Abstract
Human Securin, also called PTTG1 (pituitary tumor transforming gene 1 product), is an estrogen-regulated proto-oncogene with multifunctional properties. We characterized human full-length Securin using a variety of biophysical techniques, such as nuclear magnetic resonance, circular dichroism, and size-exclusion chromatography. Under physiological conditions, Securin is devoid of tertiary and secondary structure except for a small amount of poly-(L-proline) type II helix and its hydrodynamic characteristics suggest it behaves as an extended polypeptide. These results suggest that Securin is unstructured in solution and so belongs to the family of natively unfolded proteins. In addition, to gain structural and quantitative insight, we investigated the binding of Securin to p53. Analytical ultracentrifugation and fluorescence anisotropy studies revealed no evidence of any direct interaction between unmodified recombinant Securin and p53 in vitro.
Keywords: Securin, p53, natively unfolded, poly(L-proline) helix type II, circular dichroism spectroscopy
PTTG (pituitary tumor transforming gene product) was first isolated in rat pituitary tumor cells (Pei and Melmed 1997). Subsequently, several groups cloned the human form (hpttg) and other eukaryote homologs from different healthy tissue and tumor samples (Zhang et al. 1999b; Wang and Melmed 2000). Studies also revealed the existence of a human pituitary tumor-transforming gene family consisting of a total of three different genes (Chen et al. 2000). Human PTTG1, also called Securin, is overexpressed in the leukocytes of patients with different haematopoietic neoplasms or myelo-dysplastic syndromes, testicular tumors, germ-cell tumors of the ovary, breast carcinomas (Puri et al. 2001), and in all pituitary tumors, specifically in hormone-secreting tumors, in which invasiveness correlates with high levels of expression (Zhang et al. 1999a).
Overexpression of human PTTG1 stimulates the expression and secretion of basic fibroblast growth factor (bFGF) (Ishikawa et al. 2001), enhances cell proliferation, induces cell transformation (Pei 2001), and promotes tumor formation in nude mice (Pei and Melmed 1997). Recently, human PTTG1, or Securin, has been identified as an inhibitor of sister-chromatid separation based on its biochemical similarities with the Pds1 protein of yeast (Zou et al. 1999). Human PTTG1, or Securin, binds and inactivates Separase, a protease that cleaves the Scc1 cohesin subunit responsible for sister chromatid cohesion at the metaphase-to-anaphase transition. Securin remains attached to Separase until activation of the Anaphase-promoting complex (APC) and undergoes subsequent proteolysis in a D box-dependent manner (Zou et al. 1999). Human PTTG1 is involved in several cellular processes, such as mitosis (Ramos-Morales et al. 2000), cell cycle progression (Zou et al. 1999), DNA repair (Romero et al. 2001a), and maintenance of chromosome stability (Jallepalli et al. 2001). Human PTTG1 overexpression causes apoptosis in both a p53-dependent and p53-independent manner (Yu et al. 2000), however, the relationship between human PTTG1 and p53 is still not completely understood. Overexpression of human PTTG1 activates the expression of p53 at both the transcriptional and translational level as a result of an indirect effect that involves the regulation of c-myc expression, which then interacts with the p53 gene promoter. Overexpression of human PTTG1 also stimulates the expression of the Bax protein, a proapoptotic protein induced by p53 (Hamid and Kakar 2004). Later, an interaction between human PTTG1 and p53 was identified, which abrogates p53 transcriptional activity by blocking its ability to bind DNA (Bernal et al. 2002) and induce apoptosis. Moreover, p53 also suppresses human PTTG1 expression by repressing its transcription (Zhou et al. 2003).
The field of biochemistry has been dominated by the idea that structure is a prerequisite to function. In the last decade, however, this structure–function paradigm has been re-evaluated based on the growing evidence that many proteins are unfolded in their functional state. These proteins are referred to as “intrinsically unstructured” (Wright and Dyson 1999) or “intrinsically disordered” (Dunker et al. 2002). The advantage of lacking a folded structure, as opposed to a rigid one, may reside in the plasticity conferred by flexibility: (1) allowing binding to multiple targets with high specificity and low affinity and (2) providing the ability to overcome steric restrictions and thus enabling larger surface interactions (Wright and Dyson 1999; Dunker et al. 2002; Tompa 2002; Uversky 2002a). The current list of proteins considered either to be random coil or to contain large disordered regions consists of more than 150 entries (http://divac.ist.temple.edu/disprot/database.php).
In this study, we characterized full-length human Securin or hPTTG1 in vitro using biophysical techniques. We found that under physiological conditions, Securin exhibited properties typical of a natively unfolded protein and behaved as a random coil with a small amount of poly(L-proline) type II helix. We used analytical ultracentrifugation and fluorescence anisotropy to further characterize, both structurally and quantitatively, the previously reported interaction between Securin and p53. We were unable to detect any interaction between these two proteins when using unmodified recombinant Securin and p53 in vitro.
Results
Hydrodynamic behavior of Securin
To investigate the degree of compactness and the oligomeric state of Securin, we used size-exclusion chromatography and analytical ultracentrifugation, respectively. Purified recombinant Securin eluted from the gel-filtration column as a single peak that slightly tails off, probably due to the presence of small amounts of truncated Securin. This tailing off in the elution profile was also observed for experiments done with less amount of protein and in the presence of 6 M urea (data not shown). The apparent molecular weight of Securin calculated from Equation 1 corresponds to 105,500 (Fig. 1A ▶), a value five times larger than the theoretical value of 22,225 Da expected from its amino acid sequence. Furthermore, the experimental molecular weight obtained by mass spectroscopy (22.19 kDa; data not shown) was in agreement with the theoretical value. Such a high Mr may result from the formation of soluble aggregates, or from an extended, rather than globular, conformation. We used analytical ultracentrifugation to further investigate these two possibilities. Analysis of the data from a sedimentation equilibrium centrifugation experiment showed that the protein was monomeric with a Mr of 18,600 ± 1000 (Fig. 1B ▶). The Stokes radius of the protein was calculated from the experimental size-exclusion molecular weight as described in Uversky (1993), using Equations 2 and 3. The value of 39.7 ± 0.04 Å obtained for Securin (Mr 105,500) compared more favorably with the theoretical Stokes radius predicted for a fully unfolded protein of 22.2 kDa (41.7 ± 0.3 Å) than that predicted for the native globular monomeric form (22.4 ± 0.01 Å). Together, these results imply that Securin has the dimensions of a denatured protein and that it has an extended conformation.
Figure 1.
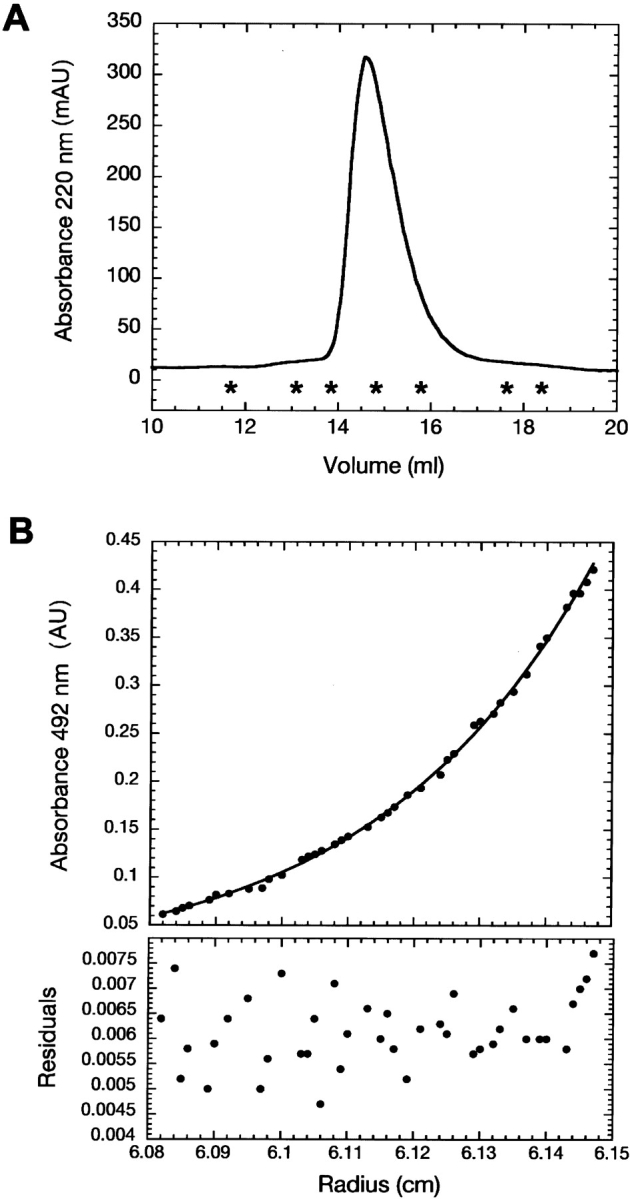
Hydrodynamic characterization of Securin. (A) Size exclusion chromatography elution profile of Securin monitored at 220 nm. Asterisks denote the positions of the molecular weight standards. (Left to right) Ferritin (440 kDa); catalase (232 kDa); aldolase (158 kDa); albumin (67 kDa); ovoalbumin (43 kDa); chymotrypsinogen (25 kDa); ribonuclease (13.7 kDa). (B) Equilibrium sedimentation experiments of fluorescein-labeled Securin at 10°C. Residuals correspond to the fitting of the data to a single exponential model to deduce the apparent molecular weight.
Secondary structure analysis of Securin
The far-UV CD spectrum of Securin (Fig. 2A ▶) lacked the typical signatures of secondary structure, exhibiting instead only a negative signal at 200 nm. The minimum at 200 nm was more pronounced than that for random coils (−5000 deg cm2 dmol−1), suggesting rather that the protein contains a small amount of poly(L-proline) type II helix (PPII). As the temperature was increased (Fig. 2A ▶), the CD spectra showed a red shift of ~2 nm, and the minimum became less negative. Also present was a well-defined isodichroic point at 212 nm, a feature described for temperature-dependent CD spectra of peptides that contain PPII structures, and attributed to the destruction of the PPII in favor of the unstructured state (Makarov et al. 1992). Dependence of the molar ellipticity as a function of temperature lacked any cooperative transition (Fig. 2B ▶). The observed linear shape of the spectrum with a slight negative slope is typical of random coil polypeptides. These results indicate that Securin does not have significant regular secondary structure, and that it lacks a compact globular structure.
Figure 2.
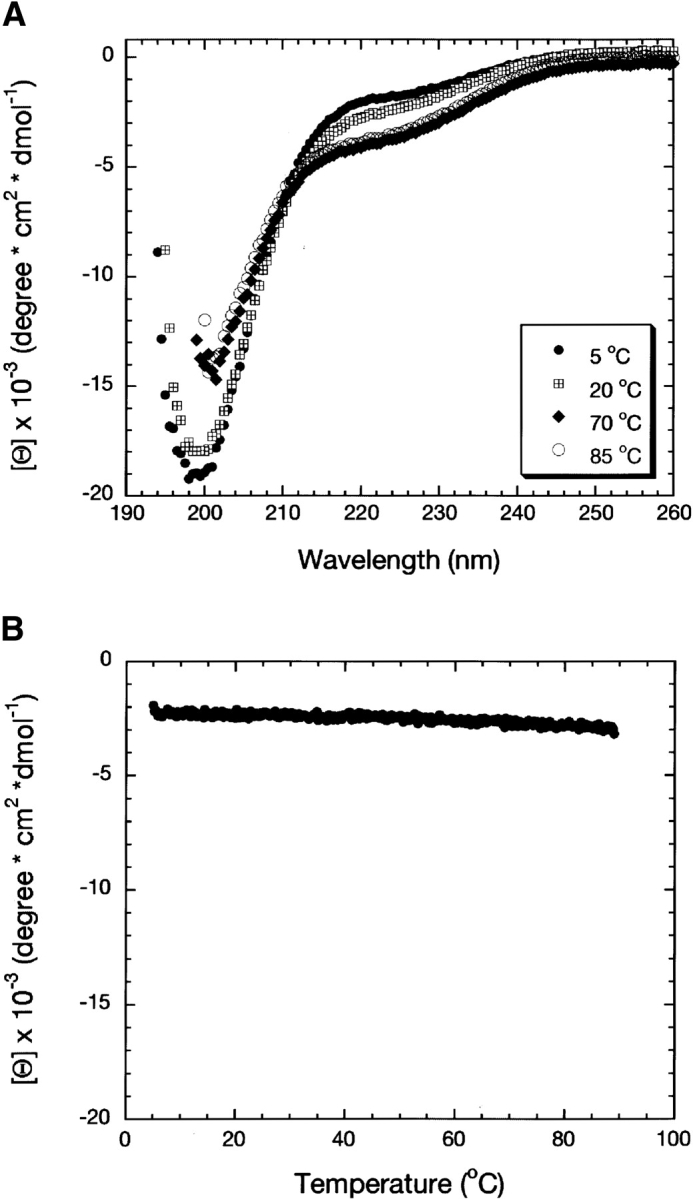
Secondary structure analysis of Securin evaluated by CD. (A) Far-UV CD spectra of Securin at different temperatures. (B) Temperature dependence of the molar ellipticity of Securin followed at 222 nm.
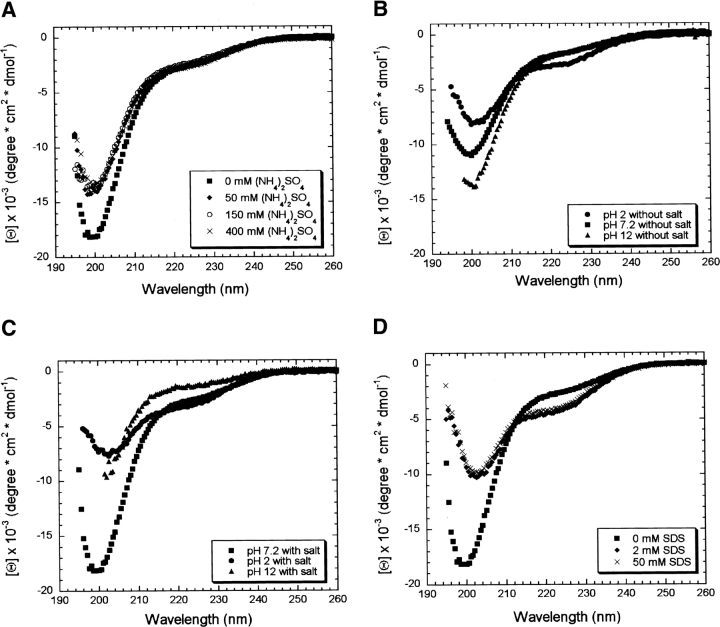
Changes in the environment can induce the formation of secondary structure in natively unfolded proteins (for review, see Uversky 2002b). Figure 3 ▶ shows the results of adding to Securin different chemical compounds known to induce secondary structure. An increase in the ionic strength by addition of (NH4)2SO4 had no significant effect on the secondary structure of Securin, although the small decrease in the intensity of the signal at 200 nm suggested a reduction in the amount of PPII (Fig. 3A ▶; Jones et al. 1994). Changes in pH in the presence or absence of salt (Fig. 3B,C ▶) had no effect on the structure of Securin. Even at extreme pHs, the overall shape of the spectra remained that of an unfolded protein. SDS (sodium dodecyl sulphate) had a more pronounced effect on Securin. The amplitude of the minimum at 200 nm decreased, reaching values close to that of a disordered polypeptide (Fig. 3D ▶). Also, the spectra showed an isodichroic point around 212 nm that has approximately the same value as that observed for its CD thermal dependence (Fig. 2A ▶), suggesting that SDS has shifted the equilibrium toward a fully unstructured state instead of promoting any secondary structure. Securin aggregated in the presence of trifluoroethanol (TFE) and trimethylamine N-oxide (TMAO), making it impossible to study their effects on its secondary structure.
Figure 3.
Effect of the solution environment on the secondary structure content of Securin evaluated by CD. (A) (NH4)2SO4; (B) pH in the absence of salt; (C) pH in the presence of salt; and (D) SDS.
Analysis of Securin using Nuclear Magnetic Resonance
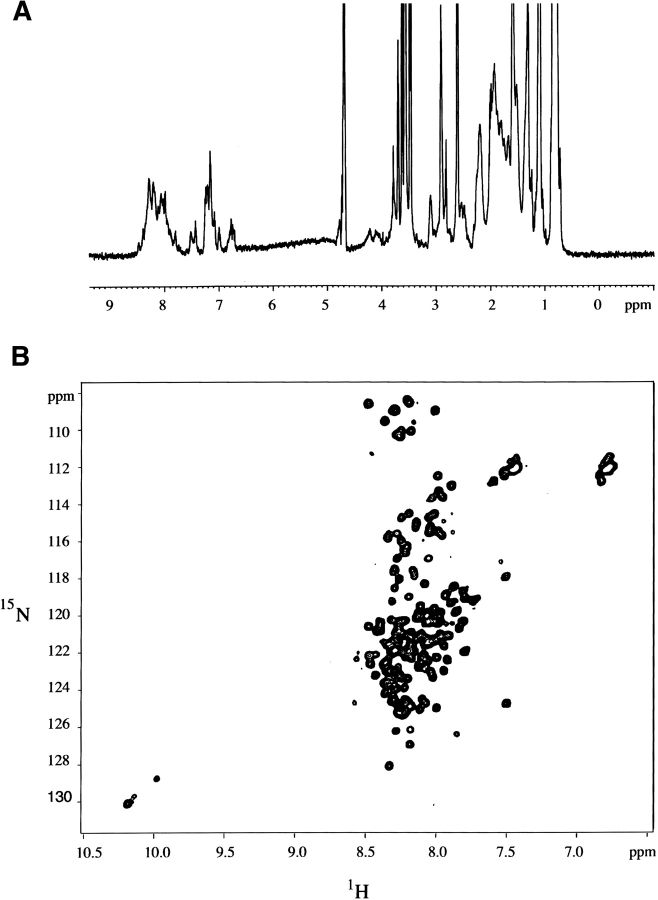
The 1H-NMR spectrum of Securin has the characteristics of a random coil (Fig. 4A ▶). The resonances are in general clustered, and not dispersed, as expected for a folded protein. Signals from methyl group protons are clustered at around 1 ppm instead of being shifted up-field to negative chemical shifts, as would be expected in the case of a folded protein. Methyl groups are clustered into three groups, those corresponding to Val, Leu, and Ile at around 0.8 ppm, those from Thr at 1.2 ppm, and Ala at 1.4 ppm. Finally, the amide group protons are also clustered at 8 ppm (Dwek 1973). The overall appearance of the proton spectrum has the hallmark of an unfolded polypeptide with discrete groups of signals at random coil values. In addition, the 1H-15N HSQC spectrum of Securin revealed limited dispersion of proton chemical shifts of only 1 ppm and a very narrow line width for all resonances as expected for highly flexible regions (Fig. 4B ▶). These observations indicate that Securin is an unfolded protein as previously described for urea denatured proteins (Kriwacki et al. 1996; Dyson and Wright 1998; Schwarzinger et al. 2000).
Figure 4.
Nuclear magnetic resonance characterization of Securin. (A) 1H NMR spectrum of Securin, and (B) 1H-15N HSQC spectrum of Securin.
Characterization of the interaction between Securin and p53
Studying the function of human Securin in tumorigenesis, Bernal and collaborators described the interaction between p53 and Securin using pull-down and coimmunoprecipitation assays. This interaction represses p53 transcriptional activity and reduces its ability to induce cell death in vivo (Bernal et al. 2002). They determined that the N terminus of Securin contains the fragment that interacts with residues 300–374 of p53, which comprises the tetramerization domain and a portion of the C terminus. We used fluorescence anisotropy and analytical ultracentrifugation to quantify and gain insight into this reported interaction; however, in all cases, we were not able to detect any binding. When using fluorescence anisotropy, addition of a final concentration of 160 μM Securin to fluorescein-labeled p53-TC (residues 293–393, tetramerization domain and C terminus) resulted in a linear increase of the fluorescence anisotropy signal of only 0.003 units, a value equal to that of the limit of detection of the instrument (0.003–0.005 units) (Fig. 5A ▶).
Figure 5.
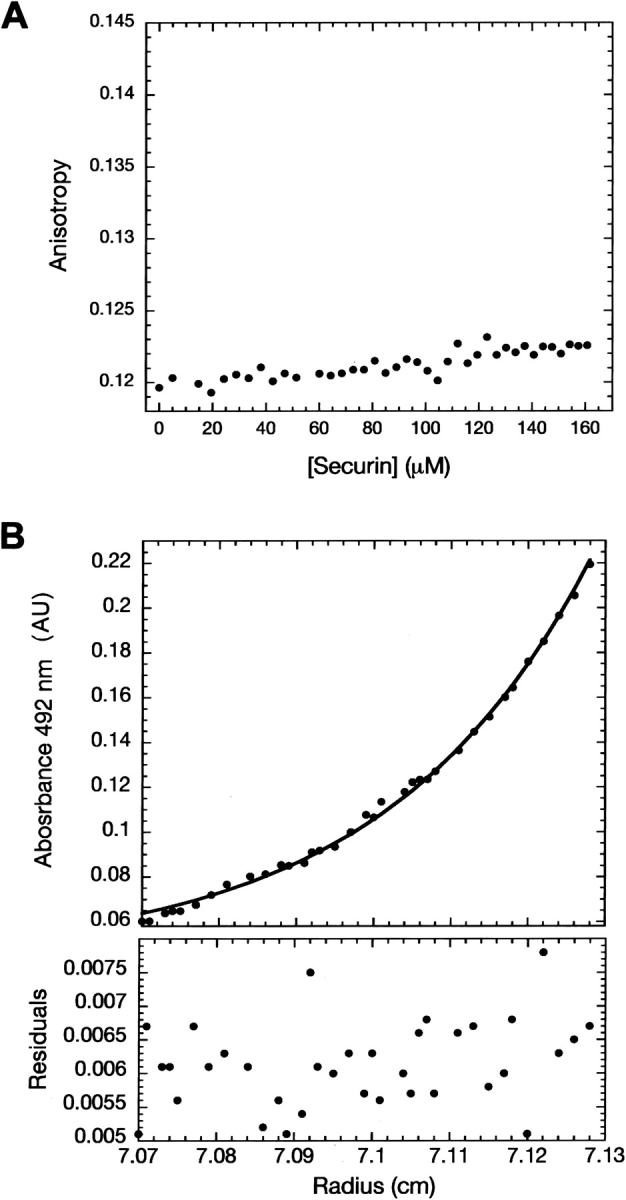
Characterization of the interaction between Securin and p53. (A) Fluorescence anisotropy experiments with fluorescein-labeled p53- TC and Securin. (B) Equilibrium sedimentation analysis with fluorescein- labeled Securin and p53 full-length quadruple mutant.
In addition, analytical ultracentrifugation experiments were set up to detect fluorescein-labeled Securin in the presence of p53 full-length quadruple mutant (Joerger et al. 2004) (p53 flQM). Data described a single species present with a Mr of 21,000 ± 1000 (Fig. 5B ▶), a value comparable to the theoretical molecular weight for Securin of 22,200 Da deduced from its amino acid sequence.
Discussion
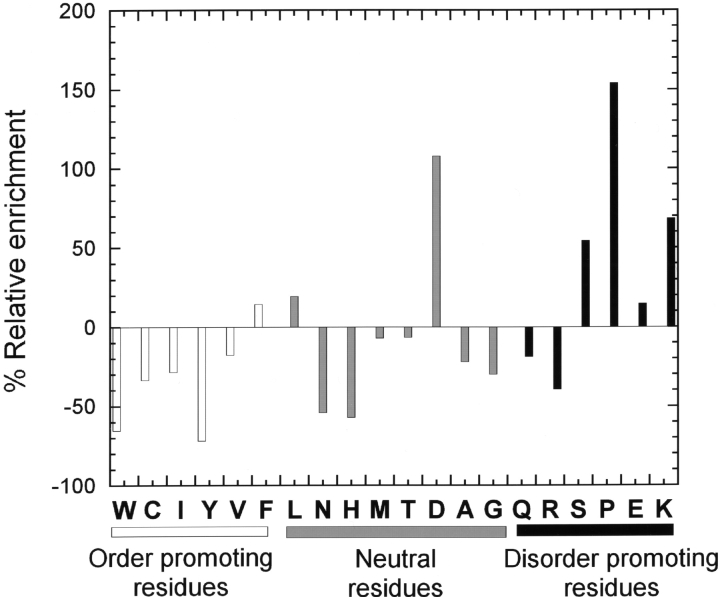
The hydrodynamic properties of Securin inferred from gel filtration and analytical ultracentrifugation show that it behaves as an extended monomeric protein. NMR and far-UV CD spectroscopy indicate that Securin is unstructured and lacks significant secondary structure content, except for a small amount of PPII. The experimental evidence presented here suggests that Securin is a natively unfolded protein. Furthermore, sequence analysis and secondary structure prediction confirm this idea. Parameters such as mean hydrophobicity <H>and mean net charge<R>, which are normally used to recognize lack of structure, were calculated as described in Uversky et al. (2000). The values for Securin are 0.45 and 0.05, respectively, which are comparable to those predicted for natively unfolded proteins of <H> 0.39 ± 0.05 and <R> 0.12 ± 0.09. The average amino acid composition of Securin indicates that it contains few order-promoting residues (W, Y, C, and I) and is enriched in disorder-promoting residues (P, S, and K) (Dunker et al. 2001). Figure 6 ▶ shows the comparison of the deviation in the amino acid composition of Securin with that of globular proteins contained in the SCOP database. Securin is depleted in most order-promoting residues and enriched as much as 2.5-, 1.7-, and 1.5-fold for the disorder-promoting residues Pro, Lys, and Ser, respectively. Furthermore, the PONDR predictor of intrinsically disordered regions (Romero et al. 1997, 2001b) predicts an overall percentage of disorder of 54%. Finally, the Psi-PRED server (http://bionf.cs.ucl.ac.uk/psiform.html) predicts 72% random coil. The N terminus of yeast Securin (Pds1) has also been shown to be natively unfolded. It is interesting that despite both yeast Pds1 and human Securin sequences being so dissimilar that it is impossible to align them, both proteins are unstructured and perform the same function (Cox et al. 2002).
Figure 6.
Amino acid deviation composition of human full-length Securin (hPTTG1) from the average amino acid composition values of globular proteins (excluding mutant, transmembrane, and coiled-coil proteins) in the SCOP data base. Columns show the relative enrichment of order-promoting (white bars) and disorder-promoting (black bars) residues.
CD spectroscopy results suggest that Securin may contain a small amount of poly(L-proline) type II helix (PPII). The spectrum, however, does not contain the typical maximum at 228 nm exhibited by PPII (Toumadje and Curtis Johnson Jr. 1995), which may be due to the fact that the amount of PPII structure is small compared with the mainly random coil conformation. The most favored residue present in PPII helices is proline, which is also the most abundant residue in Securin (12%), particularly in the C terminus of the protein. Some of the prolines present within Securin are followed by another proline, or by glycine or arginine residues, all of which have a strong propensity to form PPII structures (Stapley and Creamer 1999). The Securin N terminus contains 22/26 of the total number of basic residues in the protein, while the C terminus has 19/27 of the acidic residues. At neutral pH, both types of residues are charged, and salt can mask these charges, causing the protein to adopt a structure constrained only by the conformation of the backbone, which is the most important determinant in polyproline helices. The inability of organic solvents, pH, and electrolytes to induce any secondary structure in Securin suggests that under physiological conditions, its stable conformation is that of an extended polypeptide with stretches of polyproline helix type II.
The C terminus of Securin contains two diproline motifs PXXP, which have been shown to adopt PPII structure and are involved in SH3-ligand interactions (Yu et al. 1994). In addition, one of these PXXP motifs contains a Cdc2 phosphorylation site that may regulate hPTTG1 function, either by the phosphorylation and dephosphorylation of Ser165 (Ramos-Morales et al. 2000) or by folding upon binding, as previously observed for the binding of the KID domain of CREB to CBP (Radhakrishnan et al. 1997; Zor et al. 2002, 2004). Site-directed mutagenesis of the PXXP motifs abrogates the transforming and tumor-inducing activity of the protein (Zhang et al. 1999b).
Many cell-signaling and cancer-associated proteins have been shown to be rich in disordered regions (Iakoucheva et al. 2002). In agreement with this observation, we show here that Securin, a protein implicated in cell cycle regulation and cell transformation, is also unfolded. In addition, Securin has a destruction box (D-box) and a KEN box at the N terminus, which targets the protein for degradation by the APC. Unfolded regions are often found among domains responsible for protein degradation (Hochstrasser 1996).
Securin is highly expressed in tumors (Zhang et al. 1999a,b; Wang and Melmed 2000), but the mechanism by which it promotes tumorigenesis is unclear. A possible mechanism may result from aneuploidy caused by defective sister-chromatid separation. Bernal et al. (2002) described the interaction between Securin and p53, which in turn, blocks the specific binding of p53 to DNA, providing another potential mechanism for tumor formation. The interaction maps to the tetramerization domain and C terminus of p53 and the N terminus of Securin. Using various biophysical techniques including analytical ultracentrifugation, which is capable of detecting weak interactions (Kd ~10−3 M), and purified recombinant proteins, we could not detect any interaction between Securin and p53. The difference between our findings and those reported by Bernal may arise from the use of eukaryotic cell-based experiments rather than entirely in vitro experiments using prokaryotic recombinant proteins. Perhaps the reported binding may be mediated either by one or more proteins acting as a bridge between p53 and Securin, or by post-translational modifications in either or both proteins. In particular, there are extensive studies describing the regulation of p53 by protein–protein interactions and by covalent modification. It is noteworthy that the C terminus of p53 is the site of several post-translational modifications including phosphorylation, acetylation, SUMOylation, ubiquitination, neddylation, and glycosylation. These covalent modifications regulate p53 sequence-specific DNA binding, transactivation, tetramerization, and stability (May and May 1999). Beside the proteins involved in these post-translational modifications, the C terminus interacts with many other proteins. In particular, it associates with the DNA helicases XPB and XPD, which are subunits of TFIIH, and with proteins involved in transcription and DNA repair such as TBP, Rad51, and CSB, and with the calcium-binding protein S100B (Jayaraman and Prives 1999). The binding of S100B to p53 involves not only the C terminus, but also the tetramerization domain (Delphin et al. 1999). In contrast, the N terminus of Securin has only been shown to interact with the regulatory subunit of the DNA-dependent protein kinase (DNA-PK), resulting in the phosphorylation of Securin. The residue(s) involved in this modification have not yet been identified, but there are at least six potential motifs for DNA-PK phosphorylation (Romero et al. 2001a). The Securin N terminus also contains seven other predicted sites for Protein kinase C phosphorylation and one site for cyclic-AMP and cyclic-GMP-dependent protein kinase phosphorylation. Any of the aforementioned protein– protein interactions or post-translational modifications, or others not described to date, may mediate the binding between Securin and p53.
Materials and methods
Plasmids
The wild-type human pttg1 (encoding the pituitary tumor-transforming 1, or Securin, protein) was amplified by PCR from a human thymus cDNA library (Clontech) and cloned into the pGEMT vector (Promega). The fragment was reamplified by PCR and digested with BamHI and EcoRI. The amplified fragment was subsequently purified and ligated in frame into the plasmid pRSETHisLipoTEV (Sánchez-Puig et al. 2005) at the BamHI and EcoRI sites immediately down-stream of the TEV protease site.
Protein expression and purification
The plasmid pRSETHisLipoTEV-Securin was transformed into Escherichia coli C41 (Miroux and Walker 1996) for protein overexpression. Cultures were grown in 2 × TY medium containing 100 μg/mL ampicillin to an optical density 600 nm of ~0.8, induced with 1 mM isopropyl-1-thio-β-D-galactoside (IPTG) and further incubated for 4 h at 37°C. The protein was purified on a fast flow Ni column (QIAGEN) using a FPLC system (Pharmacia). The eluted protein was digested with His-TEV (Lucast et al. 2001) protease and subsequently reapplied onto the Ni column. The sample was further purified by anion exchange chromatography on a 20HQ POROS column using a Vision Workstation BioCAD (Applied Biosystems), to separate the fragments of proteolysis from the full-length protein, and finally purified by gel filtration on a HiLoad 26/60 Superdex 75 column (Pharmacia). 15N isotopically labeled Securin was expressed and purified in the same way as above, except that the cells were grown in minimal M9 medium supplemented with 15NH4Cl. Fluorescein-labeled Securin was produced using the Fluorescein-EX Protein labeling kit from Molecular Probes following the manufacturer’s instructions. Fluorescein-labeled p53-TC protein (residues 293–393) (Fernández-Fernández et al. 2005) was a kind gift from Dr. Rosario Fernández-Fernández (Centre for Protein Engineering, Cambridge, UK) and p53 full-length quadruple mutant protein (p53 flQM) was a kind gift from Caroline Blair (Centre for Protein Engineering, Cambridge, UK). p53 flQM is a thermostable mutant of the full-length p53 protein (Joerger et al. 2004).
Size exclusion chromatography
Purified Securin (100 μM) was injected onto an analytical Superdex 200 HR10/30 column (Pharmacia) in an ÅKTA design XT Explorer 900 Kit (Pharmacia) instrument equipped with a Monitor UV-900 detector and the Unicorn 3.10 software package. The buffer used consisted of 25 mM phosphate buffer (NaPi) (pH 7.2), 150 mM KCl, and 10 mM 2-mercaptoethanol. Molecular weight standards (Pharmacia) were run under the same conditions and their elution volumes were used to create a calibration curve (Equation 1), from which the apparent molecular weight (Mr) of Securin was calculated. The theoretical Stokes radius (Rs) of a native (RsN) and fully unfolded (RsUrea) protein were determined as described in Uversky (1993) using Equations 2 and 3, respectively.
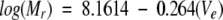 |
(1) |
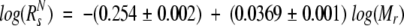 |
(2) |
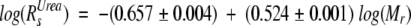 |
(3) |
Far UV circular dichroism
Temperature dependence of the ellipticity was followed with a JASCO J-720 spectropolarimeter equipped with a JASCO PTC- 348WI temperature controller. CD spectra were recorded using a 1-mm pathlength cuvette and protein concentration of 10 μM. Thermal denaturation was followed at 222 nm, with an increase of 1° per minute, a time response of 10 sec and a bandwidth of 1 nm. Circular dichroism wavelength scan measurements were followed with an AVIV 2025F stopped flow circular dichroism spectrometer equipped with a Pelletier temperature controller. CD spectra were recorded using a 1-mm pathlength cuvette at the same protein concentrations as for thermal denaturation. Scan wavelength was followed from 260 to 190 nm, with an increase of 0.5 nm per step, an averaging time of 5 sec, and a bandwidth of 1 nm. All samples were dialyzed against a buffer containing 25 mM NaPi (pH 7.2), 150 mM KCl, and 1 mM DTE.
Analytical ultracentrifugation
Experiments were performed at 10°C using a Beckman Optima XLI analytical ultracentrifuge with an An60Ti rotor. Data were collected at 492 nm in order to selectively detect the fluorescein labeled Securin. To assess the oligomeric state of Securin, we used a 5-μM fluorescein-labeled sample of Securin. When testing the interaction of fluorescein-labeled Securin with p53 flQM, the protein concentrations used were 5 μM for the former and 100 μM for the latter. The buffer used in all cases consisted of 25 mM NaPi (pH 7.2), 150 mM KCl, and 5 mM DTT. Samples were loaded in triplicate into 6-sector 12-mm pathlength cells. Scans were collected at 6-h intervals until equilibrium was reached, as judged by the fact that there were no further changes in subsequent scans. Data were analyzed using the UltraSpin software (www.mrc-cpe.cam.ac.uk/ultraspin) and the Kaleida-graph programme version 3.0.4 (Abelbeck Software) for graph plotting. Data were fitted to a single exponential model indicating equilibrium of single species of unspecified mass.
Two-dimensional Nuclear Magnetic Resonance
A sample containing purified recombinant 150 μM 15N Securin, in 25 mM NaPi (pH 7.2), 150 mM KCl, 10 mM 2-mercaptoethanol was used to acquire a 15N-HSQC spectrum on a DRX600 Bruker spectrometer at 20°C. Standard Bruker pulse sequences were used and the data was analyzed with UXNMR software.
Fluorescence anisotropy
A sample of 800 μM Securin (initial concentration) was titrated into 0.1 μM fluorescein-labeled p53-TC in a buffer consisting of 25 mM NaPi (pH 7.2), 150 mM NaCl, and 1 mM DTT at 10°C. Measurements were made in a Perkin Elmer LS55 luminescence spectrometer equipped with a Hamilton microlab dispenser controlled by laboratory software at an excitation wavelength of 480 nm and emission wavelength of 525 nm. For each data point, the mixture was incubated for 1 min with 30 sec of stirring before measuring the fluorescence.
Acknowledgments
We thank Caroline Blair for purified p53 flQM protein, Dr. Stefan M.V. Freund for the NMR experiments, Dr. Antonina Andreeva for statistics on the amino acid composition of proteins deposited on the SCOP database, and Dr. Rosario Fernández-Fernández for the fluorescein-labeled p53-TC sample and for valuable discussions and helpful comments. N.S.-P. was supported by a Gates Cambridge Trust scholarship.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051368005.
References
- Bernal, J., Luna, R., Espina, A., Lázaro, I., Ramos-Morales, F., Romero, F., Arias, C., Silva, A., Tortolero, M., and Pintor-Toro, J. 2002. Human securin interacts with p53 and modulates p53-mediated transcriptional activity and apoptosis. Nat. Genet. 32 306–311. [DOI] [PubMed] [Google Scholar]
- Chen, L., Puri, R., Lefkowitz, E., and Kakar, S. 2000. Identification of the human pituitary tumor transforming gene (hPTTG) family: Molecular structure, expression and chromosomal localization. Gene 248 41–50. [DOI] [PubMed] [Google Scholar]
- Cox, C.J., Dutta, K., Petri, E.T., Hwang, W.C., Lin, Y., Pascal, S.M., and Basavappa, R. 2002. The regions of securin and cyclin B proteins recognized by the ubiquitination machinery are natively unfolded. FEBS Lett. 527 303–308. [DOI] [PubMed] [Google Scholar]
- Delphin, C., Ronjat, M., Deloulme, J.C., Garin, G., Debussche, L., Higashimoto, Y., Sakaguchi, K., and Baudier, J. 1999. Calcium-dependent interaction of S100B with the C-terminal domain of the tumor suppressor p53. J. Biol. Chem. 274 10539–10544. [DOI] [PubMed] [Google Scholar]
- Dunker, A.K., Lawson, J.D., Brown, C.J., Williams, R.M., Romero, P., Oh, J.S., Oldfield, C.J., Campen, A.M., Ratliff, C.M., Hipps, K.W., et al. 2001. Intrinsically disordered protein. J. Mol. Graph. Model 19 26–59. [DOI] [PubMed] [Google Scholar]
- Dunker, A.K., Brown, C.J., Lawson, J.D., Iakoucheva, L.M., and Obradovic, Z. 2002. Intrinsic disorder and protein function. Biochemistry 41 6573–6582. [DOI] [PubMed] [Google Scholar]
- Dwek, R.A. 1973. Nuclear Magnetic Resonance (N.M.R) in Biochemistry. Applications to Enzyme Systems. Oxford University Press, Belfast, Northern Ireland.
- Dyson, H.J. and Wright, P.E. 1998. Equilibrium NMR studies of unfolded and partially folded proteins. Nat. Struct. Biol. 5 499–503. [DOI] [PubMed] [Google Scholar]
- Fernández-Fernández, M., Veprintsev, D.B., and Fersht, A.R. 2005. Proteins of the S100 family regulate the oligomerization of p53 tumor suppressor. Proc. Natl. Acad. Sci. 102 4735–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid, T. and Kakar, S.S. 2004. PTTG/securin activates expression of p53 and modulates its function. Mol. Cancer 3 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30 405–439. [DOI] [PubMed] [Google Scholar]
- Iakoucheva, L.M., Brown, C.J., Lawson, J.D., Obradovic, Z., and Dunker, A.K. 2002. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 323 573–584. [DOI] [PubMed] [Google Scholar]
- Ishikawa, H., Heaney, A.P., Yu, R., Horwitz, G.A., and Melmed, S. 2001. Human pituitary tumor-transforming gene induces angiogenesis. J. Clin. Endocrinol. Metab. 86 867–874. [DOI] [PubMed] [Google Scholar]
- Jallepalli, P., Waizenegger, I., Bunz, F., Langer, S., Speicher, K., Vogelstein, B., and Lengauer, C. 2001. Securin is required for chromosomal stability in human cells. Cell 105 445–457. [DOI] [PubMed] [Google Scholar]
- Jayaraman, L. and Prives, C. 1999. Covalent and noncovalent modifiers of the p53 protein. Cell. Mol. Life Sci. 55 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger, A.C., Allen, M.D., and Fersht, A.R. 2004. Crystal structure of a superstable mutant of human p53 core domain. Insights into the mechanism of rescuing oncogenic mutations. J. Biol. Chem. 279 1291–1296. [DOI] [PubMed] [Google Scholar]
- Jones, C., Mulloy, B., and Thomas, A.H. 1994. Methods in Molecular Biology. Humana Press, Totowa, NJ.
- Kriwacki, R.W., Hengst, L., Tennant, L., Reed, S.I., and Wright, P.E. 1996. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: Conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. 93 11504–11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucast, L.J., Batey, R.T., and Doudna, J.A. 2001. Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques 30 544–546, 548, 550 passim. [DOI] [PubMed] [Google Scholar]
- Makarov, A.A., Lobachov, V.M., Adzhubei, I.A., and Esipova, N.G. 1992. Natural polypeptides in left-handed helical conformation. A circular dichroism study of the linker histones’ C-terminal fragments and β-endorphin. FEBS Lett. 306 63–65. [DOI] [PubMed] [Google Scholar]
- May, P. and May, E. 1999. Twenty years of p53 research: Structural and functional aspects of the p53 protein. Oncogene 18 7621–7636. [DOI] [PubMed] [Google Scholar]
- Miroux, B. and Walker, J.E. 1996. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260 289–298. [DOI] [PubMed] [Google Scholar]
- Pei, L. 2001. Identification of c-myc as a down-stream target for pituitary tumor-transforming gene. J. Biol. Chem. 276 8484–8491. [DOI] [PubMed] [Google Scholar]
- Pei, L. and Melmed, S. 1997. Isolation and characterization of a pituitary tumor-transforming gene (PTTG). Mol. Endocrinol. 11 433–441. [DOI] [PubMed] [Google Scholar]
- Puri, R., Tousson, A., Chen, L., and Kakar, S. 2001. Molecular cloning of pituitary transforming gene 1 from ovarian tumors and its expression in tumors. Cancer Lett. 163 131–139. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan, I., Perez-Alvarado, G.C., Parker, D., Dyson, H.J., Montminy, M.R., and Wright, P.E. 1997. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: A model for activator:coactivator interactions. Cell 91 741–752. [DOI] [PubMed] [Google Scholar]
- Ramos-Morales, F., A., D., Romero, F., Luna, R., Multon, M., Pintor-Toro, J., and Tortolero, M. 2000. Cell cycle regulated expression and phosphorylation of httpg proto-oncogene product. Oncogene 19 403–409. [DOI] [PubMed] [Google Scholar]
- Romero, P., Obradovic, Z., and Dunker, A.K. 1997. Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inform. Ser. Workshop Genome Inform. 8 110–124. [PubMed] [Google Scholar]
- Romero, F., Multon, M.C., Ramos-Morales, F., Domínguez, A., Bernal, J.A., Pintor-Toro, J.A., and Tortolero, M. 2001a. Human securin, hPTTG, is associated with Ku heterodimer, the regulatory subunit of the DNA-dependent protein kinase. Nucleic Acids Res. 29 1300–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero, P., Obradovic, Z., Li, X., Garner, E.C., Brown, C.J., and Dunker, A.K. 2001b. Sequence complexity of disordered protein. Proteins 42 38–48. [DOI] [PubMed] [Google Scholar]
- Sánchez-Puig, N., Veprintsev, D.B., and Fersht, A.R. 2005. Binding of natively unfolded HIF-1α ODD Domain to p53. Mol. Cell 17 11–21. [DOI] [PubMed] [Google Scholar]
- Schwarzinger, S., Kroon, G.J., Foss, T.R., Wright, P.E., and Dyson, H.J. 2000. Random coil chemical shifts in acidic 8 M urea: Implementation of random coil shift data in NMRView. J. Biomol. NMR 18 43–48. [DOI] [PubMed] [Google Scholar]
- Stapley, B.J. and Creamer, T.P. 1999. A survey of left-handed polyproline II helices. Protein Sci. 8 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa, P. 2002. Intrinsically unstructured proteins. Trends Biochem. Sci. 27 527–533. [DOI] [PubMed] [Google Scholar]
- Toumadje, A. and Curtis Johnson Jr., W. 1995. System in has the characteristics of a poly(L-proline) II type helix. J. Am. Chem. Soc. 117 7023–7024. [Google Scholar]
- Uversky, V. 1993. Use of fast size-exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry 32 13288–13298. [DOI] [PubMed] [Google Scholar]
- ———. 2002a. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 11 739–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2002b. What does it mean to be natively unfolded? Eur. J. Biochem. 269 2–12. [DOI] [PubMed] [Google Scholar]
- Uversky, V.N., Gillespie, J.R., and Fink, A.L. 2000. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41 415–427. [DOI] [PubMed] [Google Scholar]
- Wang, Z and Melmed, S. 2000. Characterization of the murine pituitary tumor transforming gene (PTTG) and its promoter. Endocrinology 141 763–771. [DOI] [PubMed] [Google Scholar]
- Wright, P.E. and Dyson, H.J. 1999. Intrinsically unstructured proteins: Reassessing the protein structure–function paradigm. J. Mol. Biol. 293 321–331. [DOI] [PubMed] [Google Scholar]
- Yu, H., Chen, J.K., Feng, S., Dalgarno, D.C., Brauer, A.W., and Schreiber, S.L. 1994. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell 76 933–945. [DOI] [PubMed] [Google Scholar]
- Yu, R., Heaney, A.P., Lu, W., Chen, J., and Melmed, S. 2000. Pituitary tumor transforming gene causes aneuploidy and p53-dependent and p53-independent apoptosis. J. Biol. Chem. 275 36502–36505. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Horwitz, G.A., Heaney, A.P., Nakashima, M., Prezant, T.R., Bronstein, M.D., and Melmed, S. 1999a. Pituitary tumor transforming gene (PTTG) expression in pituitary adenomas. J. Clin. Endocrinol. Metab. 84 761–767. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Horwitz, G.A., Prezant, T.R., Valentini, A., Nakashima, M., Bronstein, M.D., and Melmed, S. 1999b. Structure, expression, and function of human pituitary tumor-transforming gene (PTTG). Mol. Endocrinol. 13 156–166. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Mehta, K.R., Choi, A.P., Scolavino, S., and Zhang, X. 2003. DNA damage-induced inhibition of securin expression is mediated by p53. J. Biol. Chem. 278 462–470. [DOI] [PubMed] [Google Scholar]
- Zor, T., Mayr, B.M., Dyson, H.J., Montminy, M.R., and Wright, P.E. 2002. Roles of phosphorylation and helix propensity in the binding of the KIX domain of CREB-binding protein by constitutive (c-Myb) and inducible (CREB) activators. J. Biol. Chem. 277 42241–42248. [DOI] [PubMed] [Google Scholar]
- Zor, T., De Guzman, R.N., Dyson, H.J., and Wright, P.E. 2004. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J. Mol. Biol. 337 521–534. [DOI] [PubMed] [Google Scholar]
- Zou, H., McGarry, T., Bernal, T., and Kirschnner, M. 1999. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285 418–422. [DOI] [PubMed] [Google Scholar]