Abstract
The S-adenosyl-L-methionine (SAM)-dependent methyltransferases represent a diverse and biologically important class of enzymes. These enzymes utilize the ubiquitous methyl donor SAM as a cofactor to methylate proteins, small molecules, lipids, and nucleic acids. Here we present the crystal structure of PH1915 from Pyrococcus horikoshii OT3, a predicted SAM-dependent methyltransferase. This protein belongs to the Cluster of Orthologous Group 1092, and the presented crystal structure is the first representative structure of this protein family. Based on sequence and 3D structure analysis, we have made valuable functional insights that will facilitate further studies for characterizing this group of proteins. Specifically, we propose that PH1915 and its orthologs are rRNA- or tRNA-specific methyltransferases.
Keywords: COG1092, crystal structure, methyltransferase, PH1915, S-adenosylmethionine
The S-adenosyl-L-methionine (SAM)-dependent methyl-transferases (MTases) represent a diverse and biologically important class of enzymes. These enzymes utilize the ubiquitous methyl donor SAM as a cofactor to methylate proteins, small molecules, lipids, and nucleic acids (Martin and McMillan 2002; Miller et al. 2003). SAM MTases are central to cellular biochemistry since they mediate numerous biological processes, such as protein trafficking and sorting, signal transduction, biosynthesis, metabolism, and gene expression (Martin and McMillan 2002).
A number of SAM MTase crystal structures belonging to the families of RNA, DNA, proteins, and small molecule methyltransferases have been determined. Although these families methylate different substrates, all SAM MTases contain a structurally conserved SAM-binding domain consisting of a central seven-stranded β-sheet that is flanked by three α-helices per side of the sheet (Martin and McMillan 2002). Additionally, depending on the size of the substrate, a substrate-recognition domain may be appended to the SAM-binding domain (Martin and McMillan 2002). However, this domain is highly variable at both the structural and sequence levels (Martin and McMillan 2002), which is consistent with the fact that these enzymes methylate a variety of distinct substrates.
Among the many cellular functions that are influenced by SAM MTases, the metabolism and maturation of RNAs, in particular tRNAs, are ubiquitous. The presence of modified nucleosides is a hallmark feature of tRNAs, and is associated with a range of biological functions. To date, more than 96 types of RNA nucleoside modifications have been characterized from cellular RNAs (McCloskey and Crain 1998; Rozenski et al. 1999), of which greater than 80 such modifications exist in tRNAs (Okamoto et al. 2004). Importantly, it appears that tRNA MTases play dominant roles in tRNA modifications since methylations of the ribose sugar or base are common.
Although the specific function of many methylated nucleosides remains obscure, some nucleoside modifications that are conserved at unique positions of specific tRNAs from all three phylogenetic domains of life have been well characterized. For example, the nucleosides m1G37 (Bjork et al. 1989; Brule et al. 2004), m5U54 (Davanloo et al. 1979; Kersten et al. 1981; Johansson and Bystrom 2002), and m22G10/26 (Armengaud et al. 2004) are crucial for preventing +1 frameshifting, tRNA maturation and Escherichia coli viability, and facilitating proper tRNA folding, respectively. Other conserved modifications have been shown to contribute to the maintenance of translational efficiency and fidelity, regulation of cell-cycle transitions, tRNA-protein interactions, and adaptations to cellular stresses (Hopper and Phizicky 2003).
In addition to tRNAs, another important target of SAM MTases is rRNA. However, unlike tRNA methylations, which are mostly associated with translational fidelity, rRNA methylations are mainly involved in biological processes that are environment or condition-specific. For example, deletion of E. coli RrmJ ( FtsJ ), an rRNA methyltransferase that methylates U2552 of the 23S rRNA in 50S ribosomal subunit, results in severe growth deficits at various temperatures, as well as an accumulation of ribosomal subunits at the expense of functional 70S subunits (Bugl et al. 2000; Caldas et al. 2000a,b). Another example is that of Erm from E. coli. This rRNA methyltransferase confers resistance to Macrolid, Lincosamid, and Streptogramin B (MLSB) antibiotics by methylating a specific adenine in the 23S rRNA (Maravic 2004); this methylated nucleoside sterically hinders MLSB binding to the 50S ribosomal subunit.
Members of the Cluster of Orthologous Group (COG) 1092 have been functionally annotated, on the basis of sequence homology, to be SAM-dependent MTases. However, direct functional assignment, such as substrate specificity and catalyzed reaction, has not been established. Here we present the first representative structure of this COG, as the crystal structure of PH1915 from Pyrococcus horikoshii OT3, a COG1092 member, has been determined. Furthermore, based on sequence and 3D structure analysis, we have made valuable functional insights that will facilitate further studies for characterizing these proteins. Specifically, we propose that PH1915 and its orthologs are rRNA- or tRNA-specific methyltransferases. Moreover, as members of COG1092 are present in Archea, Bacteria, and Eucarya, this family of proteins may be universally important as they may methylate nucleosides at a highly conserved position.
Results and Discussion
Sequence analysis
Sequence analysis of PH1915 indicated that it belongs to a large group of proteins all with no known function except that they are predicted to be SAM-dependent methyltransferases. These proteins all cluster into an orthologous group belonging to COG1092. A detailed PSI-BLAST analysis of these proteins revealed high sequence conservation judged by their high sequence identity which is >30%. Proteins in COG1092 were identified from the three different kingdoms, Archea, Bacteria, and Eucarya. Some of the eukaryotic homologs identified from this analysis are AP005572 from Oryza sativa, AY059153 from Arabidopsis thaliana, and three proteins from different species of Plasmodium.
Structure determination
The selenium–methionine labeled protein–crystal diffracted to 1.8 Å resolution. X-ray diffraction data collected at the selenium (Se) peak wavelength were used to obtain initial phase information by single wavelength anomalous dispersion (SAD). The phase information from the SAD experiment with the Se peak data were used to build the initial model, which consists of two molecules in the asymmetric unit. The final model of PH1915 consists of all 396 residues, and it displays overall excellent stereochemistry judged by Ramachandran plot, which shows that 99.5% of the residues are in the allowed region. The final energy minimization refinement of PH1915 yields a protein model that has an R-value of 20.7% (Rfree 24%). Data collection and refinement statistics are summarized in Table 1.
Table 1.
X-ray data collection and refinement statistics for PH1915
| X-ray data | Peak |
| Space group | P21 |
| Unit cell (Å3) | a, b, c = 70.51, 86.16, 76.69 α, β, γ = 90.0, 114.16, 90.0 |
| Resolution (Å) | 30–1.8 |
| Wavelength (λ) | 0.97934 |
| Se sites (no.) | 28 |
| Total observations (no.) | 366,357 |
| Unique reflections (no.) | 77,977 |
| Intensity (I/σ 〈I〉) | 23 (2.5) |
| Completeness (%) | 99.5 |
| Rsyma | 0.083 (0.521) |
| Rworkb | 0.207 |
| Rfree | 0.24 |
| Protein atoms (no.) | 6270 |
| Water molecules (no.) | 512 |
| RMSD bond lengths (Å) | 0.005 |
| RMSD bond angles (°) | 1.2 |
| RMSD dihedral (°) | 23.8 |
| RMSD improper (°) | 0.70 |
| Average main-chain B-factor (Å2) | 24.8 |
Numbers in parentheses refer to the highest resolution shell, 1.91–1.80 Å.
aRsym = ∑|I – 〈I〉|/∑I, where I is the observed intensity and 〈I〉 is the average intensity from multiple observations of symmetry-related reflections.
bR =∑|Fobs – Fcalc|/|Fobs|.
Overview of the structure
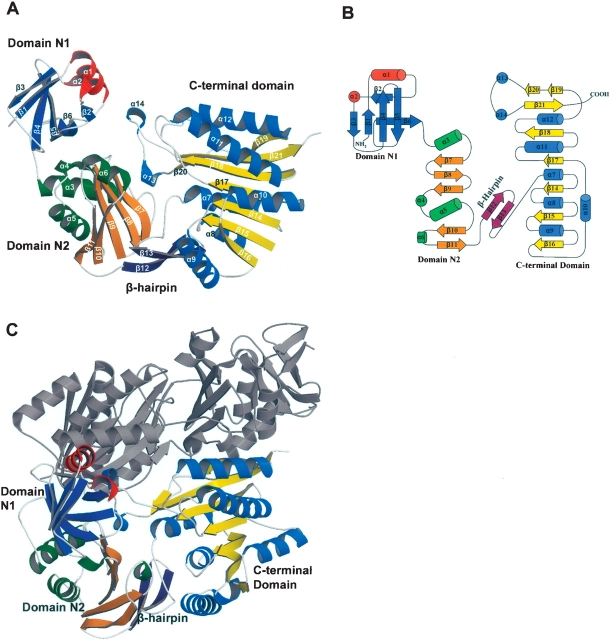
The final model of PH1915 is a tightly packed dimer in the asymmetric unit with each monomer consisting of three distinct structural domains: an N-terminal (N1, residues 1–71) α/β pseudobarrel, a middle (N2, residues 74–174) α/β domain, and a C-terminal (residues 202–396) α/β domain (Fig. 1A,B ▶). In the monomer, the N1 and N2 domains are closely linked to each other and are involved in a number of interactions, whereas the C-terminal domain is linked to the N2 domain via a β-hairpin (residues 178–193; β12 and β13). The N1 and N2 domains are predicted to form the substrate binding domain, and the C-terminal domain is predicted to bind SAM.
Figure 1.
Ribbon and topology diagrams of the PH1915 structure. (A) Ribbon diagram of the PH1915 subunit. The monomer consists of three domains: N1, colored blue and red; N2, colored orange and green; and the C-terminal domain, colored yellow and light blue; for β-strands and α-helices, respectively. The β-hairpin linking domain N2 to the C-terminal domain is colored purple. (B) Topology diagram of the PH1915 subunit. α-helices are represented as cylinders, 310 helices as circles, and β-strands as arrows. The diagram shows the arrangement of secondary structures and the three domains in the subunit. The color scheme is retained from the subunit representation in A. (C) Ribbon diagram of the PH1915 homodimer. One subunit is colored according to the representation in A and the other subunit is colored gray. The organization of the subunits results in the N1 and C-terminal domains from one subunit tightly packing with the C-terminal domain of the other subunit.
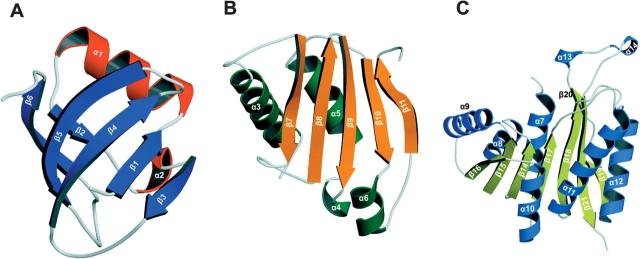
The N1 domain is composed of a mainly anti-parallel six stranded β-sheet (β1–β6; strand order β 3–1–4–5–6–2), one α-helix (α1), and a 310 helix (α2) (Fig. 2A ▶). The six stranded β-sheet adopts a pseudo-β-barrel topology and the inclusion of helix α1 completes the barrel. This N1 α/β barrel domain is analogous to the PUA domain of Pseu-doUridine Synthase and Archaeosine-specific transglycosylase (Aravind and Koonin 1999).
Figure 2.
Ribbon diagrams of the individual domains of PH1915. (A) Domain N1. α-Helices are colored red, and β-strands are colored blue. The overall topology of the N1 domain is analogous to the PUA domain. (B) Domain N2. α-Helices are colored green, whereas β-strands are colored orange. (C) C-terminal domain. α-helices are colored light blue, and β-strands are colored yellow. This C-terminal domain is reminiscent of the SAM cofactor binding domain seen in other MTases.
Domain N2 is also a mixed α/β structure; it consists of an anti-parallel five-stranded β-sheet (β7–β11, strand order β7–8–9–10–11), three α-helices (α3, α5, α6), and one 310 helix (α4) (Fig. 2B ▶). The longer two helices (α3 and α5) are found on the outer face of the β-sheet, whereas the smaller helices (α4 and α6) lie in-plane with the sheet.
The C-terminal SAM binding domain consists of a central, mainly parallel, eight-stranded β-sheet (β14–β21) that is sandwiched by three α-helices on each face of the sheet (α7–α12) (Fig. 2C ▶). The order of the β-strands is 16–15–14–17–18–21–(19–20), where all strands, except β21, are in parallel orientation; this order is analogous to the consensus SAM MTase fold (Martin and McMillan 2002). However, the C-terminal domain of PH1915 contains three notable deviations from that of a typical SAM binding domain. First, the turn, which connects β20 to β21, has been modified by the addition of two 310 helices (α13 and α14) that extend outward to form a protrusion. Second, unlike the consensus fold, which consists of a seven-stranded β-sheet (Martin and McMillan 2002), the corresponding PH1915 C-terminal domain contains eight β-strands; strands β19 and β20 are structurally derived from a larger β-strand that has been divided into two smaller strands. Finally, helix α13 is lengthened. This variation, although not conserved among DNA and RNA SAM-MTases, has only been observed in these subclasses (Martin and McMillan 2002).
Quaternary structure
The crystal structure reveals that PH1915 is a homo-dimer, and that the subunits are related by a twofold symmetry (Fig. 1C ▶). The major intersubunit contacts involve reciprocal interactions between the C-terminal domains of each subunit. In addition, the N1 and N2 may contribute to dimer stability by packing against the complementary C-terminal domain. Specifically, regions of the helices α1, α7, α12, α13, and α14, and regions of the strands β7, β19, and β20 are involved in these said interactions.
The dimer interface is mainly hydrophobic, with a composition of 65% nonpolar and 35% polar residues; however, there are also 15 hydrogen bonds formed between residues from the two subunits. Some key residues which form a network of interactions at this junction include Lys84A, Ala106A, Tyr108A, Lys361B, and Glu393B.
Structural homologies and comparisons
Dali (Holm and Sander 1995) analysis of the monomer of PH1915 did not identify any structural homologs. The proteins identified with the queried monomer are a result of conservation at the N1 and the C-terminal domains. Analysis with the individual domains revealed that the N1 domain has significant homology to the PUA domain, the N2 domain may be involved in protein–protein interactions, and the C-terminal domain has significant relationship to the SAM-dependent methyltransferase domain.
Analysis of the N1 domain identified 14 structures in the PDB with Z-scores ranging from 9.4 to 2.3. Of these 14, seven have not been functionally characterized and, hence, did not provide any useful functional information. However, the majority of the remaining Dali hits correspond to proteins that associate with either rRNAs or tRNAs. The top four hits with known functions are archaeosine tRNA–guanine transglycosylase (PDB ID 1iq8, Z-score 9.4; Ishitani et al. 2002), Nip7p 60S rRNA processing homolog (PDB ID 1t5y, Z-score 9.3; Coltri et al. 2004), tRNA pseudouridine synthase b (PDB ID 1k8w, Z-score 5.7; Hoang and Ferre-D’Amare 2001), and sulfate adenylyltransferase (PDB ID 1g8f, Z-score 5.1; Ullrich et al. 2001).
Superposition of the N1 domain of PH1915 with the four identified structural homologs indicated that the sulfate adenylyltransferase is the least similar. The C3 domain of archaeosine tRNA–guanine transglycosylase, the N-terminal domain of tRNA pseudouridine synthase b, and the C-terminal domain of Nip7p are very similar to the N1 domain of PH1915. Importantly, these superposed domains have been characterized as a PseudoUridine Synthase (PUA) domain or, in the case of pseudouridine synthase b, another RNA binding domain, the TruB domain (Hoang and Ferre-D’Amare 2001). Sequence analysis also indicated that this N-terminal region (residues 1–74) contains conserved sequence motifs that are found in the PUA domain.
The PUA domain was originally annotated by bioinformatics analysis, and has since been structurally and functionally characterized (Aravind and Koonin 1999). This PUA domain has been identified in Archea, Eucarya, and Bacteria and is characterized as being mainly important for modification of RNA molecules with a complex secondary structure. The finding that the N1 domain shares high sequence and structure similarity with the PUA domain is a good indication that protein PH1915 is an RNA binding protein.
Dali analysis with the N2 domain identified a number of structures, but with low Z-scores (Z-scores 5.2–2.0), the majority of which are involved in protein–protein interactions. MinC (Cordell et al. 2001) is the closest structural homolog to PH1915 based on the Z-score (PDB ID 1hf2, Z-score 5.2). MinC is an E. coli protein that depolymerizes a bacterial tubulin homolog, FtsZ, to inhibit cell division. However, comparative structural analysis did not show a significant match in topologies between domain N2 and MinC. Thus, no functional information was inferred from this analysis with the N2 domain.
Dali analysis of the C-terminal domain of PH1915 identified other SAM-dependent MTases with Z-scores in the high teens. The top four Dali hits with known biological function are Methanococcus jannaschii MJ0882 (PDB ID 1dus, Z-score 18.4; Huang et al. 2002), Mycobacterium tuberculosis Rv2118c (PDB ID 1i9g, Z-score 16.2; Gupta et al. 2001 E. coli HemK (PDB ID 1t43, Z-score 16.1; Yang et al. 2004), and Thermotoga maritima PrmC (PDB ID 1nv8, Z-score 16.0; Schubert et al. 2003). These methyltransferases can be broadly divided into two families, namely RNA– and protein–MTases, and includes MJ0882 (Mouaikel et al. 2003) and Rv2118c (Varshney et al. 2004) in the former and HemK (Heurgue-Hamard et al. 2002; Nakahigashi et al. 2002) and PrmC (Schubert et al. 2003) in the latter. The C-terminal domain of PH1915 superposed well with the SAM binding domain of these four proteins.
SAM methyltransferases are grouped according to the substrate they methylate. Although the core SAM domain is structurally conserved, there are additional structural elements which are variable and are indicative of the group to which these proteins belong to. For example, methyltransferases that modify proteins, such as CheR (Djordjevic and Stock 1997), PIMT (Skinner et al. 2000), and PRMT3 (Zhang et al. 2000), lack helix α10, and helix α12 is either poorly defined or absent from the SAM binding domain (Martin and McMillan 2002). In contrast to these protein MTases, the C-terminal domain of PH1915 does indeed have well-defined α10 and α12 helices. Furthermore, helix α11 is ~25% longer than other helices in the SAM binding domain of PH1915, and this characteristic is only associated with DNA– and RNA–MTases. Finally, PH1915 contains two 310 insertions (α13, α14) between β20 and β21 to form a protruding loop. This feature is analogous to Rv2118c and ErmAM/ErmC′ (Yu et al. 1997; Bussiere et al. 1998), both of which are RNA–MTases. Altogether, the C-terminal domain PH1915 incorporates many structural elements that are characteristic of the SAM binding domain of other RNA MTases.
The SAM-binding site
There are three motifs in the methyltransferase domain that are highly conserved among all MTases, as they facilitate SAM binding to the structure. These are the G-loop (motif I), the D-loop (motif II), and the P-loop (motif IV) motifs (Fauman et al. 1999; Martin and McMillan 2002). The G-loop, which is positioned between β14 and α8, is a glycine-rich motif that interacts with the amino acid portion of SAM. The D-loop contains an acidic residue, either Asp or Glu, whose side-chain hydrogen bonds with the ribose hydroxyl of SAM. This motif is located between β15 and α9. Finally, the P-loop, located at the C-terminal end of β17, is a proline-rich motif that binds with the adenine ring of SAM via hydrophobic interactions.
Although PH1915 was not crystallized with the SAM cofactor, the SAM-binding site and associated catalytic residues were inferred by the superimposition of PH1915 structural homologs containing bound SAM. From this analysis, it appears that SAM binds the PH1915 methyltransferase fold according to the consensus pattern of binding: SAM lies across the top of the MTase fold, with the ribose above the carboxyl-end of β15, the methionine moiety extending to the area between α7 and α8, and the adenine ring extending to α10 and β17. Indeed, multiple sequence alignment of 15 selected COG1092 members identified a number of conserved residues in the SAM binding domain. In the C-terminal domain of PH1915 these conserved residues are D203 and D204 from α7; G229 and F231 from the G-loop motif; D247 from the D-loop motif; N263 from α9; D291, D296, and P297 from the P-loop motif; G329 from β18; D372 from α13; and finally, Y384 and L385 from β21. In conclusion, it appears that the environment and interacting residues of PH1915 are consistent with nearly all other SAM MTases of known structure.
In summary, structural homology searches and comparisons demonstrated that the PH1915 contains three domains, two of which have been previously characterized— namely the PUA domain and the SAM MTase domain—and one that represents a novel fold. The organization of the three domains together produces a molecule that does not have obvious similarities to other proteins with known 3D structures. As such, we suggest that the 3D structure of PH1915 is novel. Moreover, comparative analysis of PH1915 structure suggests that it is a SAM-dependent RNA methyltransferase. This structure provides the first glimpse of COG1092 members and, moreover, provides a framework to deduce and assay the molecular function of this family of conserved proteins.
Materials and methods
Target selection and cloning
PH1915 (GenBank accession NP_143744) from Pyrococcus horikoshii was selected as part of our structural genomics effort. This target gene was amplified from P. horikoshii genomic DNA using primers to introduce unique cleavage sites for AceI and BglII. The fragment was subsequently cloned into p11 expression vector as previously described (Zhang et al. 2001).
Protein expression, solubility, and purification
PH1915 was expressed and purified as previously described (Zhang et al. 2001). Briefly, the expression plasmid for protein PH1915 was transformed into E. coli BL21-Gold (DE3) (Stratagene), which harbor an extra plasmid (pMgk) encoding three rare tRNAs (AGG and AGA for Arg, ATA for Ile). These E. coli cells were then cultured in 1-L Luria-Bertani media supplemented with ampicillin (100 μg/mL) and kanamycin (50 μg/mL), and incubated at 37°C with shaking until the culture reached an OD600 of 0.6–0.8. At this point the culture was induced with 0.4 mM IPTG for 3 h at 37°C with shaking and allowed to grow overnight at 15°C. Cells were harvested by centrifugation, disrupted by sonication, and the insoluble cellular material was removed by centrifugation. Protein PH1915 was purified from other contaminating proteins using Ni-NTA affinity chromatography. The protein was digested with 0.15 mg TEV protease per 20 mg purified protein for 16 h at 4°C, and then passed through an Ni-NTA column to remove both the TEV protease and cleaved histidine tags. The protein was subsequently dialyzed and stored in buffer containing 10 mM HEPES (pH 7.5) and 500 mM NaCl and quantified using an extinction coefficient of 0.475 M−1 cm−1 for contributions of Trp and Tyr at 280 nm. Selenomethionine protein was prepared as follows: PH1915 was expressed in BL21 cells (described above) in M9 minimal media (Sambrook et al. 1989) containing 0.4% (w/v) glucose as a carbon source. Once the cells reached an OD600 of 0.8, selenomethionine (SeMet), plus an amino acid cocktail (60 mg SeMet; 100 mg of lysine, threonine, and phenylalanine; and 50 mg leucine, isoleucine, and valine) were added. Fifteen minutes later the culture was induced with 0.4 mM IPTG and the cells were allowed to grow as described above for the native protein. Cell lysis and protein purification were carried out exactly as described above for native protein with the addition of 5 mM β-mercaptoethanol in all buffers.
Crystallization
The initial crystallization condition was determined with a sparse crystallization matrix (Hampton Research Crystal Screen I) at room temperature using the hanging drop-vapor diffusion technique. The optimized crystallization condition consists of 16% glycerol, 1.6 M ammonium sulfate, and 0.1 M sodium acetate at pH 3.2 (2 μL of protein solution [10 mg/mL] : 2 μL of the reservoir solution). Crystals selected for SAD data collection were flash-frozen in the crystallization buffer. X-ray diffraction data were collected at the Advanced Photon Source at Argonne National Laboratories.
X-ray diffraction and structure determination
The diffraction data were processed and scaled with the HKL2000 suite of programs (DENZO/SCALEPACK; Otwinowski and Minor 1997). Initial phases were obtained with SOLVE (Terwilliger and Berendzen 1999) using the single wavelength anomalous dispersion (SAD) phasing method with the selenium (Se) peak diffraction data. Twenty-eight selenium sites were identified using SOLVE. Subsequent electron density modification followed by initial model building were done using RESOLVE (Terwilliger 2000). Additional regions of the protein model were built with ARP/warp (Perrakis et al. 1999). Two hundred seventy-six residues of the possible 792 residues for the dimer were automatically placed using the combined model building approach. The rest of the model was built manually with O (Jones et al. 1991) and subsequently refined with CNS (Crystallography &NMR system; Brünger et al. 1998). A total of 14 rounds of model building with O and CNS refinement were conducted. Water molecules were initially picked using CNS and then manually verified in O using the following criteria: a peak of a least 2.5σ in an F0 − FC map, a peak of at least 1.0σ in a 2 F0 − FC map, and reasonable intermolecular interactions. Figures were produced with MolScript (Kraulis 1991).
Accession number
The atomic coordinates and structure factors for PH1915 (PDB ID 2AS0) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org).
Acknowledgments
This project was funded by the Ontario Research and Development Challenge Fund Protein Structure Initiative of the National Institutes of Health (GM62414 and GM62413), and W.S. is supported by the University of Toronto Fellowship. A.M.E. is the Banbury Chair of Medical Research at the University of Toronto.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051821805.
References
- Aravind, L. and Koonin, E.V. 1999. Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol. 48 291–302. [DOI] [PubMed] [Google Scholar]
- Armengaud, J., Urbonavicius, J., Fernandez, B., Chaussinand, G., Bujnicki, J.M., and Grosjean, H. 2004. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J. Biol. Chem. 279 37142–37152. [DOI] [PubMed] [Google Scholar]
- Bjork, G.R., Wikstrom, P.M., and Bystrom, A.S. 1989. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244 986–989. [DOI] [PubMed] [Google Scholar]
- Brule, H., Elliott, M., Redlak, M., Zehner, Z.E., and Holmes, W.M. 2004. Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry 43 9243–9255. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Bugl, H., Fauman, E.B., Staker, B.L., Zheng, F., Kushner, S.R., Saper, M.A., Bardwell, J.C., and Jakob, U. 2000. RNA methylation under heat shock control. Mol. Cell 6 349–360. [DOI] [PubMed] [Google Scholar]
- Bussiere, D.E., Muchmore, S.W., Dealwis, C.G., Schluckebier, G., Nienaber, V.L., Edalji, R.P., Walter, K.A., Ladror, U.S., Holzman, T.F., and Abad-Zapatero, C. 1998. Crystal structure of ErmC′, an rRNA methyltransferase which mediates antibiotic resistance in bacteria. Biochemistry 37 7103–7112. [DOI] [PubMed] [Google Scholar]
- Caldas, T., Binet, E., Bouloc, P., Costa, A., Desgres, J., and Richarme, G. 2000a. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275 16414–16419. [DOI] [PubMed] [Google Scholar]
- Caldas, T., Binet, E., Bouloc, P., and Richarme, G. 2000b. Translational defects of Escherichia coli mutants deficient in the Um(2552) 23S ribosomal RNA methyltransferase RrmJ/FTSJ. Biochem. Biophys. Res. Commun. 271 714–718. [DOI] [PubMed] [Google Scholar]
- Coltri, P.P., Guimaraes, B.G., Oliveira, C.C., and Zanchin, N.I. 2004. Expression, crystallization and preliminary X-ray analysis of the Pyrococcus abyssi protein homologue of Saccharomyces cerevisiae Nip7p. Acta Crystallogr. D Biol. Crystallogr. 60 1925–1928. [DOI] [PubMed] [Google Scholar]
- Cordell, S.C., Anderson, R.E., and Lowe, J. 2001. Crystal structure of the bacterial cell division inhibitor MinC. EMBO J. 20 2454–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davanloo, P., Sprinzl, M., Watanabe, K., Albani, M., and Kersten, H. 1979. Role of ribothymidine in the thermal stability of transfer RNA as monitored by proton magnetic resonance. Nucleic Acids Res. 6 1571–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic, S. and Stock, A.M. 1997. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure 5 545–558. [DOI] [PubMed] [Google Scholar]
- Fauman, E., Blumenthal, R., and Cheng, X. 1999. Structure and evolution of AdoMet-dependent methyltransferases, pp. 1–54. World Scientific, River Edge, NJ.
- Gupta, A., Kumar, P.H., Dineshkumar, T.K., Varshney, U., and Subramanya, H.S. 2001. Crystal structure of Rv2118c: An AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J. Mol. Biol. 312 381–391. [DOI] [PubMed] [Google Scholar]
- Heurgue-Hamard, V., Champ, S., Engstrom, A., Ehrenberg, M., and Buckingham, R.H. 2002. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J. 21 769–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang, C. and Ferre-D’Amare, A.R. 2001. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: Nucleotide flipping by an RNA-modifying enzyme. Cell 107 929–939. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1995. Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20 478–480. [DOI] [PubMed] [Google Scholar]
- Hopper, A.K. and Phizicky, E.M. 2003. tRNA transfers to the limelight. Genes & Dev. 17 162–180. [DOI] [PubMed] [Google Scholar]
- Huang, L., Hung, L., Odell, M., Yokota, H., Kim, R., and Kim, S.H. 2002. Structure-based experimental confirmation of biochemical function to a methyltransferase, MJ0882, from hyperthermophile Methanococcus jannaschii. J. Struct. Funct. Genomics 2 121–127. [DOI] [PubMed] [Google Scholar]
- Ishitani, R., Nureki, O., Fukai, S., Kijimoto, T., Nameki, N., Watanabe, M., Kondo, H., Sekine, M., Okada, N., Nishimura, S., et al. 2002. Crystal structure of archaeosine tRNA-guanine transglycosylase. J. Mol. Biol. 318 665–677. [DOI] [PubMed] [Google Scholar]
- Johansson, M.J. and Bystrom, A.S. 2002. Dual function of the tRNA (m(5)U54)methyltransferase in tRNA maturation. RNA 8 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47 110–119. [DOI] [PubMed] [Google Scholar]
- Kersten, H., Albani, M., Mannlein, E., Praisler, R., Wurmbach, P., and Nierhaus, K.H. 1981. On the role of ribosylthymine in prokaryotic tRNA function. Eur. J. Biochem. 114 451–456. [DOI] [PubMed] [Google Scholar]
- Kraulis, PJ. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24 946–950. [Google Scholar]
- Maravic, G. 2004. Macrolide resistance based on the Erm-mediated rRNA methylation. Curr. Drug Targets Infect. Disord. 4 193–202. [DOI] [PubMed] [Google Scholar]
- Martin, J.L. and McMillan, F.M. 2002. SAM (dependent) I AM: The S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12 783–793. [DOI] [PubMed] [Google Scholar]
- McCloskey, J.A. and Crain, P.F. 1998. The RNA modification database—1998. Nucleic Acids Res. 26 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D.J., Ouellette, N., Evdokimova, E., Savchenko, A., Edwards, A., and Anderson, W.F. 2003. Crystal complexes of a predicted S-adenosylmethionine-dependent methyltransferase reveal a typical AdoMet binding domain and a substrate recognition domain. Protein Sci. 12 1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel, J., Bujnicki, J.M., Tazi, J., and Bordonne, R. 2003. Sequence–structure–function relationships of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res. 31 4899–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahigashi, K., Kubo, N., Narita, S., Shimaoka, T., Goto, S., Oshima, T., Mori, H., Maeda, M., Wada, C., and Inokuchi, H. 2002. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl. Acad. Sci. 99 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto, H., Watanabe, K., Ikeuchi, Y., Suzuki, T., Endo, Y., and Hori, H. 2004. Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus. J. Biol. Chem. 279 49151–49159. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode (eds. C.W. Carter Jr. and R.M. Sweet), pp. 307–326. Academic Press, New York. [DOI] [PubMed]
- Perrakis, A., Morris, R., and Lamzin, V.S. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6 458–463. [DOI] [PubMed] [Google Scholar]
- Rozenski, J., Crain, P.F., and McCloskey, J.A. 1999. The RNA modification database: 1999 update. Nucleic Acids Res. 27 196–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schubert, H.L., Phillips, J.D., and Hill, C.P. 2003. Structures along the catalytic pathway of PrmC/HemK, an N5-glutamine AdoMet-dependent methyltransferase. Biochemistry 42 5592–5599. [DOI] [PubMed] [Google Scholar]
- Skinner, M.M., Puvathingal, J.M., Walter, R.L., and Friedman, A.M. 2000. Crystal structure of protein isoaspartyl methyltransferase: A catalyst for protein repair. Struct. Fold. Des. 8 1189–1201. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T.C. and Berendzen, J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich, T.C., Blaesse, M., and Huber, R. 2001. Crystal structure of ATP sulfurylase from Saccharomyces cerevisiae, a key enzyme in sulfate activation. EMBO J. 20 316–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney, U., Ramesh, V., Madabushi, A., Gaur, R., Subramanya, H.S., and RajBhandary, U.L. 2004. Mycobacterium tuberculosis Rv2118c codes for a single-component homotetrameric m1A58 tRNA methyltransferase. Nucleic Acids Res. 32 1018–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z., Shipman, L., Zhang, M., Anton, B.P., Roberts, R.J., and Cheng, X. 2004. Structural characterization and comparative phylogenetic analysis of Escherichia coli HemK, a protein (N5)-glutamine methyltransferase. J. Mol. Biol. 340 695–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L., Petros, A.M., Schnuchel, A., Zhong, P., Severin, J.M., Walter, K., Holzman, T.F., and Fesik, S.W. 1997. Solution structure of an rRNA methyltransferase (ErmAM) that confers macrolide-lincosamide-streptogramin antibiotic resistance. Nat. Struct. Biol. 4 483–489. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Zhou, L., and Cheng, X. 2000. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 19 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, R.G., Skarina, T., Katz, J.E., Beasley, S., Khachatryan, A., Vyas, S., Arrowsmith, C.H., Clarke, S., Edwards, A., Joachimiak, A., et al. 2001. Structure of Thermotoga maritima stationary phase survival protein SurE: A novel acid phosphatase. Structure (Camb.) 9 1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]