Abstract
Genome sequencing showed that two proteins in Mycobacterium tuberculosis H37Rv contain the metal binding motif (D/E)X2HX~100(D/E)X2H characteristic of the soluble diiron enzyme superfamily. These putative acyl-ACP desaturase genes desA1 and desA2 were cloned from genomic DNA and expressed in Escherichia coli BL21(DE3). DesA1 was found to be insoluble, but in contrast, DesA2 was a soluble protein amenable to biophysical characterization. Here, we report the 2.0 Å resolution X-ray structure of DesA2 determined by multiple anomalous dispersion (MAD) phasing from a Se-met derivative and refinement against diffraction data obtained on the native protein. The X-ray structure shows that DesA2 is a homodimeric protein with a four-helix bundle core flanked by five additional helices that overlay with 192 structurally equivalent amino acids in the structure of stearoyl-ACP Δ9 desaturase from castor plant with an rms difference 1.42 Å. In the DesA2 crystals, one metal (likely Mn from the crystallization buffer) was bound in high occupancy at the B-site of the conserved metal binding motif, while the A-site was not occupied by a metal ion. Instead, the amino group of Lys-76 occupied this position. The relationships between DesA2 and known diiron enzymes are discussed.
Keywords: desaturase, Mycobacterium tuberculosis, X-ray crystallography
The acyl-ACP desaturases are one of the major functional classes of soluble diiron enzymes (Fox et al. 2004). Genome sequencing has suggested that Mycobacterium tuberculosis and related organisms also contain acyl-ACP desaturase-like proteins (Cole et al. 1998). This finding has intriguing biochemical implications, as all mycobacteria have an outer cell wall containing an extensively modified long-chain fatty acid complex called mycolic acid (Barry et al. 1998). This waxy coating provides a protective barrier against macrophage attack, desiccation, water-soluble antibiotics, and other antimicrobial agents. A widely attributed hypothesis is that the biosynthesis of mycolic acid requires desaturases to form precursors with position-specific double bonds (Yuan et al. 1995; Dubnau et al. 2000). Subsequent modifications of the double bonds allow position-specific cyclopropanation, epoxidation, hydroxylation, methoxylation, and ketonization. Therefore, desaturation is likely an essential step in the biosynthesis of structurally and chemically diverse mycolic acids.
Here we report studies of the DesA1 and DesA2 proteins from M. tuberculosis H37Rv produced in Escherichia coli, annotated as putative acyl-ACP desaturases. DesA1 was not expressed as a soluble protein in E. coli, but DesA2 was obtained in sufficient quantities to initiate further studies. The X-ray structure of DesA2 supports assignment to the acyl-ACP desaturase structural family, but also reveals several differences with known diiron enzymes. Notably, the structure reveals that the Nζ group of a Lys residue occupies the position expected to contain a metal ion of the diiron center. Furthermore, two extended sequences of disordered residues apparently occupy an interface between subunits of the otherwise well ordered 2.0 Å resolution structure. These differences introduce new questions about structural variations within the Pfam family assigned to the acyl-ACP desaturases.
Results
Phylogenetic analysis
Figure 1 ▶ compares the amino acid sequences of DesA2 (annotated desA2, rv1094) and DesA1 (desA1, rv0824c) from M. tuberculosis H37Rv with the stearoyl-acyl carrier protein Δ9 desaturase (Δ9D) from castor (Ricinus communis; Shanklin and Somerville 1991) and the model plant Arabidopsis thaliana (The Arabidopsis Genome Initiative 2000). The DesA2 protein is 275 residues long, while DesA1 and the two plant desaturases are ~350 residues long. The gray shading shows residues from the metal binding motif (D/E)X2HX~100(D/E)X2H found in all representatives of the soluble diiron superfamily (Fox et al. 1994). The arrow in Figure 1 ▶ shows a conserved Trp residue implicated in electron transfer in other diiron enzymes, while boxes indicate other conserved residues whose positions and identities will be addressed after presentation of the DesA2 structure.
Figure 1.

Sequence alignment of acyl-ACP desaturases. Putative desaturases from M. tuberculosis H37Rv are aligned with the stearoyl-ACP Δ9 desaturases from castor (Ricinus communis) and Arabidopsis thaliana. The alignment was produced using CLUSTAL W (Thompson et al. 1994) as implemented in LaserGene (version 5.52, DNASTAR). Conserved residues are identified by boxes, residues lining the proposed substrate binding tunnel of Δ9D are identified by•, diiron cluster ligands are identified by gray shading, residues providing hydrogen bonds to the Nɛ2 positions of the His ligands are identified by ♦, and Trp residues implicated in electron transfer are identified by the arrow.
Cloning and expression
The DesA1 and DesA2 proteins were expressed in E. coli BL21(DE3) with the natural N-terminal Met residue as the first amino acid under control of the T7 RNA polymerase promoter. Small-scale expression testing showed that both DesA1 and DesA2 represented up to ~30% of the total protein based on visual inspection of denaturing electrophoresis gels. However, analysis of the small-scale expression also showed that only DesA2 was expressed as a soluble protein under the conditions investigated. Consequently, the remainder of this work is focused on DesA2. The production-level expression of DesA2 and selenomethionine-labeling were done in shaken flask culture, and the labeling experiment was done using a metabolic inhibition approach (Doublie 1997). Both unlabeled and labeled expression experiments gave similar yields of cells.
Purification
The purification procedure used for DesA2 consisted of ion exchange and gel filtration chromatographies, and gave ~13 mg of purified protein per g of wet cells. Upon the basis of calibrated gel filtration measurements, the elution time observed for DesA2 was consistent with the protein being a dimer in solution. MALDI-TOF mass spectral analysis revealed that the unlabeled, purified protein had molecular masses of 31,138 m/z and 31,345 m/z, which closely matched the values calculated from the gene sequence for protein lacking and retaining the N-terminal methionine, respectively. Furthermore, ESI-MS analysis of the selenomethionine-labeled DesA2 indicated that >95% substitution at the three internal methionine sites was achieved. The purified DesA2 exhibited no significant optical chromophores in the visible range, such as might be expected from an oxo-bridged diiron center near 340 nm (Fox et al. 1994). Furthermore, colorimetric metal analysis revealed substoichiometric quantities of Fe, ~0.2 mol per mol of polypeptide, compared to the anticipated ~2 mol per mol of polypeptide expected for a diiron enzyme such as Δ9D (Fox et al. 1993).
Crystallization and structure statistics
Crystals of DesA2 grew from light precipitate as elongated hexagonal bipyramids which attained a size of 0.7 × 0.2 × 0.2 mm within 24 h. They belong to space group P6522, with unit-cell parameters a=b=65.8 Å, c=292.0 Å. Table 1 shows the data collection and processing statistics obtained for DesA2, while Table 2 shows the refinement statistics. The final R-factor and Rfree were 0.206 and 0.236 for the 24,420 unique reflections observed between 56.8 and 2.0 Å (96.8% complete). The bond lengths and bond angles of the refined atoms had rms deviation from the ideal values of 0.022 Å and 1.63°, respectively.
Table 1.
Data collection statistics
| Peak | Edge | Remote | Native | |
| Wavelength (Å)a | 1.2661 | 1.2660 | 1.2861 | 1.2861 |
| Total reflections | 165,831 | 162,522 | 172,596 | 251,492 |
| Unique reflections | 17,440 | 17,261 | 18,143 | 25,801 |
| Resolution (Å) | 2.35 | 2.35 | 2.32 | 2.00 |
| Rmerge (%)b,c | 0.090 (0.249) | 0.108 (0.323) | 0.087 (0.287) | 0.088 (0.340) |
| Completeness (%) | 98.4 (91.8) | 98.2 (86.2) | 99.9 (98.4) | 96.8 (75.2) |
| Average I/σ | 23.8 (3.1) | 23.4 (3.3) | 26.4 (3.3) | 33.8 (2.6) |
aAPS, Advanced Photon Source at Argonne National Laboratory (Argonne, IL).
bRmerge=(|Ihkl − I|/(Ihkl)), where the average intensity I is taken over all symmetry equivalent measurements, and Ihkl is the measured intensity for a given reflection.
cStatistics for the highest-resolution bin.
Table 2.
Refinement statistics
| Resolution | 56.8 3 2.0 Å |
| Space group | P6522 |
| Unit cell dimensions | a = b = 65.8 Å , c = 292.0 Å |
| Reflections: total (unique) | 24,420 (1,305) |
| Protein atomsa | 1917 |
| Solvent atomsb | 109 |
| Rwork (%)c | 20.6 |
| Rfree (%)c | 23.6 |
| Ramachandran analysis (%) | |
| Most favored | 97.2 |
| Additionally allowed | 2.8 |
| Generously allowed | 0 |
| Disallowed | 0 |
| rms deviation | |
| Bond lengths (Å) | 0.022 |
| Bond angle (deg) | 1.633 |
a These include multiple conformations for Arg-184.
b These include water molecules, 1 Mn atom, and 1 ethylene glycol.
cR-factor=(∑|Fo − Fc|/∑|Fo|) × 100 where Fo is the observed structure-factor amplitude and Fc is the calculated structure-factor amplitude.
Protein structure
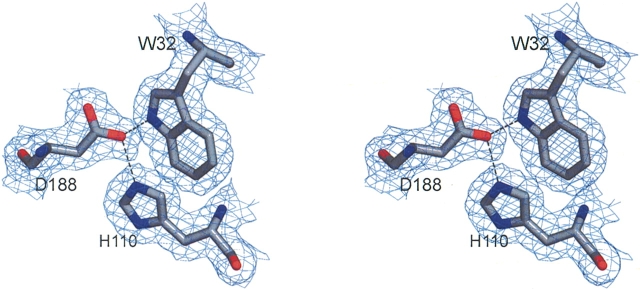
Figure 2 ▶ shows representative electron density of Trp-32, Asp-188, and His-110. This triad of residues is conserved in diiron proteins (Trp-62 in Δ9D, see Fig. 1 ▶, Trp-48 in ribonucleotide reductase) (Nordlund and Eklund 1993) and has been implicated in electron transfer reactions of ribonucleotide reductase by mutagenesis and catalytic studies (Sahlin et al. 1994; Bollinger et al. 1998; Baldwin et al. 2001). In the DesA2 crystal structure, the Nɛ1 of Trp-32 is within 3.0 Å of OΔ2 from Asp-188, while OΔ2 of Asp-188 is also within 2.8 Å of Nɛ2 from His-110.
Figure 2.
A stereoview of representative electron density. The map was calculated with coefficients of the form 2Fo-Fc and was contoured at 1 σ. The region in the protein around conserved residues Trp-32, Asp-188, and His-110 is shown. Figure was prepared with the program MacPyMOL (DeLano 2003).
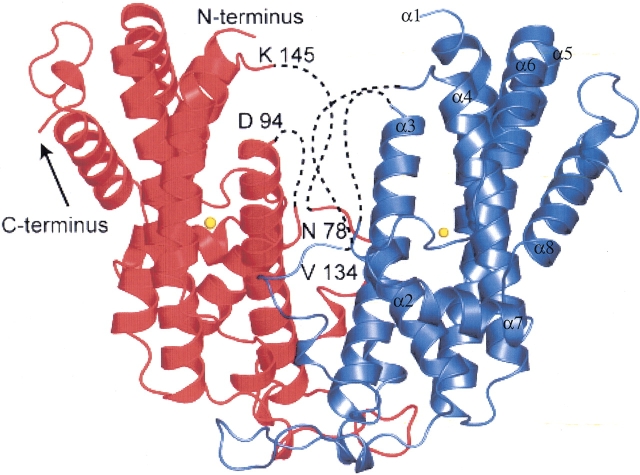
Figure 3 ▶ shows a ribbon diagram of the X-ray structure of DesA2. Positions of the N- and C-terminal, bound metal, and internal disordered regions are shown. The protein has α2 quaternary structure and is assembled from nine α-helical segments that have connectivity similar to that observed in the castor Δ9D (Lindqvist et al. 1996). In DesA2, α-helix-1 extends from Ala-9 to Glu-29 with a bend between Leu-14 and Glu-15. The residues Asp-30 to Pro-62 form an extended region that includes numerous intersubunit contacts from residues His-35 to Gly-50. α-helix-2 continues from Arg-63 to Asn-78. Residues Leu-79 to Glu-93 are not observed. α-helix-3 continues from Trp-96 to Thr-121, while Arg-122 to His-134 forms an extended region. Residues Gln-135 to Glu-144 are not observed, while Lys-145 to Thr-147 is an extended region. α-helix-4 continues from Gln-148 to Glu-172, and is followed by a three-residue turn consisting of lle-173 to Glu-175. α-helix-5 continues from Pro-176 to Tyr-206, and is followed by a single residue turn provided by Thr-207. α-helix-6 continues from Arg-208 to Asp-221 and is followed by an extended region from Leu-222 to Tyr-232. α-helix-7 continues from Arg-233 to Ala-242 and is followed by an extended region from Gly-243 to Gly-246. α-helix-8 continues from Lys-247 to Trp-261 and is followed by an extended region from Gly-262 to Thr-274. The C-terminal residue Gly-275 was not observed.
Figure 3.
Ribbon diagram of DesA2. Residues Met-1 to Ala-7, Leu-79 to Glu-93, and Gln-135 to Glu-144 were not observed. The position of N-terminal Asp-8 is indicated. Hypothetical connectivity indicated by dotted lines. The C-terminal residue Gly-275 was not observed. The location of a single metal ion per protein subunit is also indicated. The α-helical secondary structure elements described in the Results section are marked on one side of the dimer.
Metal binding site
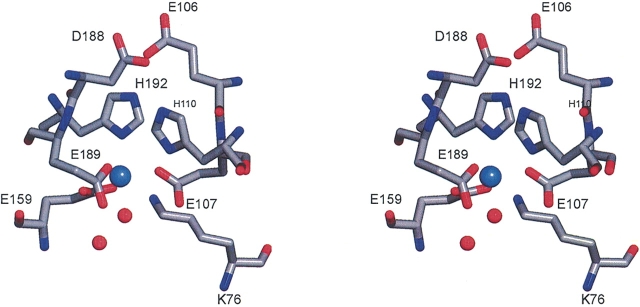
As shown by the primary sequence alignment of Figure 1 ▶, the following residues from DesA2 align with the metal binding ligands of plant Δ9D: Lys-76, Glu-107, His-110, Glu-159, Glu-189, and His-192. Figure 4 ▶ shows the putative metal-binding site of DesA2. In this crystal form, a single metal bound is almost certainly Mn2+ from the crystallization buffer since X-ray fluorescence scans of crystals of both the native and Se-met substituted proteins failed to show the presence of iron. The protein-derived ligands to the metal are NΔ1 of His-192 (2.2 Å between the metal and ligand atom), Oɛ1 and Oɛ2 of Glu-159 (interatomic distances of 2.3 Å and 2.4 Å, respectively), Oɛ2 of Glu-107 (2.2 Å from metal), and Oɛ2 of Glu-189 (2.1 Å from metal). The electron density for the sixth position is somewhat extended and can be modeled as two water molecules where the first is 2.2 Å from the metal center and the second is 2.4 Å from the atom directly coordinated to the metal. The NΔ1 of His-192 and the oxygen from the water occupy axial positions of a coordination sphere with distorted octahedral geometry. In addition, Glu-106 and Asp-188 align with acidic residues that provide conserved hydrogen bonds to the Nɛ2 atoms of the His ligands in other diiron enzymes. Thus Oɛ1 of Glu-106 is within 2.5 Å of Nɛ2 from His-192, while Oɛ1 of Asp-188 is within 2.8 Å of Nɛ2 from His-110.
Figure 4.
A stereo representation of the metal bound in DesA2 and closely associated residues. Ligands to the metal are Glu-107, Glu-159, Glu-189, and His-192. Glu-107, His-110, and Glu-189 are within hydrogen bonding distance of Nζ of Lys-76.
The Nζ of Lys-76 is located in the approximate position expected for the second metal of a diiron center (Fig. 4 ▶). This atom is 3.8 Å from the bound Mn2+, 3.0Å from NΔ1 of His-110, 2.9 Å from Oɛ1 of Glu-107, and 2.7 Å from Oɛ1 of Glu-189, and 3.2 Å from the water bound to the Mn2+. These hydrogen-bond donors provide a distorted square pyramidal arrangement around Nζ of Lys-76.
Discussion
Acyl-ACP desaturase superfamily
The acyl-ACP desaturases are members of Pfam PF03405, which presently contains 78 nonredundant examples. These enzymes were first identified in plants, and the majority of efforts have focused on stearoyl-ACP Δ9 desaturase (Δ9D). This enzyme has been cloned from the castor plant (Ricinus communis; Shanklin and Somerville 1991) and expressed in E. coli (Fox et al. 1993; Hoffman et al. 1995). The X-ray structure of the castor enzyme was solved at ~2.4 Å resolution (Lindqvist et al. 1996; Moche et al. 2003). The fold of the central α-helical bundle has high similarity to ribonucleotide reductase (Nordlund and Eklund 1993), methane monooxygenase (Rosenzweig et al. 1993; Elango et al. 1997), toluene/o-xylene monooxygenase (Sazinsky et al. 2004), and ferretin (Takagi et al. 1998). In these proteins, the aspartate, glutamate, and histidine residues of the conserved metal binding motif (D/E)X~40(D/E)X2HX~60 (D/E)X~40(D/E)X2H provide ligands to the catalytically essential diiron center.
Genome sequencing revealed three genes with similarity to known fatty acid desaturases (Cole et al. 1998). One of these, DesA3, was assigned to be an integral membrane desaturase and was more recently characterized by heterologous expression in E. coli to function as a stearoyl-CoA Δ9 desaturase (Phetsuksiri et al. 2003). Thus DesA3 appears to be functionally analogous to the intensively studied mammalian stearoyl-CoA desaturase (Ntambi 1995), which has a number of important physiological roles in cells (Ntambi 1995; Ntambi et al. 2002). DesA3 was recently shown to be essential for the viability of M. tuberculosis, and was identified as a target of the long-known anti-tuberculosis drug isoxyl (Phetsuksiri et al. 2003).
Two other genes from M. tuberculosis H37Rv, desA1 and desA2, were annotated to encode putative acyl-ACP desaturase homologs DesA1 and DesA2 (Cole et al. 1998). Other organisms identified by Pfam analysis to have these putative desaturases include Mycobacterium tuberculosis strain CDC1551, Mycobacterium avium sub-species paratuberculosis, Mycobacterium leprae, Mycobacterium bovis, Mycobacterium smegmatis, Streptomyces colieocolor, and Streptomyces avermitilis. Albeit with the limited number of examples available, the DesA1 proteins have ~82% amino acid sequence identity, while the DesA2 proteins have ~75% amino acid sequence identity. In contrast, the mycobacterial proteins share only ~20% identity with the plant acyl-ACP desaturases. DesA1 is ~338 residues long, which is closer in size to the plant acyl-ACP desaturases (~350 residues). Moreover, DesA1 has a Glu-76/Asp-77 pattern in the diiron sequence motif at the FeA ligand, which is consistent with that observed in the plant acyl-ACP desaturases (Glu-105/Glu-106 in mature castor Δ9D, but Glu-130/Asp-131 in the full-length A. thaliana protein; see Fig. 1 ▶). In contrast, DesA2 is only 275 residues long and has the Lys-76/Asp-77 motif pattern at the FeA ligand.
DesA2 structure
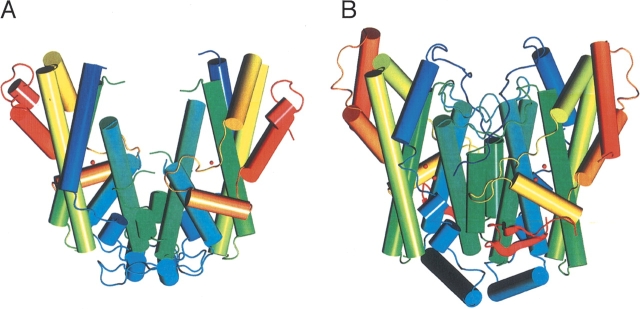
Figure 5 ▶ shows a schematic comparison of DesA2 and Δ9D. Visual inspection reveals that these two structures are related, even though DesA2 is a considerably smaller protein. Indeed, when superimposed with Align (Cohen 1997) the tertiary structures of DesA2 and castor Δ9D have an rms difference between 192 structurally equivalent amino acids of only 1.42 Å . Even more striking is the similarity in the DesA2 structure (Lys-145 to Leu-263) following the regions of disorder compared to Δ9D (Glu-182 to Val-310), where the rms difference between 114 structurally equivalent amino acids is 1.23 Å . This demonstrates the remarkable structural similarity between the plant and bacterial proteins in light of their limited sequence identity of ~24%.
Figure 5.
Comparison of X-ray structures. DesA2 (A; this work) and Δ9D (B; Lindqvist et al. 1996).
There are disordered residues in both the DesA2 and Δ9D structures, including Met-1 to Ala-7, Leu-79 to Glu-93, Gln-135 to Glu-144, and Gly-275 for DesA2 and Ala-1 to Pro-17 and Glu-338 to Ala-345 for Δ9D, with the majority of the disordered residues in DesA2 located at the interface between the subunits. Δ9D has a nonhomologous loop (Phe-18 to Ile-37) that folds over the amino acids Leu-172 to Glu-182. These latter amino acids correspond in the structural alignment to where the disordered residues Gln-135 to Glu-144 will likely be in the DesA2 structure. DesA2 has no features structurally equivalent with helix 5 (Gly-75 to Glu-89) of Δ9D. Instead, DesA2 has a loop from Phe-40 to Ser-59 that directly interacts with the other monomer, providing a potential contribution to stability of the homodimer. Likewise, DesA2 lacks the helix 9 of Δ9D (Asp-161 to Gly-176) but instead has an extended loop from Asp-125 to Val-134 that is followed by disorder from Gln-135 to Glu-144. As mentioned previously, Δ9D is also a larger protein than DesA2, with significant differences occurring at the C terminus due to the additional residues. Thus the DesA2 structure ends with a short helix (Pro-267 to Thr-274) while Δ9D has an extended helix (Ser-317 to Arg-336) followed by two small β-sheet motifs (Thr-350 to Pro-352 and Gln-360 to Lys-362).
Metal binding motif and disposition of metal center in DesA2
The identity and spacing of the first ligand provided by the diiron metal binding motif are variable among the known diiron enzymes. A variety of studies of soluble diiron enzymes have implicated the FeA site, bound by this first, variable ligand, to have a leading role in catalysis including proximity to the Tyr-122 radical (Nordlund and Eklund 1993), stabilization of peroxodiiron(III) state (Baldwin et al. 2001) in ribonucleotide reductase, and alterations of product distributions for regiospecific toluene hydroxylation upon mutagenesis of immediately adjacent residues (Mitchell et al. 2002). Since all sequences of DesA2 available from clinical isolates of M. tuberculosis and other organisms have a Lys residue at the position corresponding to Lys-76, it is likely that this amino acid residue has a unique role in the presently unknown function of DesA2.
The DesA2 crystal structure has only one metal bound per subunit (likely Mn2+ from the crystallization buffer), while the position comparable to that occupied by a second metal in other diiron enzymes is occupied by the Nζ atom of Lys-76. A Lys amino group has not been observed as a ligand in an iron-containing protein, and this residue would be expected to be a poor ligand at physiologic pH or the slightly acidic conditions of the crystallization experiment. One disordered sequence of DesA2 begins immediately after Lys-76, which may be attributed to a rearrangement caused by Nζ of Lys-76 occupying the putative second metal binding site. Also, sequence alignments indicate that this region is shorter in DesA2, suggesting the possibility of an altered route for access to the metal center in DesA2. It is plausible that substrate binding or other presently unknown biochemical factors unique to mycobacteria may assist in assembly of a dimetallic site in this protein. This functional relationship is also suggested by the conservation of a hydrogen-bonding pathway involving Trp-32, Asp-188, and putative metal ligand His-110 that has been implicated in electron transfer in ribonucleotide reductase (Fig. 2 ▶). However, it is also possible that DesA2 may have evolved away from a function requiring a bimetallic center to some other function, which remains to be determined.
Role of desaturases in Mycobacteria
Acyl-ACPs are the only relevant physiological substrates of Δ9D (Broadwater et al. 1998; Haas and Fox 1999; Lyle et al. 2003), and many aspects of catalysis likely arise from the influence of extensive enzyme-substrate interactions (Bloch 1969; Fox et al. 2004). Figure 2 ▶ shows the positions of 14 residues identified to line the bent tunnel in Δ9D, presumably at the correct depth from the surface to positions C–9 and C–10 of the 18:0 moiety of 18:0-ACP for reaction (Lindqvist et al. 1996). DesA2 has a different residue than the other proteins at 11 of 14 of these positions, including changes in size and polarity. Moreover, DesA2 has two well-ordered residues placed over and into an internal cavity near the putative metal binding site (Val-224 and Leu-225). Access to this cavity would apparently require conformational change, perhaps induced by substrate binding, metal incorporation, or some other effect.
Mass spectral analysis of mycolic acids from M. tuberculosis H37Rv suggested that 24:0 is desaturated to cis-Δ5-24:1, chain-elongated, and then desaturated to form cis-Δ3, cis-Δ15-34:2 as intermediates of mycolic acid biosynthesis (Watanabe et al. 2002). There is also evidence that AcpM acts as the specialized long acyl chain carrier in this process. AcpM shares similarity in primary sequence and tertiary structure with other known ACPs (Holak et al. 1988; Kim and Prestegard 1989; Crump et al. 1997; Xu et al. 2001; Li et al. 2003). However, AcpM contains an elongated C-terminal (~35 amino acids), which forms a flexible extension from the conserved fold of the ACP (Wong et al. 2002). Although the function of this unique extension is not known, these 35 amino acids are postulated either to sequester the long acyl chain or to mediate interactions with specific enzymes (Wong et al. 2002). The potential involvement of acyl-AcpM desaturases in mycolic acid biosynthesis implies differences in the architecture of the substrate binding channel used for a cis-Δ5-24:0 or a cis-Δ15-34:1-Δ3 desaturation compared to a cis-18:0-Δ9 desaturation. It also implies differences in the protein interactions required for formation of the enzyme-substrate complex due to the structural features of AcpM. The differences in structure observed for DesA2 and Δ9D, particularly the extent of disorder in the intersubunit region, which also closely approaches the metal center region, are a tantalizing indication that structural differences may be associated with functional differences.
Conclusions
The present crystallographic results provide evidence that DesA2 is structurally related to the plant acyl-ACP desaturases. However, the differences in the variable FeA region and the extent of disorder in the intersubunit region observed with DesA2 suggest different capabilities for this mycobacterial variant. As recent gene disruption studies have suggested that DesA2 is essential for the viability of pathogenic mycobacteria (gene disruption studies and attempts to introduce conditional mutations in DesA2 indicate that loss of function of either of these proteins is associated with an inability to recover viable mycobacteria; M. Jackson and P.J. Brennan, unpubl.), further studies of the unique properties of this putative desaturase are clearly needed.
Materials and methods
Materials
M. tuberculosis H37Rv total genomic DNA was obtained from the Tuberculosis Research Materials Facility at Colorado State University (Prof. J. Belisle, Director, NIH NIAD NO1AI75320). Vent DNA polymerase was from New England Biolabs. Oligonucleotide primers were obtained from Integrated DNA Technologies. The E. coli strain DH5α [sup E44ΔlacU169(φ80lacZΔM15)hsdR17recA1 endA1 gyrA96 thi-1 relA1] was used for cloning purposes. The E. coli strain BL21(DE3) [F− ompT hsdSB (rB−mB−) gal dcm (DE3)] was used as the protein expression host. Gateway Entry and Destination vectors and recombinase were from Invitrogen.
Cloning methods
M. tuberculosis H37Rv genomic DNA was used as a template for PCR. For the rv0824c gene (DesA1), the forward oligonucleotide primer contained the sequence 5′-atggcacagaaacctgtcgct gatg, which began with the annotated start codon. The reverse primer contained the sequence 5′-ctagcccgtgacgaattgcttgagg. For the rv1094 gene (DesA2), the forward oligonucleotide primer contained the sequence 5′-atggcacagaaacctgtcgctgatg, which began with the annotated start codon. The reverse primer contained the sequence 5′-ctagcccgtgacgaattgcttgagg. The PCR contained 10% dimethyl sulfoxide (DMSO) and consisted of 29 cycles of melt, anneal, and extend at temperatures of 94°C, 59°C, and 72°C, respectively. The resulting DNA fragments were purified by gel electrophoresis and extracted using a QIAquik Gel Extraction Kit (QIAGEN). The attB DNA recombination sequences were introduced into the 5′ and 3′ ends of the amplified genes by PCR. For the rv0824c gene, the forward primer 5′-ggggacaagtttgtacaaaaaagcaggctgaaggagatatacatatgg cacagaaacctgtcgctg and the reverse primer 5′-ggggaccactttgta caagaaagctgggtctagcccgtgacgaattgcttgagg were used. For the rv1094 gene, the forward primer 5′-ggggacaagtttgtacaaaaaag caggctgaaggagatatacatatggcacagaaacctgtcgctg and the reverse primer 5′-ggggaccactttgtacaagaaagctgggtctagcccgtgacgaattgctt gagg were used. The resulting ~1-kb DNA fragments were purified by gel electrophoresis followed by extraction with the QIAquik kit (QIAGEN). The fragments were inserted into a Gateway Entry vector (pDONR221) containing attP sites using the BP Clonase enzyme mix. This reaction created entry plasmids containing attL sites flanking both ends of the cloned gene sequence. CaCl2 competent E. coli DH5α (Sambrook et al. 2001) were transformed by heat shock with 1 μL of the BP reaction, and the transformation mixture was plated onto Luria-Bertani agar plates containing 50 μg/mL of kanamyacin. Plasmids were isolated from kanamyacin-resistant transformants using a QIAprep Spin MiniPrep kit (QIAGEN). The LR Clonase enzyme mix was used in a recombination reaction between the attL sites of the entry clones and the attR sites of a Gateway pDEST-14 vector. This recombination created an expression plasmid with the gene of interest under control of the viral T7 RNA polymerase promoter. E. coli DH5α cells were transformed with the reaction mixture and plated onto Luria-Bertani agar plates containing 200 μg/mL ampicillin. Plasmids from ampicillin-resistant transformants were purified using a QIAprep Spin Mini-Prep kit. Big Dye DNA sequencing (version 3.1, Applied Biosystems) performed at the University of Wisconsin Biotechnology Center was used to verify the coding sequence of the expression plasmids. The sequence-verified expression plasmids were named pKLDES1-a and pKLDES2-a.
Expression experiments
E. coli BL21(DE3) was transformed by electroporation with 1 μL of ~100 μg/mL plasmid DNA and plated onto Luria-Bertani agar plates (Sambrook et al. 2001) containing 200 μg/mL ampicillin. After ~16 h, a single colony was aseptically transferred to a sterile culture tube containing 2 mL of Luria-Bertani medium supplemented with 200 μg/mL ampicillin. The culture was grown with shaking at 37°C until the OD600 reached ~0.2; then 500 μL of this culture was used to inoculate 2 mL of Luria-Bertani medium containing 400 μg/mL ampicillin. Initial expression tests were performed by adding IPTG (final concentration of 1 mM) to the 2-mL cultures when the OD600 reached ~0.5. These cultures were analyzed by denaturing gel electrophoresis for protein expression after ~3 h. For scale-up, the 2-mL culture was grown without addition of IPTG to an OD600 ~0.5, and 250 μL was used to inoculate each of four 2-L flasks containing 500 mL of Luria-Bertani medium supplemented with 400 μg/mL ampicillin. The cultures were grown at 37°C until the OD600 reached ~0.6. At this point, the growth temperature was reduced to 25°C and the culture was induced by addition of IPTG to a concentration of 1 mM. At induction, the cultures were supplemented with Casamino acids (Difco) to a concentration of 0.5 g/L and with FeSO4 to a concentration of 80 μM. The induced culture was grown for ~12 h and harvested by centrifugation for 15 min at 4°C and 4400g in a Beckman J-6B centrifuge equipped with a JS-5.2 rotor (Beckman). This procedure gave ~1.5 g/L wet cell paste.
For selenomethionine labeling, the transformation and growth of a 3-mL culture in Luria-Bertani medium were performed as described above for the 2-mL culture. After the 3-mL starter culture reached an OD600 of ~0.2, 750 μL was used to inoculate each of four 2-L flasks containing 500 mL of a chemically defined medium (Studts and Fox 1999) and 200 μg/mL ampicillin. Each 500-mL culture was supplemented with 500 μL of a solution of 5 mg/mL thiamine. The cultures were grown with shaking at 37°C for ~8 h, after which the temperature was reduced to 23°C for ~10 h. When the culture reached OD600 ~0.4, the temperature was increased to 30°C and the following amino acids were added to the cultures: 100 μg/mL lysine, threonine, and phenylalanine, 100 mg/mL leucine, isoleucine, and valine, and 50 mg/mL selenomethionine. The cultures were induced after 20 min with 1 mM IPTG, and the medium was supplemented with 80 μM of FeSO4 and 5 g/L glucose. The induced culture was grown for ~6 h at 30°C and harvested as described above. This procedure gave ~1.5 g/L wet cell paste.
Purification of DesA2
All purification steps were performed at 4°C. The cell paste from 2 L of culture medium (~6 g) was thawed in a 50-mL stainless steel beaker and resuspended in 25 mL of 25 mM MOPS, pH 7.0. The cells were incubated on ice for 10 min after the addition of 50 mg each of lysozyme, RNase, and DNase to the cell suspension. The cell mixture was sonicated for a total of 1 min using 10-sec pulses (Fisher Model 550 Sonic Dismembrator, 1/8-inch horn, 45% maximum output). During sonication, the temperature of the cell suspension was maintained below 10°C by placing the beaker in an ice water bath containing NaCl. The sonicated cell suspension was centrifuged for 1 h at 39,000g to remove cell debris. The supernatant was diluted to 100 mL with 25 mM MOPS, pH 7.0 and loaded onto a Fast Flow Q-Sepharose 16/10 (Pharmacia LKB Biotechnology) equilibrated in 25 mM MOPS, pH 7.0. The column was washed with 60 mL of 25 mM MOPS, pH 7.0 and the protein was eluted at 7.6 cm/h in a 0.6-L linear gradient from 0 to 0.5M NaCl in 25 mM MOPS, pH 7.0. Column fractions were analyzed by SDS-PAGE, and peak fractions were pooled and concentrated by ultrafiltration (YM30 membrane, Amicon). The concentrated peak fractions were loaded onto a HiPrep 26/60 Sepharacyl S-100 column (Pharmacia LKB Biotechnology) equilibrated with 25 mM MOPS, pH 7.0, containing 25 mM NaCl and eluted at 6.8 cm/h. Peak fractions were analyzed by SDS-PAGE, pooled, and concentrated by ultrafiltration. After concentration to ~30 mg/mL, the purified DesA2 was drop-frozen in liquid nitrogen and stored at ~80°C.
Protein determinations
The identity of the purified protein was confirmed by mass determinations at the Mass Spectrometry Facility at the University of Wisconsin Biotechnology Center using an Applied Biosystems MDS Sciex API 365 LC/MS/MS triple quadrupole spectrometer equipped with a Perkin Elmer ABI 140D HPLC inlet system and operated in negative ionization mode. The concentration of purified DesA2 was determined by absorbance at 280 nm using ɛ280=50,550 M−1 cm−1 calculated from the amino acid content using DNASTAR (version 5.52).
Crystallization
Before crystallization trials, DesA2 was dialyzed in a Slide-ALyzer dialysis cassette (Pierce) in 25 mM HEPES, pH 7.0 containing 25 mM NaCl for 20 h at 4°C. Crystals of DesA2 were grown by the method of hanging drop vapor diffusion at room temperature. Typically, 2 μL of DesA2 at 10 mg/mL was combined with an equal volume of 17% methyl ether polyethylene glycol, 10 mM MnCl2, 20 mM Bis Tris propane, pH 7.0, and 30 mM sodium acetate, pH4.5. For low-temperature data collection, crystals were flash frozen in liquid N2 using the crystallization mother liquor plus 15% ethylene glycol and an incubation time of 10 sec. The selenomethionine-substituted DesA2 was crystallized in a similar manner.
Structure determination
MAD data were collected at three wavelengths on a single frozen crystal at beamline 32-ID of the Structural Biology Center at the Advanced Photon Source (Argonne, IL). For the peak, remote, and inflection point wavelengths, 180 images were recorded on a Mar Research CCD detector using 0.5° oscillations and 2-sec exposure times at a crystal-to-detector distance of 180 mm and a temperature of −160°C. The diffraction data were processed and scaled using the HKL 2000 software suite (Otwinowski and Minor 1997). The Friedel differences in the reference data set (peak) were locally scaled to remove systematic errors. A high-resolution native data set was collected for refinement of the final model. A preliminary X-ray fluorescence scan revealed the absence of iron in both the native and Se-Met crystals. SOLVE (Terwilliger 2002) was utilized to locate three Se sites in the asymmetric unit and to calculate phases using the 20–2.35 Å MAD data. The Se sites were employed to calculate initial phases, which were improved by the solvent flattening program RESOLVE (Terwilliger 2003). An initial model was built with ARP/wARP (Terwilliger 2003) and improved by manual adjustment with TURBO FRODO (Roussel and Cambillau 1991). After initial rounds of maximum likelihood refinement with the program REFMAC (Murshudov et al. 1997), the native structure was determined by molecular replacement utilizing the program MOLREP (Vagin and Teplyakov 2000) starting from the model obtained from the MAD structural solution. The final structural model was obtained from alternating rounds of model building and maximum likelihood refinement. Coordinates for the crystal structure and structure factors have been deposited in the Protein Data Bank with accession number 1ZA0.
Acknowledgments
The National Institutes of Health supported this work (GM-50853 to B.G.F. and AR-35186 to I.R). K.S.L. was a trainee of the NIH Institutional Molecular Biophysics Pre-Doctoral Training Grant T32 GM-08293. We thank the Mycobacterium Structural Genomics Consortium for their useful public Web site (http://www.doe-mbi.ucla.edu/TB/), Dr. Patrick J. Brennan (Colorado State University) for enlightening discussions, and Dr. Mary Jackson (Institute Pasteur) for the contribution of various proteins and enzymes to mycolic acid biosynthesis and viability of mycobacteria.
Abbreviations
A cis -Δ5–24:1 fatty acid is a C24 molecule with one cis double bond at the C–5 position
a cis-Δ3, cis-Δ15-34:2 fatty acid is a C34 molecule with two cis double bonds at the C–3 and C–15 positions
IPTG, isopropyl-β-D-thiogalactopyranoside
SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis
OD600, optical density at 600 nm
MAD, multiple anomalous dispersion
rms, root mean square.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041288005.
References
- The Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. [DOI] [PubMed] [Google Scholar]
- Baldwin, J., Voegtli, W.C., Khidekel, N., Moénne-Loccoz, P., Krebs, C., Pereira, A.S., Ley, B.A., Huynh, B.H., Loehr, T.M., Riggs-Gelasco, P.J., et al. 2001. Rational reprogramming of the R2 subunit of Escherichia coli ribonucleotide reductase into a self-hydroxylating monooxygenase. J. Am. Chem. Soc. 123 7017–7030. [DOI] [PubMed] [Google Scholar]
- Barry 3rd, C.E., Lee, R.E., Mdluli, K., Sampson, A.E., Schroeder, B.G., and Slayden, R.A. 1998. Mycolic acids: Structure, biosynthesis, and physiological functions. Prog. Lipid Res. 37 143–179. [DOI] [PubMed] [Google Scholar]
- Bloch, K. 1969. Enzymatic synthesis of monounsaturated fatty acids. Acc. Chem. Res. 2 193–202. [Google Scholar]
- Bollinger, Jr., J.M., Krebs, C., Vicol, A., Chen, S., Ley, B.A., Edmondson, D.E., and Huynh, B.H. 1998. Engineering the diiron site of Escherichia coli ribonucleotide reductase protein R2 to accumulate an intermediate similar to Hperoxo, the putative peroxodiiron(III) complex from the methane monooxygenase catalytic cycle. J. Am. Chem. Soc. 120 1094–1095. [Google Scholar]
- Broadwater, J.A., Ai, J., Loehr, T.M., Sanders-Loehr, J., and Fox, B.G. 1998. Peroxodiferric intermediate of stearoyl-acyl carrier protein Δ9 desaturase: Oxidase reactivity during single turnover and implications for the mechanism of desaturation. Biochemistry 37 14664–14671. [DOI] [PubMed] [Google Scholar]
- Cohen, G.H. 1997. ALIGN: A program to superimpose protein coordinates, accounting for insertions and deletions. J. App. Cryst. 30 1160–1161. [Google Scholar]
- Cole, S.T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S.V., Eiglmeier, K., Gas, S., Barry 3rd, C.E., et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393 537–544. [DOI] [PubMed] [Google Scholar]
- Crump, M.P., Crosby, J., Dempsey, C.E., Parkinson, J.A., Murray, M., Hopwood, D.A., and Simpson, T.J. 1997. Solution structure of the actinorhodin polyketide synthase acyl carrier protein from Streptomyces coelicolor A3(2). Biochemistry 36 6000–6008. [DOI] [PubMed] [Google Scholar]
- DeLano, W.L. 2003. MacPyMOL, 0.95 ed. DeLano Scientific LLC, San Carlos, CA.
- Doublie, S. 1997. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276 523–530. [PubMed] [Google Scholar]
- Dubnau, E., Chan, J., Raynaud, C., Mohan, V.P., Laneelle, M.A., Yu, K., Quemard, A., Smith, I., and Daffe, M. 2000. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol. Microbiol. 36 630–637. [DOI] [PubMed] [Google Scholar]
- Elango, N., Radhakrishnan, R., Froland, W.A., Wallar, B.J., Earhart, C.A., Lipscomb, J.D., and Ohlendorf, D.H. 1997. Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b. Protein Sci. 6 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, B.G., Shanklin, J., Somerville, C., and Münck, E. 1993. Stearoyl acyl carrier protein Δ9 desaturase from Ricinus communis is a diiron-oxo protein. Proc. Natl. Acad. Sci. 90 2486–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, B.G., Shanklin, J., Ai, J., Loehr, T.M., and Sanders-Loehr, J. 1994. Resonance Raman evidence for an Fe-O-Fe center in stearoyl-ACP desaturase. Primary sequence identity with other diiron-oxo proteins. Biochemistry 33 12776–12786. [DOI] [PubMed] [Google Scholar]
- Fox, B.G., Lyle, K.S., and Rogge, C.E. 2004. Reactions of the diiron enzyme stearoyl-ACP desaturase. Acc. Chem. Res. 37 421–429. [DOI] [PubMed] [Google Scholar]
- Haas, J.A. and Fox, B.G. 1999. The role of hydrophobic partitioning in substrate selectivity and turnover of the Ricinus communis stearoyl-ACP Δ9 desaturase. Biochemistry 38 12833–12840. [DOI] [PubMed] [Google Scholar]
- Hoffman, B.J., Broadwater, J.A., Johnson, P., Harper, J., Fox, B.G., and Kenealy, W.R. 1995. Lactose fed-batch overexpression of recombinant metalloproteins in Escherichia coli BL21(DE3): Process control yielding high levels of metal incorporated, soluble protein. Protein Expression Purif. 6 646–654. [DOI] [PubMed] [Google Scholar]
- Holak, T.A., Kearsley, S.K., Kim, Y., and Prestegard, J.H. 1988. Three dimensional structure of acyl carrier protein determined by NMR pseudoenergy and distance geometry calculations. Biochemistry 27 6135–6142. [DOI] [PubMed] [Google Scholar]
- Kim, Y. and Prestegard, J.H. 1989. A dynamic model for the structure of acyl carrier protein in solution. Biochemistry 28 8792–8797. [DOI] [PubMed] [Google Scholar]
- Li, Q., Khosla, C., Puglisi, J.D., and Liu, C.W. 2003. Solution structure and backbone dynamics of the holo form of the frenolicin acyl carrier protein. Biochemistry 42 4648–4657. [DOI] [PubMed] [Google Scholar]
- Lindqvist, Y., Huang, W., Schneider, G., and Shanklin, J. 1996. Crystal structure of Δ9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other diiron proteins. EMBO J. 15 4081–4092. [PMC free article] [PubMed] [Google Scholar]
- Lyle, K.S., Haas, J.A., and Fox, B.G. 2003. Rapid-mix and chemical quench studies of ferredoxin-reduced stearoyl-acyl carrier protein desaturase. Biochemistry 42 5857–5866. [DOI] [PubMed] [Google Scholar]
- Mitchell, K.H., Studts, J.M., and Fox, B.G. 2002. Combined participation of effector protein binding and hydroxylase active site residues provide toluene 4-monooxygenase regiospecificity. Biochemistry 41 3176–3188. [DOI] [PubMed] [Google Scholar]
- Moche, M., Shanklin, J., Ghoshal, A., and Lindqvist, Y. 2003. Azide and acetate complexes plus two iron-depleted crystal structures of the diiron enzyme Δ9 stearoyl-acyl carrier protein desaturase. J. Biol. Chem. 278 25072–25080. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., and Dodson, E.J. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53 240–255. [DOI] [PubMed] [Google Scholar]
- Nordlund, P. and Eklund, H. 1993. Structure and function of the Escherichia coli ribonucleotide reductase protein R2. J. Mol. Biol. 232 123–164. [DOI] [PubMed] [Google Scholar]
- Ntambi, J.M. 1995. The regulation of stearoyl-CoA desaturase (SCD). Prog. Lipid Res. 34 139–150. [DOI] [PubMed] [Google Scholar]
- Ntambi, J.M., Miyazaki, M., Stoehr, J.P., Lan, H., Kendziorski, C.M., Yandell, B.S., Song, Y., Cohen, P., Friedman, J.M., and Attie, A.D. 2002. Loss of stearoyl-CoA desaturase-1 function protects mice against adiposity. Proc. Natl. Acad. Sci. 99 11482–11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Phetsuksiri, B., Jackson,M., Scherman, H.,McNeil, M., Besra, G.S., Baulard, A.R., Slayden, R.A., DeBarber, A.E., Barry 3rd, C.E., Baird, M.S., et al. 2003. Unique mechanism of action of the thiourea drug isoxyl on Mycobacterium tuberculosis. J. Biol. Chem. 278 53123–53130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- A.C., Frederick, C.A., Lippard, S.J., and Nordlund, P. 1993. Crystal structure of a bacterial non-haem iron hydroxylase that catalyses the biological oxidation of methane. Nature 336 537–543. [DOI] [PubMed] [Google Scholar]
- Roussel, A. and Cambillau, C. 1991. Silicon graphics geometry partners directory. Silicon Graphics, Mountain View, CA.
- Sahlin, M., Lassmann, G., Pötsch, S., Slaby, A., Sjöberg, B.-M., and Gräslund, A. 1994. Tryptophan radicals formed by iron/oxygen reaction with Escherichia coli ribonucleotide reductase protein R2 mutant Y122F. J. Biol. Chem. 269 11699–11702. [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. 2001. Molecular cloning: A laboratory manual, 3d ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Sazinsky, M.H., Bard, J., Di Donato, A., and Lippard, S.J. 2004. Crystal structure of the toluene/o-xylene monooxygenase hydroxylase from Pseudomonas stutzeri OX1. J. Biol. Chem. 279 30600–30610. [DOI] [PubMed] [Google Scholar]
- Shanklin, J. and Somerville, C. 1991. Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc. Natl. Acad. Sci. 88 2510–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studts, J.M. and Fox, B.G. 1999. Application of fed-batch fermentation to the preparation of isotopically labeled- or selenomethionine-labeled proteins. Protein Expression Purif. 16 109–119. [DOI] [PubMed] [Google Scholar]
- Takagi, H., Shi, D., Ha, Y., Allewell, N.M., and Theil, E.C. 1998. Localized unfolding at the junction of three ferritin subunits. A mechanism for iron release? J. Biol. Chem. 273 18685–18688. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C. 2002. Automated structure solution, density modification and model building. Acta Crystallogr. D Biol. Crystallogr. 58 1937–1940. [DOI] [PubMed] [Google Scholar]
- ———. 2003. SOLVE and RESOLVE: Automated structure solution and density modification. Methods Enzymol. 374 22–37. [DOI] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, A. and Teplyakov, A. 2000. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 56 1622–1624. [DOI] [PubMed] [Google Scholar]
- Watanabe, M., Aoyagi, Y., Mitome, H., Fujita, T., Naoka, H., Ridell, M., and Minnikin, D.E. 2002. Location of functional groups in mycobacterial meromycolate chains; the recognition of new structural principles in mycolic acids. Microbiology 148 1881–1902. [DOI] [PubMed] [Google Scholar]
- Wong, H.C., Liu, G., Zhang, Y.-M., Rock, C.O., and Zheng, J. 2002. The solution structure of acyl carrier protein from Mycobacterium tuberculosis. J. Biol. Chem. 277 15874–15880. [DOI] [PubMed] [Google Scholar]
- Xu, G.Y., Tam, A., Lin, L., Hixon, J., Fritz, C.C., and Powers, R. 2001. Solution structure of B. subtilis acyl carrier protein. Structure 9 277–287. [DOI] [PubMed] [Google Scholar]
- Yuan, Y., Lee, R.E., Besra, G.S., Belisle, J.T., and Barry 3rd, C.E. 1995. Identification of a gene involved in the biosynthesis of cyclopropanated mycolic acids in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. 92 6630–6634. [DOI] [PMC free article] [PubMed] [Google Scholar]