Abstract
This work combines two well-established technologies to generate a breakthrough in protein production and purification. The first is the production of polyhydroxybutyrate (PHB) granules in engineered strains of Escherichia coli. The second is a recently developed group of self-cleaving affinity tags based on protein splicing elements known as inteins. By combining these technologies with a PHB-specific binding protein, a self-contained protein expression and purification system has been developed. In this system, the PHB-binding protein effectively acts as an affinity tag for desired product proteins. The tagged product proteins are expressed in E. coli strains that also produce intracellular PHB granules, where they bind to the granules via the PHB-binding tag. The granules and attached proteins can then be easily recovered following cell lysis by simple mechanical means. Once purified, the product protein is self-cleaved from the granules and released into solution in a substantially purified form. This system has been successfully used at laboratory scale to purify several active test proteins at reasonable yield. By allowing the bacterial cells to effectively produce both the affinity resin and tagged target protein, the cost associated with the purification of recombinant proteins could be greatly reduced. It is expected that this combination of improved economics and simplicity will constitute a significant breakthrough in both large-scale production of purified proteins and enzymes and high-throughput proteomics studies of peptide libraries.
Keywords: protein purification, protein expression, polyhydroxybutyrate, intein, self-cleaving affinity tag
Advances in protein expression systems have made possible the production of virtually any peptide product in at least one of a variety of host cells. Once expressed, however, these products must be purified to allow their study or use. Thus, the rapid and economical purification of recombinant proteins represents a persistent challenge in the field of biotechnology. Protein purification typically involves several chromatographic steps, each of which must be individually optimized for each product protein. Each step can be costly and time-consuming, and inevitably decreases the final yield of the product (Freitag and Horvath 1996). This complexity can delay research on new proteins at the laboratory scale, and is becoming more significant with the completion of several genome sequencing projects. In the large-scale manufacture of recombinant proteins for industrial and therapeutic use, downstream purification is very costly and can account for up to 80% of the total production cost (Hearn and Acosta 2001). Although these costs may be acceptable for high-value therapeutics, the development of generic biologics will demand more cost-effective manufacturing strategies. The development of simple and reliable methods for protein purification, which can be applied to arbitrary products at laboratory to manufacturing scales is therefore an important goal in bioseparations technology development.
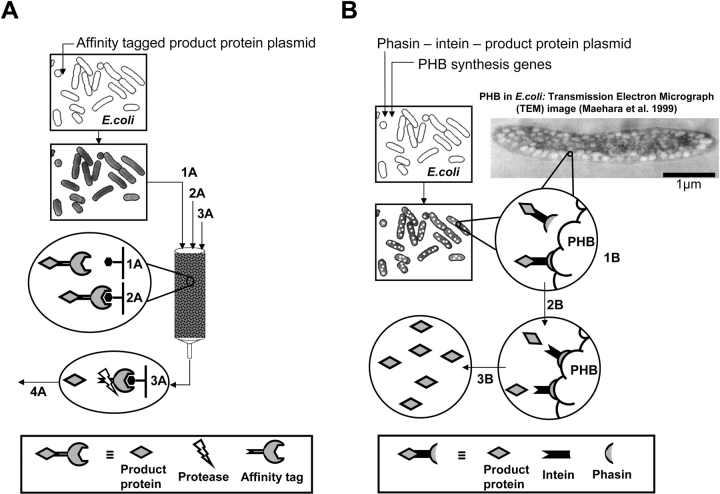
One method that can simplify the purification of virtually any protein is the addition of an affinity tag sequence to the target protein gene (LaVallie and McCoy 1995). This gene fusion results in the expression of an affinity-tagged target protein that can easily be purified by exploiting the highly selective binding characteristics of the tag (Ford et al. 1991). Once the fusion protein is purified, the tag can be enzymatically removed by one of a number of protease enzymes (Fig. 1A ▶). This method thus provides a simple and reliable method for the purification of native target proteins, and has become a core technology in bioseparations at the research scale (Hearn and Acosta 2001; Terpe 2003). A recent advance in this field has been the development of self-cleaving affinity tags (Chong et al. 1997). These tags, based on a recently discovered self-splicing protein element known as an intein, eliminate the need for proteolytic processing of the purified fusion protein to acquire a native product. Furthermore, intein-based tags allow the cleaving reaction to take place on the affinity matrix itself, greatly simplifying the overall purification method (Wood et al. 2000; Xu and Evans 2001). The development of pH and temperature-controllable inteins has further improved the simplicity and economic advantages of this technology (Southworth et al. 1999; Wood et al. 1999).
Figure 1.
Comparison of conventional affinity-based protein purification (A) and protein purification using an intein tag and affinity to PHB (B). (A) Conventional affinity-based protein purification: cells containing a plasmid for expression of the affinity tag-product protein fusion are induced and harvested. The cell pellet is resuspended, lysed, and passed over an affinity resin (1A). The column is then washed to rinse away impurities (2A). The fusion is retrieved from the column by addition of excess affinity tag or a displacing substitute and a protease is typically added to cleave off the product protein from the affinity tag (3A). A separation step (4A) salvages the protease and separates the product protein. (B) PHB-intein method of affinity-based protein purification: cells containing two plasmids, one for biosynthesis of PHB granules and another for expression of the phasin-intein tagged product protein, are grown to produce PHB and express the affinity fusion. Harvested cells are lysed and centrifuged to separate soluble components (1B). The insoluble PHB granules with the PHB-bound fusion protein are washed and resuspended in a cleavage-inducing buffer for release of the product protein (2B). A final centrifugation separates the PHB granules and associated proteins from the cleaved product protein, leaving only the product protein in the soluble fraction (3B).
A remaining limitation to the use of self-cleaving affinity tags is the high cost of the affinity resins that are typically used in these separations. Although there are several binding chemistries available, the affinity resins typically used with inteins cost over $1000 per L of bed volume, and in many cases have low binding capacity for the tagged fusion protein (based on product literature from New England Biolabs). This difficulty might be alleviated through the use of a suitable naturally occurring material that can be easily produced in a granular form by simple expression systems. These characteristics are satisfied by polyhydroxyalkanoic acids (PHA). These polymers are believed to be intracellular aliphatic carbon storage reserves, and are produced as granular inclusion bodies in many bacteria (Anderson and Dawes 1990). The PHA polymer consists of repeating units with the general form -[O-CH(R)(CH2)xCO]n-, the most common of which is polyhydroxybutyrate (PHB) -[O-CH(CH3)CH2CO]n- (McCool and Cannon 1999). Most importantly, PHB polymer granules have been produced in a wide variety of protein expression systems through simple genetic modification (Anderson and Dawes 1990). These systems include many bacterial and yeast systems, including Escherichia coli (Fidler and Dennis 1992) and Saccharomyces cerevisiae (Leaf et al. 1996), as well as transgenic plant cells (John and Keller 1996; Hahn et al. 1999). The macroscopic size and relatively high density of the granules allows them to be easily recovered by a variety of mechanical means following cell lysis, suggesting that they may be useful as an affinity carrier for tagged proteins.
In this study we use the unique properties of PHB in combination with a self-cleaving intein tag to create a simple, economic alternative for conventional affinity-based protein purification (Fig. 1B ▶). In this system, intracellular PHB granules are produced by E. coli cells to act as an affinity matrix for a coexpressed tagged protein. The affinity tag is derived from a class of PHB regulatory proteins known as phasins, which have been shown to exhibit strong and specific binding to the surface of the granules. During the fermentation, both granules and tagged proteins are coproduced in the E. coli cells. The tagged proteins adhere to the surface of the granules via the phasin tag. Once the cells are lysed, the granules are recovered and cleaned by repeated centrifugation and resuspension. The desired target protein is then released from the granules by pH-induced self-cleaving of the intein tag, allowing the affinity-purified native product to be easily separated from the granules and cleaved tags by a final centrifugation step. Tests on several target proteins indicate that this system is capable of providing highly purified active proteins at reasonable yields. Furthermore, the simple mechanical recovery of the PHB granules suggests a variety of means for trivial scaleup and transfer to other protein expression systems. The simplicity and self-contained nature of this system promise a significant breakthrough in the production of purified recombinant proteins for research and commercial use.
Results
Production of PHB granules with associating phasin in expression strains
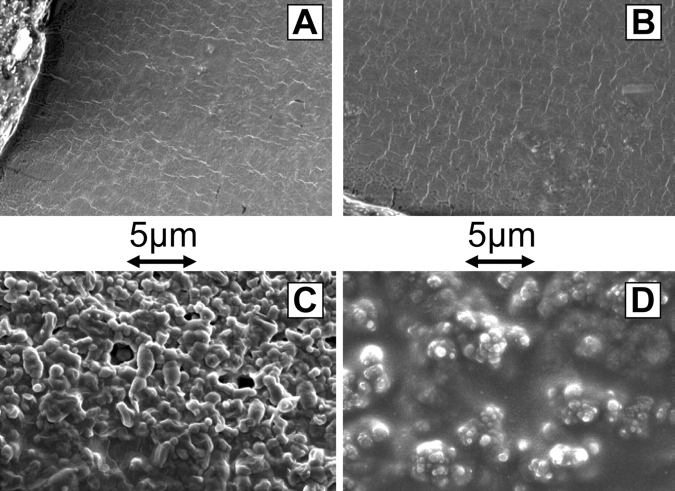
Three enzymes, α-ketothiolase (encoded by the PhaA gene), a stereo-specific reductase (PhaB), and PHA synthase (PhaC), are necessary for transforming meta-bolic acetyl CoA to PHB and are encoded on plasmid pJM9131 (Zhang et al. 1994). Following published procedures for producing PHB in E. coli XL1-Blue (Pieper-Furst et al. 1995; Wieczorek et al. 1995; Maehara et al. 1999), several E. coli laboratory strains were transformed with pJM9131 and grown for 30 h in LB medium supplemented with 2% sodium lactate as a carbon source for PHB synthesis. Scanning electron microscopy images of iridium-coated dried cell lysates indicate the presence of granules of the expected size (~100–700 nm) and characteristic shape absent in controls (Fig. 2 ▶). This result is similar to the SEM images published previously for Alcaligenes eutrophus (Doi 1990), and is in agreement with transmission electron micrographs previously published for PHB production in E. coli XL1-Blue (Lee 1996). The E. coli strains XL1-Blue, ER2566, BL21 (DE3), and BLR (DE3) all successfully produced PHB granules when transformed with pJM9131 (Fig. 2 ▶; data not shown). However, to assure strong expression of tagged product proteins from the pET-21 vector, BLR (DE3) carrying the T7 RNA polymerase gene was chosen as the host strain for subsequent expression and purification experiments.
Figure 2.
Scanning electron micrograph (SEM) images showing PHB granule synthesis in BLR (DE3) and XL1-Blue strains. All samples were grown for 30 h, lysed, dried, and iridium coated. (A) BLR strain carrying pJM9131 (PHB biosynthesis plasmid) grown in LB media. (B) BLR strain carrying a control ampicillin-resistant plasmid grown in lactate-supplemented LB media. (C) BLR strain carrying pJM9131 (PHB biosynthesis plasmid) grown in lactate-supplemented LB media. (D) XL1-Blue strain carrying pJM9131 (PHB biosynthesis plasmid) grown in lactate-supplemented LB media.
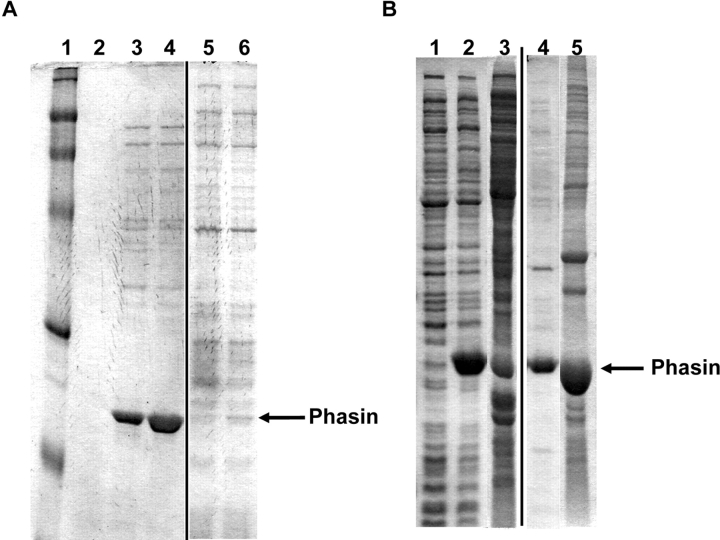
Affinity of the phaP-encoded phasin protein to intracellular PHB granules was examined by expression of the phasin in the presence and absence of coexpressed PHB granules in E. coli cells. SDS-PAGE analysis indicated that phasin expression for 2 h at 37°C produced a highly soluble protein in the absence of pJM9131 (Fig. 3A ▶). However, in strains transformed with pJM9131 and grown for 30 h to produce PHB granules in addition to phasin, the phasin was displaced from the soluble fraction of the lysate to the insoluble pellet (Fig. 3B ▶). An earlier time point of these double transformants shows that the phasin remains in the soluble fraction prior to PHB production regardless of the presence of pJM9131. This result demonstrates phasin affinity to PHB and is consistent with previously published observations (Wieczorek et al. 1995).
Figure 3.
SDS-PAGE results for phasin affinity to PHB. (A) BLR strain carrying phaP gene (plasmid pET/phaP) induced for 0.5 and 2 h at 37°C. Lane 1, molecular weight marker. Lane 2, preinduction whole-cell lysate. Lanes 3 and 4, soluble fractions of cell lysates at 0.5- and 2-h inductions, respectively. Lanes 5 and 6, insoluble fractions corresponding to lanes 3 and 4. (B) BLR strain carrying the phaP gene (plasmid pET/phaP) and PHB biosynthesis genes (plasmid pJM9131) grown and induced for 8 and 30 h. Lane 1, preinduction whole-cell lysate. Lanes 2 and 3, soluble fractions after 8 and 30 h, respectively. Lanes 4 and 5, insoluble fractions corresponding to lanes 2 and 3. Note the displacement of phasin from the soluble fraction (B, lane 2) to the insoluble fraction (B, lane 5) in the presence of PHB (after 30 h of growth).
Purification of maltose binding protein
The maltose binding protein (MBP) was chosen as the initial test protein for our purification scheme. Expression tests indicated that although the phasin alone has high affinity for PHB, fusion proteins of the phasin with the intein and various product proteins had noticeably lower affinity (data not shown); this led to leakage of the phasin-tagged precursors during the purification procedure, resulting in unacceptable losses in yield. Therefore, multiple phasins, separated by flexible linker peptides, were included in the binding tag to enhance fusion affinity to PHB and improve recovery. Ultimately, three phasins were combined with an engineered mini-intein and the maltose binding domain (MBD) to form PPPI:M (Phasin-Phasin-Phasin-Intein:MBD). The intein is the previously reported ΔI-CM mini-intein, engineered from the splicing domain of the Mycobacterium tuberculosis (Mtu) recA intein to self-cleave upon application of a pH or temperature shift (Wood et al. 1999). C-terminal cleavage of the intein after expression of PPPI:M releases the maltose binding protein (M) from the triple-phasin–intein (PPPI) complex. The PPPI:M fusion-protein gene was inserted into the T7 expression vector pET 21(+) to form pET/PPPI:M.
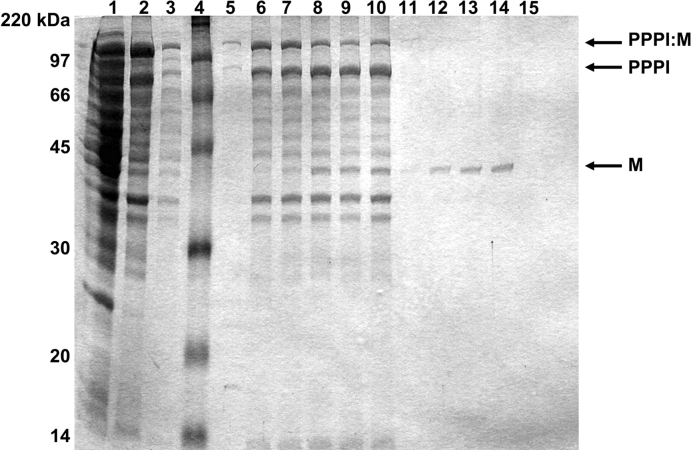
Double transformants carrying PHB biosynthesis genes (pJM9131) and the PPPI:M expression plasmid (pET/PPPI:M) were grown for 30 h in lactate-supplemented medium to produce PHB granules, at which point overexpression of the PPPI:M fusion protein was induced by IPTG addition. After an additional 4 h of incubation the cells were recovered by centrifugation and lysed by sonication into a pH 8.5 buffer. It has been shown (Wood et al. 1999) that the intein cleaving reaction is suppressed at this pH, allowing the precursor to be stabilized in an uncleaved form during subsequent granule wash steps. The soluble and insoluble fractions of the resulting cell lysates were separated by centrifugation and analyzed by SDS-PAGE (Fig. 4 ▶, lanes 1,2). The insoluble pellet, containing the PHB granules and any bound proteins, was washed several times by repeated centrifugation and resuspension in fresh pH 8.5 buffer. The pH was then shifted to 6.0 in the final wash to initiate the intein self-cleavage reaction (Fig. 4 ▶, lanes 3,5). Unclarified samples (including both soluble and insoluble material) were collected during the cleaving reaction and analyzed for cleavage product formation (Fig. 4 ▶, lanes 7–10). Each of these samples was then clarified by centrifugation and the corresponding supernatant was analyzed to detect cleaved soluble product proteins (Fig. 5 ▶, lanes 11–14). The results indicate that during incubation over 25 h at 20°C the PPPI:M fusion protein cleaves to yield PPPI and M. As expected, PPPI is retained in the insoluble phase with the PHB granules, while M (MBP) is released into the soluble fraction. Activity of the purified MBP was subsequently confirmed by its affinity for maltose resin (Fig. 4 ▶, lane 15). Similar results were obtained for the double phasin construct of PPI:M. The total MBP yield from this shake-flask experiment was 36.2 mg of MBP per L of culture (~3.35 mg per g of dry cell weight). Yields from similar experiments using PPI:M also fell within the range of 35–40 mg per L of culture.
Figure 4.
Maltose binding protein (MBP) purification: BLR strain double transformed with pJM9131 and pET/PPPI:M, grown for 24 h at 37°C in lactate-supplemented media and IPTG-induced for an additional 4 h at the same temperature. Lane 1, supernatant fraction of cell lysate. Lane 2, insoluble fraction of cell lysate. Lanes 3 and 5, decanted wash. Lane 4, molecular weight markers. Lane 6, post-wash pellet. Lanes 7–10, insoluble fraction for the cleavage time course after 1, 3, 20, and 25 h, respectively. Lanes 11–14, soluble fractions corresponding to lanes 7–10. Lane 15, supernatant from lane 14 after addition of maltose resin and centrifugation.
Figure 5.
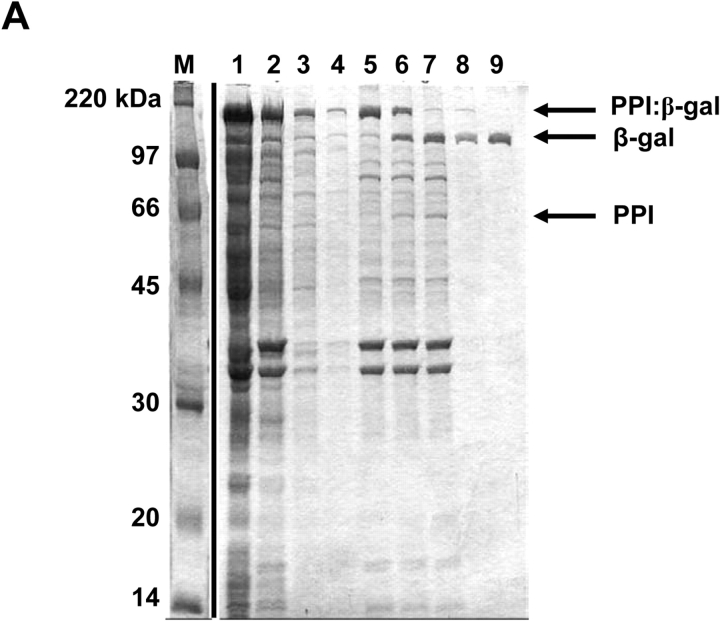
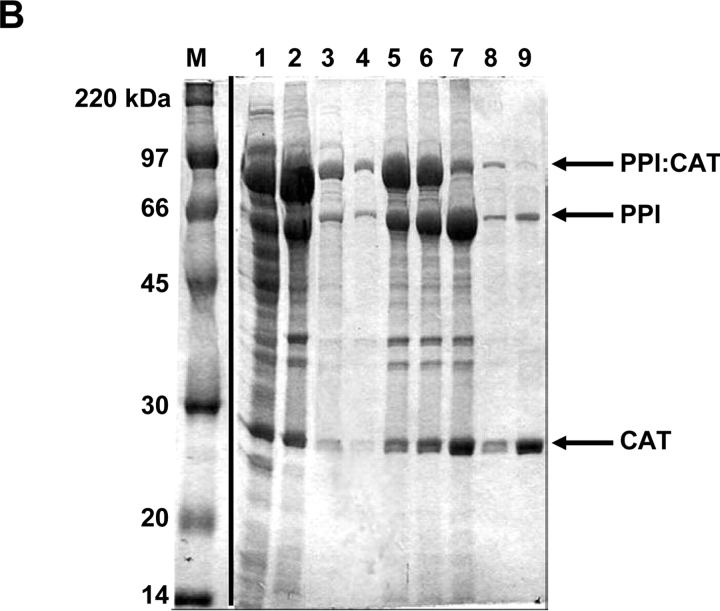
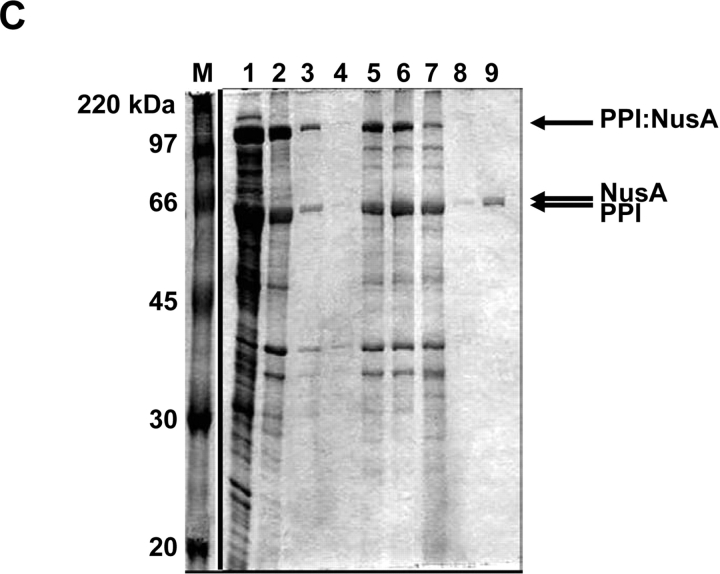
Purification of additional test proteins. (A) β-Galactosidase, (B) chloramphenicol acetyltransferase (CAT), (C) NusA protein. Proteins were expressed and purified as described in text. Lanes: M, molecular weight marker—same for all gels. Lane 1, supernatant fraction of cell lysate. Lane 2, insoluble fraction of cell lysate. Lanes 3 and 4, decanted wash supernatants. Lane 5, post-wash pellet. Lane 6 and 7, insoluble fraction for the cleavage time course after 2 and 30 h, respectively. Lanes 8 and 9, soluble fraction for the cleavage time course after 2 and 30 h, respectively.
Purification of other proteins
In addition to the maltose binding protein, several other target proteins were subsequently tested in our system using a two-phasin tag (Fig. 5 ▶, data not shown). These included the E. coli β-galactosidase enzyme (PPI:β-gal; Fig. 5A ▶), the chloramphenicol acetyltransferase (CAT) enzyme (PPI:CAT; Fig. 5B ▶), and the large and highly soluble NusA protein (PPI:NusA; Fig. 5C ▶). The fermentation, induction, and granule washing steps were similar to those described for MBP. Samples taken during the cleaving reaction indicated that each product protein was successfully purified at reasonable yield. In Figure 5 ▶, lane 9 of each gel represents the corresponding purified protein (β-gal, CAT, and NusA) with typical yields of 30 to 40 mg per L of culture (Table 1). Furthermore, an ONPG assay on the purified β-gal fraction (Fig. 5A ▶, lane 9) verified high yield and activity levels after purification (Table 1).
Table 1.
Quantification and activity assay of purified proteins
| Purified protein concentration | |||
| Target protein | mg/liter culture | mg/g dry cell weighta | Activity |
| MBP | 36.3 ± 2.2 | 3.35 ±0.2 | Affinity to Maltose resin |
| β-Galactosidase | 39.6 | 3.67 | 91.0 units/mg purified lysatec |
| CAT | 86.0b | 7.96 | N/A |
| NusA | 34.3 ±2.8 | 3.17 ±0.26 | N/A |
a Approximate cell pellet weight for 1-mL culture: 27.0 mg. Approximate dry cell weight: 10.8 mg.
b This protein content includes the impurities (PPI) shown in lane 9 of Fig. 5B ▶.
cUnit definition: One unit will hydrolyze 1.0 μmol of o-nitrophenyl β-D-galactoside (ONPG) to o-nitrophenol and D-galactose per minute.
It was observed that the purified CAT protein included significant impurities arising from cleaved PPI leaching off of the granules into the soluble fraction (Fig. 5B ▶, lane 9). It is likely that this arises from the high levels of overexpression of the PPI:CAT fusion relative to the other proteins tested, resulting in saturation of the available PHB granule surface area. This result suggests an upper limit of ~5 to 10 mg of purified protein per gram dry cell weight for this method. However, it has been reported that granule size and morphology can be modified by expression levels of phasin protein (Maehara et al. 1999). This implies that significant improvements in yield might be achieved through the optimization of fusion protein expression levels relative to PHB production, and this work is currently in progress.
Discussion
Considerations for efficient purification of general proteins
Over the course of this work, several fermentation and purification conditions were explored with a variety of test proteins. It was observed that PHB production is generally adequate after 30 h of incubation at 37°C, and that expression of the tagged fusion protein should generally be repressed as much as possible until the end of the fermentation. Leaky expression of the tagged protein can allow prematurely cleaved phasin tags to accumulate in the cells, substantially decreasing the effective capacity of the granules for the target protein. This effect is clearly visible with the MBP purification (Fig. 4 ▶, lane 6), but is greatly reduced for β-galactosidase (Fig. 4A ▶, lane 5). Although fermentation conditions can be optimized to minimize this effect for each protein, it appears that the identity of the protein itself can influence its tendency for premature cleaving.
The successful purification of a target protein using this method is contingent on several additional factors. Obviously, the solubility of the tagged target protein plays a significant role, since proper folding and activity of the phasin tag and intein are critical to the purification process. In experiments where PPI:X fusion proteins were expressed in the absence of PHB production, it was observed that the solubility of the fusion generally reflected the solubility of the native product protein (X). Some of these data indicate that the addition of the phasin tags can somewhat decrease solubility in some proteins, although in many cases a highly soluble and active precursor can be produced easily. More importantly, addition of the phasin tag does not seem to affect the overall expression levels of any proteins tested. Finally, target proteins with specific affinity for PHB granules should also be avoided. For example, it has been reported that the β-lactamase enzyme exhibits selective affinity for PHB granules, and therefore competes with phasins in binding to PHB granules (Maehara et al. 1999). Although fusions of phasin– intein–β-lactamase could be recovered from cell lysates in our laboratory, much of the cleaved tag is displaced into the final soluble phase while a substantial amount of the cleaved β-lactamase remains bound to the PHB granules (data not shown).
An additional significant parameter that can affect the performance of this system is the salt concentration of the washing and cleaving buffers. Low-salt buffers (0 to 50 mM) increased protein yield for less soluble proteins (CAT, GFP, and β-lactamase) at the expense of purity (data not shown). In this case, leaching of nonphasin PHB-associated proteins into the final supernatant fraction was increased. High-salt buffers (150 to 1000 mM) reduced nonphasin impurities and increased yield for more soluble proteins (MBP, NusA, and β-gal) but also increased displacement of the phasin–intein domain into the final soluble fraction. An intermediate salt content maintained the phasin–intein fraction bound to PHB granules, reduced nonphasin impurity leakage into the soluble phase, and improved yield. Changes in other buffer components, such as concentrations of EDTA or DTT, or the buffering agents themselves, had minimal effects on system performance. This is typical of intein-based purification processes, and suggests that this system will be applicable to a wide variety of target proteins.
Advantages in comparison with other methods
This method has significant economic advantages in comparison with other affinity-tag systems because it eliminates the need for protease treatment to deliver a native product and it eliminates the cost associated with the affinity matrix itself. The conventional chitin resin associated with commercially available intein systems costs $192 per 100 mL (New England Biolabs), and has a nominal capacity of ~2 mg of target protein per mL of bed volume. Although this resin can be used several times, a regeneration procedure is required between each purification. With the PHB system described here, the initial cost of the resin is reduced to that of a simple carbon source supplement, and the resulting granules are effectively disposable.
The approach presented here also offers mild conditions for recovery of proteins and is largely independent of the product protein. The results presented in Figures 4 ▶ and 5 ▶ for MBP, β-gal, CAT, and NusA were obtained using the same conditions, expression variables, and buffer components. Risk of degradation and irreversible damage to the target protein is reduced, since this process includes no pH extremes, proteases, or denaturing reagents. Furthermore, the relatively mild mechanical separations associated with the process are unlikely to compromise the function of the cleaved target protein. This was verified with activity assays for both the maltose binding protein and β-galactosidase enzyme. In the case of poorly expressed proteins, where the ratio of desired target protein to contaminant is lower, this system has the usual advantages of general affinity separations. Because the contaminant proteins do not interact with the PHB, they are unlikely to impact the overall yield of the target. Indeed, lower expression of target protein may in some cases increase recovery due to reduced saturation of the PHB granule surface with uncleaved fusion protein.
Although experiments presented here were carried out in E. coli, this method is applicable to protein purification in any protein expression system capable of synthesizing PHB. These include Ralstonia eutropha (York et al. 2001), Paracoccus denitrificans (Maehara et al. 1999), Baccillus megaterium (McCool and Cannon 1999), Rhodococcus ruber (Pieper-Furst et al. 1995), cotton (John and Keller 1996), and many others (Madison and Huisman 1999). These organisms could each offer specific advantages in expressing recombinant proteins, such as reducing inclusion body formation, improving glycosylation, or increasing fermentation cell density as demonstrated for Ralstonia eutropha (Srinivasan et al. 2002). Hence, the potential of the method presented here and its impact on protein purification encompasses genetic and experimental advances made in working with numerous organisms. In addition to a number of options for expression host organisms, a variety of carbon sources can also be used as the feed for PHB production. Although lactate was used as the supplemental carbon source in this case, other carbon sources such as glucose, sucrose, fructose, xylose, and whey have also been used for producing PHAs (Zhang et al. 1994; Ahn et al. 2000).
Future improvements
The data presented in this study are derived from simple proof-of-principle experiments, and generally indicate reasonable purity at relatively low concentration. It is anticipated that substantial purity and yield at high concentration will require significant optimization with regard to process design and operation, and may require additional subsequent steps. In cases where extremely high purity is necessary, as with pharmaceutical products, additional purification of the cleaved product protein will undoubtedly be required. We therefore expect that this system will find use in the large-scale production of inexpensive proteins with moderate purity requirements, or as a first-capture step to simplify the early purification of more highly specialized products. In either case, the simplicity of the system is likely to find utility in the production and purification of a variety of products.
Product protein yield is dependent on PHB granule production and morphology. PHB granules should be large enough to separate by trivial mechanical means and yet small enough to offer the maximum surface area for capturing and binding tagged product protein fusions. It has been speculated that phasin expression is correlated to an increase in number and decrease in size of PHB granules (Maehara et al. 1999). It is likely that independent control of PHB production and phasin- tagged fusion expression will allow significant yield increases over the initial results reported here.
Other factors which could improve the yield include: phasin–intein linker length and phasin structure. Depending on the mechanism and conformation of phasin association with PHB, which is still unclear, varying the linker length can lead to closer packing of fusion molecules on the PHB surface. The phasin sequence used here contains 191 amino acids with 46 alanine residues. In addition, of the 28 C-terminal amino acids, 14 are alanine and 7 are threonine residues. Perhaps a smaller phasin could be engineered to provide the same or greater affinity for PHB and hence reduce the total fusion size for higher efficiency.
We have introduced a protein purification process in which the host cells, in this case E. coli, produce an affinity tagged product protein and an easily recovered affinity carrier. The self-cleaving capability of the affinity tag allows the simple recovery of a native target protein at reasonable yield and very low cost. This simple technique is likely to be portable to numerous other expression hosts and is a significant advance in the large-scale production of recombinant protein products.
Materials and methods
Bacterial strains, constructs, and standard genetic manipulations
E. coli strains XL1-Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(TetR)]) from Stratagene; ER2566 (F− lamda− fhuA2 [Ion] ompT lacZ::T7 gene1 gal sulA11 D(mcrC-mrr)114::IS10 R(mcr- 73::miniTn10—TetS)2 R(zgb-210::Tn10)(TetS) endA1[dcm]) from NEB; BL21 (DE3) (F− ompT hsdISB (rB−mB−) gal dcm(DE3)) and BLR (DE3) (F− ompT hsdSB (rB−mB−) gal dcm (DE3) Δ(srl-recA)306::Tn10 (TetR)) from Novagen were used for cloning and expression using standard techniques (Sambrook and Russell 2001). Plasmids pJM9131 (KanR) containing the phaCAB operon for PHB biosynthesis and phaK (CamR) containing the phasin phaP gene were kindly provided by Professor Douglas Dennis (Arizona State University, West Campus, Phoenix) and are described elsewhere (Slater et al. 1992; Zhang et al. 1994; Kidwell et al. 1995). Plasmid pET-21(+) (AmpR) from Novagen featuring the T7lac promoter was used for expression and modified by adding a PCR amplified product to include a ribosome binding site and the maltose binding domain (from the pMAL plasmid, New England Biolabs) between the BamHI and EcoRI sites. After sequence and expression verification for the maltose binding domain (MBD), MBD was replaced by a phasin sequence using NdeI (introduced by the MBD PCR) and EcoRI. The phasin was followed by this linker sequence: AACAATAACAACAACCTCGGGATCGAGGGAAGGAT TTCAGAATTC. An additional phasin with two flanking NdeI sites was PCR amplified and inserted upstream of the initial phasin. In the case of the triple phasin constructs PCR amplification was again used to generate a third phasin with two flanking EcoRI sites for insertion downstream of the first phasin. PCR amplifications were carried out such that the linker sequence mentioned above followed each inserted phasin in the final construct. The mutated and evolved mini-intein from M. tuberculosis (Mtu) recA was digested out of a previous plasmid pMΔI †T-CM (Wood et al. 1999) using EcoRI and BsrGI and was inserted downstream of the phasin sequences. The maltose binding domain or other target protein domains, NusA, β-gal, and CAT, were PCR amplified flanked by BsrGI and HindIII or NotI and inserted downstream from the intein. The NusA gene came from the pET-43.1 vector available from Novagen. β-Gal was PCR amplified from E. coli chromosome and CAT from the phaK plasmid carrying the CamR gene.
Media, expression and PHB generation
Strains carrying pJM9131 and producing PHB were diluted 100:1 from overnight cultures and grown for 30 h at 37°C (unless otherwise noted) in Luria-Bertani medium (1% Bacto tryptone, 0.5% yeast extract, and 1% NaCl) supplemented with 2% sodium lactate and 50 μg/mL kanamycin. In the case of double transformants carrying a modified pET-21 vector expressing a fusion protein (such as pET/PPPI:M) the media was additionally supplemented with ampicillin (100 μg/mL). All growth steps were carried out in shake flasks or 5-mL test tubes in a Labline orbital shaker at 300 rpm. Isopropyl-β- D-thiogalactopyranoside (IPTG) was added for inductions and cultures grown for an additional 4 to 8 h at 37°C or 20°C as indicated, at which point the cells were harvested by centrifugation (5000g, 10 min, 4°C).
Scanning electron micrographs
Modifying a previously described method (Doi 1990), 1-mL samples grown as described above (in LB+2% lactate for 30 h) were resuspended in 100 μL lysozyme-containing lysis buffer (10 mM Tris-HCl, 10 mM CaCl2, 0.5 mg/mL lysozyme) before adding 100 μL of an alkaline-SDS solution (0.4 M NaOH, 2% SDS). Four 15-sec sonications were carried out on ice allowing the samples to cool intermediately. Samples were dried on carbon tab specimen mounts (Ted Pella) and sputtered with a 2-nm layer of iridium before being examined using a Philips XL30 FEG-SEM under a 5-KeV beam.
Purification and SDS-PAGE analysis
Harvested cell pellets from 1-mL samples were resuspended in 300 μL modified lysis buffer (20 mM Tris, 20 mM Bis, 50 mM NaCl, 1 mM DTT, 2 mM EDTA, 0.25 mg/100 mL lyzozyme, at pH 8.5) and disrupted by ultrasonic disruption at 4°C. Lysed cells were spun in a bench-top centrifuge at 14,000g for 10–30 min at 4°C. Supernatant was then discarded and the cells resuspended in a wash buffer (20 mM Tris, 20 mM Bis, 50 mM NaCl, 1 mM DTT, 2 mM EDTA, at pH 8.5). Resuspended pellet was centrifuged at 14,000g for 10–30 min at 4°C and the wash discarded. This wash step was repeated as necessary. In the last wash cycle, the pellet was resuspended in a cleavage buffer (20 mM Tris, 20 mM Bis, 50 mM NaCl, 1 mM DTT, 2 mM EDTA, at pH 6.5 or pH 6.0) and centrifuged at 14,000g for 10–30 min at 4°C and the supernatant discarded. This was to ensure homogenous pH throughout the pellet and the tube. The pellet was resuspended again in the cleavage buffer and left to rest at room temperature (18–23°C) for cleavage. At each time point a total solution fraction was taken and the sample centrifuged at 14,000g for 10–30 min at 4°C to take a supernatant (soluble) fraction. Samples were resuspended after taking the supernatant time point and left to rest at 18–23°C for the cleavage to continue to completion. Samples were analyzed by 12% SDS-PAGE followed by staining with Coomassie Brilliant Blue G-250.
Protein content quantification and β-gal activity assay
Protein concentrations were measured using the Bradford method (Ausubel 1998). β-Galactosidase activity assay based on activity with o-Nitrophenyl-β-D-galactopyranoside (ONPG) was measured by the β-gal Activity Assay kit by Stratagene.
Acknowledgments
We thank Professor Douglas Dennis for plasmid pJM9131 (containing the phaCAB operon) and plasmid phaK (coding for the phaP gene). This work was partially supported by the National Science Foundation Graduate Student Fellowship and Army Research Office grant W911NF-04-1-0056.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041296305.
References
- Ahn, W.S., Park, S.J., and Lee, S.Y. 2000. Production of Poly(3-hydroxybutyrate) by fed-batch culture of recombinant Escherichia coli with a highly concentrated whey solution. Appl. Environ. Microbiol. 66 3624–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, A.J. and Dawes, E.A. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54 450–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M. 1998. Protein analysis. In Current protocols in molecular biology (eds. F.M. Ausubel et al.), Section 10.11.14. John Wiley & Sons, New York.
- Chong, S., Mersha, F.B., Comb, D.G., Scott, M.E., Landry, D., Vence, L.M., Perler, F.B., Benner, J., Kucera, R.B., Hirvonen, C.A., et al. 1997. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene 192 271–281. [DOI] [PubMed] [Google Scholar]
- Doi, Y. 1990. Fermentation and analysis of microbial polyesters. In Microbial polyesters, p. 156. VCH, New York.
- Fidler, S. and Dennis, D. 1992. Polyhydroxyalkanoate production in recombinant Escherichia coli. FEMS Microbiol. Rev. 9 231–235. [DOI] [PubMed] [Google Scholar]
- Ford, C.F., Suominen, I., and Glatz, C.E. 1991. Fusion tails for the recovery and purification of recombinant proteins. Protein Expr. Purif. 2 95–107. [DOI] [PubMed] [Google Scholar]
- Freitag, R. and Horvath, C. 1996. Chromatography in the downstream processing of biotechnological products. Adv. Biochem. Eng. Biotechnol. 53 17–59. [DOI] [PubMed] [Google Scholar]
- Hahn, J.J., Eschenlauer, A.C., Sleytr, U.B., Somers, D.A., and Srienc, F. 1999. Peroxisomes as sites for synthesis of polyhydroxyalkanoates in transgenic plants. Biotechnol. Prog. 15 1053–1057. [DOI] [PubMed] [Google Scholar]
- Hearn, M.T. and Acosta, D. 2001. Applications of novel affinity cassette methods: Use of peptide fusion handles for the purification of recombinant proteins. J. Mol. Recognit. 14 323–369. [DOI] [PubMed] [Google Scholar]
- John, M.E. and Keller, G. 1996. Metabolic pathway engineering in cotton: Biosynthesis of polyhydroxybutyrate in fiber cells. Proc. Natl. Acad. Sci. 93 12768–12773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidwell, J., Valentin, H.E., and Dennis, D. 1995. Regulated expression of the Alcaligenes eutrophus pha biosynthesis genes in Escherichia coli. Appl. Environ. Microbiol. 61 1391–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVallie, E.R. and McCoy, J.M. 1995. Gene fusion expression systems in Escherichia coli. Curr. Opin. Biotechnol. 6 501–506. [DOI] [PubMed] [Google Scholar]
- Leaf, T.A., Peterson, M.S., Stoup, S.K., Somers, D., and Srienc, F. 1996. Saccharomyces cerevisiae expressing bacterial polyhydroxybutyrate synthase produces poly-3-hydroxybutyrate. Microbiology 142(Pt 5) 1169–1180. [DOI] [PubMed] [Google Scholar]
- Lee, S.Y. 1996. Bacterial Polyhydroxyalkanoates. Biotechnol. Bioeng. 49 1–14. [DOI] [PubMed] [Google Scholar]
- Madison, L.L. and Huisman, G.W. 1999. Metabolic engineering of poly(3- hydroxyalkanoates): From DNA to plastic. Microbiol. Mol. Biol. Rev. 63 21–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara, A., Ueda, S., Nakano, H., and Yamane, T. 1999. Analyses of a polyhydroxyalkanoic acid granule-associated 16-kilodalton protein and its putative regulator in the pha locus of Paracoccus denitrificans. J. Bacteriol. 181 2914–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCool, G.J. and Cannon, M.C. 1999. Polyhydroxyalkanoate inclusion body-associated proteins and coding region in Bacillus megaterium. J. Bacteriol. 181 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper-Furst, U., Madkour, M.H., Mayer, F., and Steinbuchel, A. 1995. Identification of the region of a 14-kilodalton protein of Rhodococcus ruber that is responsible for the binding of this phasin to polyhydroxyalkanoic acid granules. J. Bacteriol. 177 2513–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Slater, S., Gallaher, T., and Dennis, D. 1992. Production of poly-(3-hydroxybutyrate- co-3-hydroxyvalerate) in a recombinant Escherichia coli strain. Appl. Environ. Microbiol. 58 1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southworth, M.W., Amaya, K., Evans, T.C., Xu, M.Q., and Perler, F.B. 1999. Purification of proteins fused to either the amino or carboxy terminus of the Mycobacterium xenopi gyrase A intein. Biotechniques 27 110–120. [DOI] [PubMed] [Google Scholar]
- Srinivasan, S., Barnard, G.C., and Gerngross, T.U. 2002. A novel highcell- density protein expression system based on Ralstonia eutropha. Appl. Environ. Microbiol. 68 5925–5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpe, K. 2003. Overview of tag protein fusions: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 60 523–533. [DOI] [PubMed] [Google Scholar]
- Wieczorek, R., Pries, A., Steinbuchel, A., and Mayer, F. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J. Bacteriol. 177 2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, D.W., Wu, W., Belfort, G., Derbyshire, V., and Belfort, M. 1999. A genetic system yields self-cleaving inteins for bioseparations. Nat. Biotechnol. 17 889–892. [DOI] [PubMed] [Google Scholar]
- Wood, D.W., Derbyshire, V., Wu, W., Chartrain, M., Belfort, M., and Belfort, G. 2000. Optimized single-step affinity purification with a selfcleaving intein applied to human acidic fibroblast growth factor. Biotechnol. Prog. 16 1055–1063. [DOI] [PubMed] [Google Scholar]
- Xu, M.Q. and Evans Jr., T.C. 2001. Intein-mediated ligation and cyclization of expressed proteins. Methods 24 257–277. [DOI] [PubMed] [Google Scholar]
- York, G.M., Junker, B.H., Stubbe, J.A., and Sinskey, A.J. 2001. Accumulation of the PhaP phasin of Ralstonia eutropha is dependent on production of polyhydroxybutyrate in cells. J. Bacteriol. 183 4217–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., Obias, V., Gonyer, K., and Dennis, D. 1994. Production of polyhydroxyalkanoates in sucrose-utilizing recombinant Escherichia coli and Klebsiella strains. Appl. Environ. Microbiol. 60 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]