the catalytic domain and dockerin have been found including CBDs, immunoglobulin-like domains, and SLH domains (Table 2).
TABLE 2.
Cellulosomal subunits of mesophilic Clostridia
| Cellulosomal enzyme of mesophilic Clostridium | Function | Mol mass (kDa) | Modular structurea |
|---|---|---|---|
| C. cellulovorans | |||
| ExgS | Exoglucanase | 80 | GH48-DS1 |
| EngH | Endoglucanase | 79 | GH9-CBM3-DS1 |
| EngK | Endoglucanase | 97 | CBM4-Ig-GH9-DS1 |
| EngL | Endoglucanase | 58 | GH9-DS1 |
| ManA | Mannanase | 47 | DS1-GH5 |
| EngM | Endoglucanase | 96 | CBM4-Ig-GH9-DS1 |
| EngE | Endoglucanase | 112 | (SLH)3-GH5-X-DS1 |
| EngY | Endoglucanase | 80 | CBM2-GH9-DS1 |
| EngB | Endoglucanase | 49 | GH5-DS1 |
| PelA | Pectate lyase | 94 | X-CBD2-GPL9-DS1 |
| XynA | Xylanase | 57 | GH11-DS1-CE4 |
| C. acetobutylicum | |||
| CelF | Exoglucanase | 81 | GH48-DS1 |
| CelA | Endoglucanase | 54 | GH5-DS1 |
| CelH | Endoglucanase | 80 | GH9-CBM3-DS1 |
| EngA | Endoglucanase | 67 | GH44-DS1 |
| CelG | Endoglucanase | 77 | GH9-CBM3-DS1 |
| CelL | Endoglucanase | 60 | GH9-DS1 |
| ManA | Mannanase | 47 | GH5-DS1 |
| CAC0919 | Sialidase | 91 | GH74-DS1 |
| CelE | Endoglucanase | 96 | CBM3-Ig-GH9-DS1 |
| CAC3469 | Endoglucanase | 110 | (SLH)3-GH5-X-DS1 |
| C. cellulolyticum | |||
| CelF | Exoglucanase | 78 | GH48-DS1 |
| CelC | Endoglucanase | 51 | GH8-DS1 |
| CelG | Endoglucanase | 80 | GH9-CBM3-DS1 |
| CelE | Endoglucanase | 97 | CBM4-Ig-GH9-DS1 |
| CelH | Endoglucanase | 83 | GH9-CBM3-DS1 |
| CelJ | Endoglucanase | 85 | GH9-CBM3-DS1 |
| ManK | Mannanase | 48 | DS1-GH5 |
| CelD | Endoglucanase | 63 | GH5-DS1 |
| CelA | Endoglucanase | 50 | GH5-DS1 |
| CelM | Endoglucanase | 58 | GH9-DS1 |
| C. josui | |||
| CelD | Exoglucanase | 80 | GH48-DS1 |
| CelB | Endoglucanase | 51 | GH8-DS1 |
| CelE | Endoglucanase | 81 | GH9-CBM3-DS1 |
| AgaA | α-Galactosidase | 52 | GH27-DS1 |
The modular structures of cellulosomal subunits are indicated by the following abbreviations: GH, glycosyl hydrolase; Ig, immunoglobulin-like module; DS1, dockerin domain type 1; X, unknown domain; CE4, carbohydrate family 4 (acetyl xylan esterase); GPL, polysaccharide lyase family 9 (pectate lyase). See reference 57 for a list of C. thermocellum enzymes.
Usually the dockerin interacts specifically with its cognate cohesin present on the scaffoldin (40). A more complex situation has been observed with the cellulosome system of R. flavefaciens (54). In this case, the putative scaffolding protein ScaA lacks an identifiable cellulose binding module, and its dockerin interacts with the cohesins present in a putative surface-binding protein, ScaB. The cohesins of ScaA interacted with EndB, but not with XynE or CesA, which had dockerins closely related to but different from EndB. XynE and CesA did bind to alternative electrophoretic protein bands of R. flavefaciens, suggesting that the dockerin specificities of EndB were different from those of XynE and CesA and that there may exist another scaffoldin that has cohesins that can bind XynE and CesA. The results suggest the possibility that the other protein band that binds XynE and CesA is a fragment of ScaA or that a completely different scaffolding from ScaA exists. There has been no confirmed existence to date of two different scaffoldins being present in a cell. However, it would not be surprising if such a situation does exist.
Although cellulosomes were thought initially to degrade only cellulose, it is evident that cellulosomes contained a variety of degradative enzymes that could attack cellulose, hemicelluloses, chitin, and pectin (63) (Table 2). Thus, more recently, the concept of the cellulosome has changed from a cellulase complex to an enzyme complex that can degrade various plant cell wall materials. The term “cellwallase” has been used when natural substrates have been used (43). The types of cellulosomal enzymes that have been found are listed for the mesophilic clostridia C. cellulovorans, Clostridium josui, and C. cellulolyticum in Table 2. Similar types of enzymes have been described for the thermophile C. thermocellum (57).
CELLULASES
The cellulases, which comprise the major fraction of the cellulosomal enzymes, usually fall into three families of glycosyl hydrolases, consisting of family 5 and 9 endoglucanases and family 48 exoglucanases (Table 2). The family 9 members are especially interesting, since not only can they cleave cellulose fibers internally, but they can also then processively degrade the polymer from the point of cleavage (21). This type of activity also appears to be true for a family 48 enzyme CelF from C. cellulolyticum (53). However, since this activity of CelF was observed with acid-swollen cellulose, it is still uncertain whether this processive function will occur with natural substrates that contain crystalline cellulose.
A modification of a cellulase gene has been obtained through DNA shuffling techniques. In this case, more heat-stable enzymes were derived by shuffling genes that coded for two homologous endoglucanases, EngB and EngD, from Clostridium cellulovorans (44). The parental enzymes were active at 45°C but not at 55°C. The recombinant enzymes were stable at 55°C and as active as the parental EngB enzyme. Most of the amino acid residues of the recombinant enzymes were derived from EngB, but the critical amino acids for thermostability were derived primarily from EngD. Thus, this technique can be used to modify cellulase genes and select for various recombinants by suitable screening procedures (41).
HEMICELLULASES AND OTHER ENZYMES
The cellulosomal hemicellulases include xylanase and mannanase, which occur frequently in the cellulosome (Table 2). The xylanases are members of family 11 of glycosyl hydrolases, and mannanase is a member of family 5. A pectate lyase of family 9 has been observed in cellulosomes (63), and these findings indicate that cellulosomes can degrade cellulose, hemicellulose, and pectin—all components of plant cell walls.
A cellulosomal chitinase has been reported for C. thermocellum (67). This suggests the possibility that cellulosomes not only degrade lignocelluloses, but also degrade chitin, which occurs in fungi and which comprises the exoskeleton of many insects. Since many of the cellulolytic microorganisms reside in soil, they may also use chitin found in dead insects and fungi as a carbon and nitrogen source. Thus, the enzymatic composition of the cellulosome may reflect the environmental substrates that are available.
HETEROGENEITY OF CELLULOSOMES
The large number of enzymes that are present in the cellulosomal fraction and the limited number of cohesins on the scaffoldin suggested the possibility of a heterogeneous population of cellulosomes. This was shown to be the case with Clostridium papyrosolvens (52) and in C. cellulovorans (43). The C. papyrosolvens cellulosomes were separated by anion-exchange chromatography into seven distinct populations. The only common subunit present in all of the separated populations was the 125-kDa scaffoldin. The different subunits in these populations represented the presence of a large number of different cellulosomal enzymes. Furthermore, when these populations were examined by transmission electron microscopy, each population had a distinct morphology, again indicating a difference in subunit composition of these complexes (51). It seems clear at this time that cellulosomes are heterogeneous in enzymatic subunit composition and that in nature this may help the bacteria to degrade a complex substrate such as plant cell walls very efficiently.
CLUSTERS OF CELLULOSOMAL GENES
Since the cellulosome consists of a large number of subunits that are probably expressed simultaneously, it was of interest to see whether the genes were present in a large operon or whether they existed in a number of smaller operons. The analysis of the cellulosomal genes has revealed in C. cellulovorans and C. cellulolyticum that a large cluster of genes does occur in both species. In C. cellulovorans, there are nine genes related to the cellulosome present in a large gene cluster (64). In C. cellulolyticum, the gene cluster is even larger (6). From genome analysis, C. acetobutylicum also contains a cellulase gene cluster (47). An interesting observation with the C. cellulovorans gene cluster is that a transposase gene is present at its 3′ end (64). This finding suggests the possibility that the gene cluster had been introduced into C. cellulovorans by horizontal gene transfer.
Although several of the cellulosomal genes are clustered, there are a large number of cellulosomal genes that are not linked, and several occur as monocistronic genes in C. cellulovorans (24, 64). In the case of C. thermocellum, a large gene cluster of cellulosomal genes does not occur (57).
REGULATION OF EXPRESSION OF CELLULOSOMAL GENES
Expression of cellulosomal genes.
There has been little study of the regulation of expression of the cellulosomal genes. A recent analysis of the expression of the genes in the large gene cluster of C. cellulovorans has revealed that several operons exist (24). Transcripts that contained mRNA for cbpA and exgS and for cbpA, exgS, engH, and engK were observed as well as a transcript encoding engK, hbpA, and engL. Monocistronic mRNAs for manA and engM were also identified. For cellulosomal genes not linked with the large gene cluster, monocistronic mRNAs were revealed for engE, xynA, and pelA. The promoters for cbpA, engE, manA, and hbpA exhibited a strong similarity to the σA consensus promoter sequence of gram-positive bacteria.
An analysis of the regulation of the celS gene of C. thermocellum indicated that the amount of CelS produced was less in cultures grown on cellobiose than on cellulose (16). These studies also indicated that the amount of celS mRNA was inversely proportional to the growth rate under carbon and nitrogen limitations. The promoters as determined by primer extension analysis revealed the presence of two promoters for the celS gene, one similar to the σA promoter and the other to the σB promoter of Bacillus subtilis. The regulation of celS is related to the growth rate, and the expression of celS appears to be complicated by the presence of two promoters that can be expressed either at the log phase or under certain stress conditions.
In one interesting case, the C. acetobutylicum genome contains the genes for cellulosomal components, including the scaffolding protein and cellulases, and produces a cellulosome-like complex, which for some unknown reason cannot degrade crystalline cellulose (55, 56).
Further analysis of the expression of cellulase and hemicellulase genes should provide useful information for better understanding of regulatory elements, coordinate expression of genes, substrate induction, and more efficient production of these enzymes by genetic manipulations.
DESIGNER CELLULOSOMES
In vitro constructs.
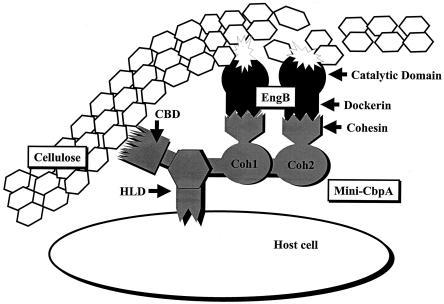
There has been a recent interest in constructing designer mini-cellulosomes (17, 42, 45, 46) for several purposes, including studying the functions of cohesins, analyzing synergy between various cellulosomal enzymes, and improving the efficiency of cellulosomes (Fig. 2). The recent studies were based on the successful construction of mini-cellulosomes (13) and ideas behind the development of designer cellulosomes (1).
FIG. 2.
Schematic representation of a mini-cellulosome. This model mini-cellulosome consists of a mini-CbpA containing a CBD; an HLD, which has been postulated in the role of binding the cellulosome to the cell surface (27); and two cohesins, to which are attached two molecules of cellulosomal enzyme (coded for by the engB gene) through its dockerin domain.
The groundwork for designing mini-cellulosomes with specific functions originated with the observation of synergistic action on crystalline cellulose when endoglucanase E (EngE) from C. thermocellum was bound to CipA (9). In further studies of cohesin-dockerin interaction, it was found that a C. thermocellum mini-CipA containing cohesin I and II domains interacted well with enzymes containing dockerin I and II (26, 33). In the latter study, there was a specific interaction between the cohesins and their cognate dockerins. It was also found that mini-CipAs containing CBM increased the activity, that synergism occurred when the enzyme was associated with mini-CipA, and that the presence of the CBM was critical for maximizing the activity. In a further development of this approach, a mini-CipA was constructed that contained a CBM and two cohesins: one from C. thermocellum and one from C. cellulolyticum. Since there is no cross-species interaction between cohesins and dockerins (49), chimeric cellulosomes with precise enzymatic composition and activity were constructed by adding cellulosomal enzymes from the two bacterial species (18).
These types of experiments have led recently to a very elegant study in which mini-scaffoldins were constructed with a CBD and neighboring cohesins that had divergent specificity. These mini-scaffoldins were combined with two cellulases, each containing a dockerin that interacted with one of the divergent cohesins (17). A combination of 75 different chimeric mini-cellulosomes was constructed. These studies showed that the formation of chimeric mini-cellulosomes contributed to higher activity on crystalline cellulose than free enzymes, that the CBD contributed to higher activity by bringing the enzymes closer to the substrate, that the composition of the neighboring enzymes had a positive effect on activity, and in some cases, that one of the enzymes predominated over the other.
In studies with mini-cellulosomes with defined enzymatic compositions, the different combinations of cellulosomal enzymes present in mini-cellulosomes resulted in significant synergy. Cellulases could act synergistically (45), and a combination of cellulases and xylanases (46) also contributed to a synergistic degradative effect on substrates. There was an optimum ratio of the cellulases for obtaining maximum synergy. Interestingly, the sequential addition of xylanase and cellulase to the substrate did not enhance the synergistic effect, whereas the simultaneous addition of xylanase and cellulase did improve the activity on corn fiber. These results suggested that the structure of the corn fiber allowed both enzymes to function simultaneously and that there was no removal of xylanase prior to attack by the cellulase.
Although synergistic effects were obtained with mini-cellulosomes, the native cellulosomes still showed much higher activity than a combination of mini-cellulosomes. Therefore, there is much to be learned about the combination of enzymes that leads to maximum activity against natural substrates. It has also been shown that there is synergy between cellulosomes and noncellulosomal enzymes for the degradation of corn fiber (28). It is therefore likely in nature that microorganisms produce not only cellulosomes but also a battery of noncellulosomal lignocellulolytic enzymes that act in concert with the cellulosome to attack complex substrates.
In vivo constructs.
There is current interest in developing in vivo systems in various bacteria in order to use relatively inexpensive biomass or agricultural wastes as a substrate to obtain the production of valuable products. One example of this type of research is to insert cellulosomal genes into organisms that already are able to produce valuable products, such as ethanol, butanol, or amino acids. If these organisms can now use biomass for their substrate, there may be considerable savings in the costs of media. Cellulosomes are important in that they can degrade crystalline forms of cellulose, which are recalcitrant to many enzymes that can degrade soluble or amorphous forms of cellulose. Furthermore, cellulosomes can degrade hemicelluloses and pectin, two other major components of plant cell wall materials.
There has been one report of designer cellulosomes being expressed in vivo in C. acetobutylicum. Although C. acetobutylicum has genes that code for enzymes and cellulosomes and produces an extracellular complex of >665 kDa, it cannot grow on cellulosic materials. By construction of a mini-CipA gene coding for CBD-HD1-HD2-Coh1-HD3-Coh2-HD4 with a mass of 89.5 kDa, insertion of this gene into C. acetobutylicum, and growth on cellobiose and crystalline cellulose, a cellulose-bound protein fraction was obtained that had a mass of about 665 kDa and contained a protein of about 250 kDa (56). A sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the 665-kDa complex revealed the presence of mini-CipA and Cel48A. The recombinant mini-cellulosome could not degrade Avicel or bacterial cellulose. However, the evidence was indicative of in vivo formation of a recombinant mini-cellulosome.
The in vivo formation of a recombinant cellulosome suggests that it will be possible in the future to insert cellulosomal genes into hosts that produce valuable products and that these hosts will produce cellulosomes and grow on inexpensive plant cell wall materials. Thus, the in vivo formation of cellulosomes could lead to great savings in the production of valuable materials that are currently being produced on high-cost substrates.
FUTURE STUDIES WITH CELLULOSOMES
Several aspects of cellulosomes require further inquiry. These include the following.
(i) How do cohesins and dockerins interact at the molecular level?
Although crystals of cohesin (58) have been obtained, crystallization of cohesin-dockerin complexes apparently has not been achieved. This could lead to an interesting explanation of how cellulosomal enzymes are bound to the scaffolding protein.
(ii) How is the extracellular cellulosome assembled?
Since all of the subunits of the complex have signal peptides and are secreted, how is each subunit formed into its final mature conformation, and how do the enzymatic subunits find their way to the cohesin sites on the scaffolding protein?
(iii) How do polycellulosomes form?
Since there are large masses of cellulosomes on the surface of the cell, are cellulosomes linked, or are they just aggregated? There is a hint that scaffolding proteins may interact, thus linking cellulosomes into polycellulosomes.
(iv) What are the mechanisms for binding the cellulosome to the cell surface?
Since all scaffolding proteins do not contain dockerin 2, which has been implicated in the case of C. thermocellum for binding the cellulosome to the cell surface through its interaction with Olp proteins that contain SLHs, there must be other modes for cell surface binding of cellulosomes. Are HLDs involved in binding cellulosomes to cell surfaces?
(v) What regulates the expression of cellulosomal genes?
There is evidence that different polymeric substrates induce different cellulosomal enzymes. An understanding of regulation is necessary to construct strains that can maximally utilize various substrates.
(vi) Can we develop designer cellulosomes that are more efficient than current native cellulosomes?
We will have to determine the combination of cellulosomal enzymes with the highest synergy activity. The use of scaffolding proteins with high affinity for the substrate and a combination of synergistic cellulosomal enzymes may increase the overall efficiency of the complex.
(vii) How do cellulosomes attack natural substrates such as plant cell walls, which are complex materials with various polymers and compounds?
Do cellulosomes attack the polymers in a sequential fashion, e.g., pectin, hemicelluloses, and finally celluloses? Can the use of cellulosomes be used to analyze plant cell wall structure?
(viii) Can we in fact construct different bacterial strains that can utilize plant cell wall materials as an inexpensive growth substrate?
Preliminary experiments suggest that cellulosomes can be constructed in vivo, and this should lead to the utilization of designer cellulosomes and metabolically engineered hosts for the formation of many valuable products.
(ix) Are cellulosomes a common feature of anaerobic cellulolytic bacteria?
There are numerous reports of dockerin-like domains, scaffolding-like proteins, and extracellular complexes or protuberances. When these reports are verified with solid evidence for the presence of cellulosomes, they may turn out to be a common feature of anaerobic degradative bacteria. Also it is likely that many environmental niches will be filled by cellulosome-producing organisms ranging from thermophilic to psychrophilic bacteria, from rumen to soil bacteria, and bacteria under various salt and pH conditions. Will they all be strictly anaerobic bacteria, or will some facultative anaerobic bacteria also produce cellulosomes?
CONCLUSIONS
Studies of cellulosomes indicate that cellulosomes are complex and heterogeneous in subunit composition. Cellulosomes are also efficient degraders of plant cell wall materials based on their content of diverse enzymes. Although the studies reported give an overall picture of the cellulosome, there is still much to be learned about their structure, function, regulation, assembly, and mechanism of action. Furthermore, if genetic manipulations are to be carried out, genetic systems will have to be developed that can modify the host cell and their cellulosomes. The ultimate goal will be to construct high-expression hosts with designer cellulosomes that can utilize various types of inexpensive biomass very efficiently.
Acknowledgments
The research reported from our laboratory was supported in part by grant DE-DDF03-92ER20069 from the U.S. Department of Energy.
REFERENCES
- 1.Bayer, E. A., E. Morag, and R. Lamed. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 12:378-386. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., E. Setter, and R. Lamed. 1985. Organization and distribution of the cellulosome in Clostridium thermocellum. J. Bacteriol. 163:552-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer, E. A., L. J. W. Shimon, Y. Shoham, and R. Lamed. 1998. Cellulosomes—structure and ultrastructure. J. Struct. Biol. 124:221-234. [DOI] [PubMed] [Google Scholar]
- 4.Bayer, E. A., Y. Shoham and R. Lamed. 2001. Cellulose-decomposing bacteria and their enzyme systems. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. Springer-Verlag, New York, N.Y. [Online.] http://141.150.157.117.8080/prokPUB/chaprender/jsp/showchap.jsp?chapnum=297.
- 5.Beguin, P., and P. M. Alzari. 1998. The cellulosome of Clostridium thermocellum. Biochem. Soc. Trans. 26:178-185. [DOI] [PubMed] [Google Scholar]
- 6.Belaich, A., G. Parsiegla, L. Gal, C. Villard, R. Haser, and J.-P. Belaich. 2002. Cel9M, a new family 9 cellulase of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 184:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, E., W. A. Jones, D. T. Jones, and D. R. Woods. 1990. Sequencing and expression of a cellodextrinase (ced1) gene from Butyrivibrio fibrisolvens H17c cloned in Escherichia coli. Mol. Gen. Genet. 223:310-318. [DOI] [PubMed] [Google Scholar]
- 8.Boraston, A. B., B. W. McLean, J. M. Kormos, M. Alam, N. R. Gilkes, C. A. Haynes, P. Tomme, D. G. Kilburn, and R. A. Warren. 1999. Carbohydrate-binding modules: diversity of structure and function, p. 202-211. In H. J. Gilbert, G. J. Davies, B. Henrissat, and B. Svensson (ed.), Recent advances in carbohydrate bioengineering. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 9.Ciruela, A., H. J. Gilbert, B. R. S. Ali, and G. P. Hazlewood. 1998. Synergistic interaction of the cellulosome integrating protein (CipA) from Clostridium thermocellum with a cellulosomal endoglucanase. FEBS Lett. 422:221-224. [DOI] [PubMed] [Google Scholar]
- 10.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 1999. A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family 9 glycosyl hydrolase. J. Bacteriol. 181:6720-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding, S.-Y., E. A. Bayer, D. Steiner, Y. Shoham, and R. Lamed. 2000. A scaffoldin of the Bacteriodes cellulosolvens cellulosome that contains 11 type II cohesins. J. Bacteriol. 182:4915-4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding, S.-Y., M. T. Rincon, R. Lamed, J. C. Martin, S. I. McCrae, V. Aurilia, Y. Shoham, E. A. Bayer, and H. J. Flint. 2001. Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183:1945-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doi, R. H., M. A. Goldstein, J. S. Park, C.-C. Liu, Y. Matano, M. Takagi, S. Hashida, F. C.-F. Foong, T. Hamamoto, I. Segel, and O. Shoseyov. 1993. Structure and function of the subunits of the Clostridium cellulovorans cellulosome, p. 43-52. In K. Shimada, K. Ohmiya, Y. Kobayashi, S. Hoshino, K. Sakka, and S. Karira (ed.), Genetics, biochemistry and ecology of lignocellulose degradation. Uni Publishers Co., Ltd., Tokyo, Japan.
- 14.Doi, R. H., J.-S. Park, C.-C. Liu, L. M. Malburg, Y. Tamaru, A. Ichi-ishi, and A. lbrahim. 1998. Cellulosome and noncellulosomal cellulase of Clostridium cellulovorans. Extremophiles 2:53-60. [DOI] [PubMed] [Google Scholar]
- 15.Doi, R. H., and Y. Tamaru. 2001. The Clostridium cellulovorans cellulosome: an enzyme complex with plant cell wall degrading activity. Chem. Rec. 1:24-32. [DOI] [PubMed] [Google Scholar]
- 16.Dror, T. W., E. Morag, A. Rolider, E. A. Bayer, R. Lamed, and Y. Shoham. 2003. Regulation of the cellulosomal celS (cel48A) gene of Clostridium thermocellum is growth rate dependent. J. Bacteriol. 185:3042-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fierobe, H.-P., E. A. Bayer, C. Tardif, M. Czjzek, A. Mechaly, A. Belaich, R. Lamed, Y. Shoham, and J.-P. Belaich. 2002. Degradation of cellulose substrates by cellulosome chimeras. J. Biol. Chem. 277:49621-49630. [DOI] [PubMed] [Google Scholar]
- 18.Fierobe, H.-P., A. Mechaly, C. Tardif, A. Belaich, R. Lamed, Y. Shoham, J.-P. Belaich, and E. A. Bayer. 2001. Design and production of active cellulosome chimeras: selective incorporation of dockerin-containing enzymes into defined functional complexes. J. Biol. Chem. 276:21257-21261. [DOI] [PubMed] [Google Scholar]
- 19.Fujino, T., P. Béguin, and J.-P. Aubert. 1993. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J. Bacteriol. 175:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gal, L., S. Pages, C. Gaudin, A. Belaich, C. Reverbel-Leroy, C. Tardif, and J.-P. Belaich. 1997. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ. Microbiol. 63:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudin, C., A. Belaich, S. Champ, and J.-P. Belaich. 2000. CelE, a multidomain cellulase from Clostridium cellulolyticum: a key enzyme in the cellulosome? J. Bacteriol. 182:1910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerngross, U., M. P. M. Romaniec, T. Kobayashi, N. S. Huskisson, and A. L. Demain. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8:325-334. [DOI] [PubMed] [Google Scholar]
- 23.Goldstein, M., M. Takagi, S. Hashida, O. Shoseyov, R. H. Doi, and I. H. Segel. 1993. Characterization of the cellulose-binding domain of the Clostridium cellulovorans cellulose-binding protein A (CbpA). J. Bacteriol. 175:5762-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han, S. O., H. Yukawa, M. Inui, and R. H. Doi. 2003. Transcription of Clostridium cellulovorans cellulosomal cellulase and hemicellulase genes. J. Bacteriol. 185:2520-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakiuchi, M., A. Isui, K. Suzuki, T. Fujino, E. Fujino, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1998. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J. Bacteriol. 180:4303-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kataeva, I., G. Guglielmi, and P. Beguin. 1997. Interaction between Clostridium thermocellum endoglucanase CelD and polypeptides derived from the cellulosome-integrating protein CipA: stoichiometry and cellulolytic activity of the complexes. Biochem. J. 326:617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosugi, A., K. Murashima, Y. Tamaru, and R. H. Doi. 2002. Cell surface anchoring role of N-terminal surface layer homology domains of Clostridium cellulovorans EngE. J. Bacteriol. 184:884-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosugi, A., K. Murashima, and R. H. Doi. 2002. Characterization of two noncellulosomal subunits, ArfA and BgaA, from Clostridium cellulovorans that cooperate with the cellulosome in plant cell wall degradation. J. Bacteriol. 184:6859-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamed, R., and E. A. Bayer. 1988. Cellulosome from Clostridium thermocellum. Methods Enzymol. 160:472-481. [Google Scholar]
- 30.Lamed, R., J. Naimark, E. Morgenstern, and E. A. Bayer. 1987. Specialized surface structures in cellulolytic bacteria. J. Bacteriol. 169:3792-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamed, R., E. Setter, and E. A. Bayer. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156:828-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leibovitz, E., and P. Béguin. 1996. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178:3077-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leibovitz, E., and P. Beguin. 1998. Comparison of two scaffolding polypeptides for the integration of different proteins in synthetic complexes derived from the Clostridium thermocellum cellulosome. Enzyme Microb. Technol. 22:588-593. [Google Scholar]
- 34.Leibovitz, E. H. Ohayon, P. Gounon, and P. Béguin. 1997. Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J. Bacteriol. 179:2519-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemaire, M., I. Miras, P. Gounon, and P. Beguin. 1998. Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology 144:211-217. [DOI] [PubMed] [Google Scholar]
- 36.Lemaire, M., H. Ohayon, P. Gounon, T. Fujino, and P. Beguin. 1995. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177:2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lynd, L. R., P. J. Weimer, W. H. van Zyl, and I. S. Pretorius. 2002. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66:506-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayer, F., M. P. Coughlan, Y. Mori, and L. G. Ljungdahl. 1987. Macromolecular organization of the cellulolytic enzyme complex of Clostridium thermocellum as revealed by electron microscopy. Appl. Environ. Microbiol. 53:2785-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miron, J., D. Ben-Ghedalia, and M. Morrison. 2001. Adhesion mechanisms of rumen cellulolytic bacteria. J. Dairy Sci. 84:1294-1309. [DOI] [PubMed] [Google Scholar]
- 40.Morag, E., E. A. Bayer, and R. Lamed. 1990. Relationship of cellulosomal and noncellulosomal xylanases of Clostridium thermocellum to cellulose-degrading enzymes. J. Bacteriol. 172:6098-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murashima, K., and R. H. Doi. 2003. Selection of heat stable Clostridium cellulovorans cellulases after in vitro recombination. Methods Mol. Biol. 230:231-237. [DOI] [PubMed] [Google Scholar]
- 42.Murashima, K., C.-L. Chen, A. Kosugi, Y. Tamaru, R. H. Doi, and S. L. Wong. 2002. Heterologous expression of Clostridium cellulovorans engB, using protease-deficient Bacillus subtilis, and preparation of active recombinant cellulosomes. J. Bacteriol. 184:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Determination of subunit composition of Clostridium cellulovorans cellulosomes that degrade plant cell walls. Appl. Environ. Microbiol. 68:1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Thermostabilization of cellulosomal endoglucanase EngB from Clostridium cellulovorans by in vitro DNA recombination with non-cellulosomal endoglucanase EngD. Mol. Microbiol. 45:617-626. [DOI] [PubMed] [Google Scholar]
- 45.Murashima, K., A. Kosugi, and R. H. Doi. 2002. Synergistic effects on crystalline cellulose degradation between cellulosomal cellulases from Clostridium cellulovorans. J. Bacteriol. 184:5088-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murashima, K., A. Kosugi, and R. H. Doi. 2003. Synergistic effects of cellulosomal xylanase and cellulases from Clostridium cellulovorans on plant cell wall degradation. J. Bacteriol. 185:1518-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nölling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, GTC Sequencing Center Production, Finishing, and Bioinformatics Teams, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohara. H., S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 2000. Characterization of the cellulolytic complex (cellulosome) from Ruminococcus albus. Biosci. Biotechnol. Biochem. 64:254-260. [DOI] [PubMed] [Google Scholar]
- 49.Pages, S., A. Belaich, J.-P. Belaich, E. Morag, R. Lamed, Y. Shoham, and E. A. Bayer. 1997. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29:517-527. [PubMed] [Google Scholar]
- 50.Pagès, S., A. Bélaïch, H. P. Fierobe, C. Tardif, C. Gaudin, and J.-P. Bélaïch. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin- containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pohlschröder, M., E. Canale-Parola, and S. B. Leschine. 1995. Ultrastructural diversity of the cellulase complexes of Clostridium papyrosolvens C7. J. Bacteriol. 177:6625-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pohlschroder, M., S. B. Leschine, and E. Canale-Parola. 1994. Multicomplex cellulase-xylanase system of Clostridium papyrosolvens C7. J. Bacteriol. 176:70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reverbel-Leroy, C., S. Pages, A. Belaich, J.-P. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rincon, M. T., S.-Y. Ding, S. I. McCrae, J. C. Martin, V. Aurilia, R. Lamed, Y. Shoham, E. A. Bayer, and H. J. Flint. 2003. Novel organization and divergent dockerin specificities in the cellulosome system of Ruminococcus flavefaciens. J. Bacteriol. 185:703-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabathe, F., A. Belaich, and P. Soucaille. 2002. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 217:15-22. [DOI] [PubMed] [Google Scholar]
- 56.Sabathé, F., and P. Soucaille. 2003. Characterization of the CipA scaffolding protein and in vivo production of a minicellulosome in Clostridium acetobutylicum. J. Bacteriol. 185:1092-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwarz, W. H. 2001. The cellulosome and cellulose degradation by anaerobic bacteria. Appl. Microbiol. Biotechnol. 56:634-649. [DOI] [PubMed] [Google Scholar]
- 58.Shimon, L. J. W., E. A. Bayer, E. Morag, R. Lamed, S. Yaron, Y. Shoham, and F. Frolow. 1997. The crystal structure at 2.15 A resolution of a cohesin domain of the cellulosomes from Clostridium thermocellum. Structure. 5:381-390. [DOI] [PubMed] [Google Scholar]
- 59.Shoseyov, O., and R. H. Doi. 1990. Essential 170 kDa subunit for degradation of crystalline cellulose by Clostridium cellulovorans cellulase. Proc. Natl. Acad. Sci. USA 87:2192-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shoseyov, O., M. Takagi, M. Goldstein, and R. H. Doi. 1992. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A (CbpA). Proc. Natl. Acad. Sci. USA 89:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sleat, R., R. A. Mah, and R. Robinson. 1984. Isolation and characterization of an anaerobic, cellulolytic bacterium, Clostridium cellulovorans sp. nov. Appl. Environ. Microbiol. 48:88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tamaru, Y., and R. H. Doi. 1999. Three surface layer homology domains at the N terminus of the Clostridium cellulovorans major cellulosomal subunit EngE. J. Bacteriol. 181:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tamaru, Y., and R. H. Doi. 2001. Pectate lyase A, an enzymatic subunit of the Clostridium cellulovorans cellulosome. Proc. Natl. Acad. Sci. USA 98:4125-4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamaru, Y., S. Karita, A. Ibrahim, H. Chan, and R. H. Doi. 2000. A large gene cluster for the Clostridium cellulovorans cellulosome. J. Bacteriol. 182:5906-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Warren, R. A. J. 1996. Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 50:183-212. [DOI] [PubMed] [Google Scholar]
- 66.Weyer, E. R., and L. F. Rettger. 1927. A comparative study of six different strains of the organism commonly concerned in large-scale production of butyl alcohol and acetone by the biological process. J. Bacteriol. 14:399-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zverlov, V. V., K.-P. Fuchs, and W. H. Schwarz. 2002. Chi18A, the endochitinase in the cellulosome of the thermophilic, cellulolytic bacterium Clostridium thermocellum. Appl. Environ. Microbiol. 68:3176-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]