Abstract
l-Arginine (l-Arg) has been widely used as an enhancer of protein renaturation. The mechanism behind its action is still not fully understood. Using hen egg white lysozyme as a model protein, we present data that clearly demonstrate the suppression of the aggregation of denatured protein by l-Arg. By chemical modification of free cysteines, a series of unfolded lysozyme species were obtained that served as models for unfolded and intermediate states during the process of oxidative refolding. An increased equilibrium solubility of unfolded species and intermediates in the presence of l-Arg seems to be its major mechanism of action.
Keywords: refolding, aggregation, l-arginine, lysozyme
The heterologous expression of recombinant proteins has become one of the very essential techniques of life science research. The high-yield production of recombinant proteins in bacteria often leads to the formation of insoluble inclusion bodies, i.e., the desired product is obtained in an inactive form. This requires the solubilization of the misfolded protein entrapped within the inclusion bodies, usually with a denaturing agent such as urea or guanidinium chloride (GuHCl), followed by refolding.
Refolding conditions have to be carefully chosen in order to obtain satisfactory amounts of active protein. For quite some time it has been known that the addition of certain small organic molecules may significantly enhance the yield of the refolding process, and in many cases the denaturation and refolding of inclusion body protein only becomes practical by making use of this effect (for recent reviews on this topic see, e.g., De Bernardez Clark et al. 1999; Tsumoto et al. 2003; Fahnert et al. 2004; Lange and Rudolph 2005).
The efficiency of the refolding process is determined by the competition between productive refolding and unproductive side reactions, i.e., the formation of misfolded species and the aggregation of denatured protein (Goldberg et al. 1991; Kiefhaber et al. 1991). Refolding additives may act on either one or all of the reactions involved, i.e., they may facilitate the refolding of a protein in question by stabilizing its native state or accelerating the kinetics of the “correct” folding reaction, as well as by suppressing unspecific aggregation of the unfolded polypeptide and/or intermediates on the folding pathway. It has been suggested that amino acids (arginine, proline, lysine) (Rudolph and Fischer 1990; Samuel et al. 2000), polyamines (putrescine, spermidine, and spermine) (Kudou et al. 2003), and mild detergents (Tandon and Horowitz 1988; Wetlaufer and Xie 1995; Krause et al. 2002), but also the denaturing agents GuHCl and urea themselves (Orsini and Goldberg 1978), act as suppressors of aggregation, while substances like sugars, polyalcohols, and ammonium sulfate improve refolding yields by stabilizing the native conformation of proteins (Sawano et al. 1992; Michaelis et al. 1995).
l-Arginine (l-Arg) is the most basic natural amino acid with a pI of about 10.8, and a derivative of the denaturing agent guanidine. l-Arginine monohydrochloride (l-ArgHCl) has been widely used as an additive in protein refolding. It was, e.g., effective in improving the yield of human plasminogen activator (Rudolph and Fischer 1990), recombinant Fab-fragments (Buchner and Rudolph 1991), immunotoxins (Brinkmann et al. 1992), functional single-chain antibody fragments (Tsumoto et al. 1998), Interleukin-21 (Asano et al. 2002), human matrix metalloproteinase-7 (Oneda and Inouye 1999), and recombinant human neurotrophins (Suenaga et al. 1998; Rattenholl et al. 2001). It was found to be the most effective amino acid in suppressing the aggregation of lysozyme after heat-induced denaturation (Shiraki et al. 2002). In spite of the widespread application of l-ArgHCl as an effective refolding additive, the mechanism behind its action still remains somewhat unclear. Only recently, Arakawa and Tsumoto (2003) found that it had no significant effect on the thermal stability of RNAse A and hen egg white lysozyme, but improved the reversibility of the respective thermal transitions. This tentatively suggests that l-ArgHCl acted as suppressor of aggregation. In earlier studies, l-ArgHCl had even been found to slightly destabilize RNAse A (Lin and Timasheff 1996), as well as cytochrome c (Taneja and Ahmad 1994).
The equilibrium folding/unfolding of hen egg white lysozyme and its renaturation by oxidative refolding have been extensively investigated (Saxena and Wetlaufer 1970; Acharya and Taniuchi 1982; Radford et al. 1992; Roux et al. 1999). Upon renaturation at high protein concentrations, lysozyme is susceptible to aggregation of misfolded protein, and refolding yields drop dramatically (Goldberg et al. 1991). The presence of l-ArgHCl has been shown to improve the process (Hevehan and De Bernardez Clark 1997; Armstrong et al. 1999; Ho et al. 2003). While the improvement of the refolding yield of lysozyme by l-ArgHCl is well documented, the mechanism of its action remains elusive, in line with what was said above for the refolding of proteins in general. In this work we present data that clearly demonstrate the suppression of aggregation of denatured lysozyme by l-ArgHCl. Furthermore, to expand on this result we set out to investigate the equilibrium solubility of denatured lysozyme under refolding conditions, and in particular the dependency of this solubility on the concentration of l-ArgHCl. Refolding of the denatured lysozyme was blocked by chemical modification of the cysteines with iodoacetamide, iodoacetic acid, or glutathione, yielding a series of unfolded lysozyme species that were intended to serve as models for unfolded and intermediate states during the process of oxidative refolding.
Results
Renaturation of lysozyme
Denatured-reduced lysozyme can be renatured (i.e., re-folded and reoxidized) in the presence of GSSG and GSH (Saxena and Wetlaufer 1970). Along with other additives, l-ArgHCl has been reported to effectively enhance the oxidative refolding of lysozyme (Hevehan and De Bernardez Clark 1997). As a starting point for our work, we reexamined this effect. Denatured-reduced lysozyme was renatured in buffer containing l-ArgHCl in concentrations ranging from 0 to 0.9 M. The recovery of native structure was monitored by measuring the protein’s enzymatic activity (Fig. 1A ▶). The renaturation yield was significantly improved by the addition of increasing concentrations of l-ArgHCl. In our hands, we found a linear increase in the recovery of activity up to 87% in the presence of 0.9 M l-ArgHCl; while in its absence, on average, the renaturation yield was only 23% (Fig. 1B ▶). At low concentrations of l-ArgHCl, a considerable variability in final yield was observed, ranging, e.g., from 10% to 35% in the absence of l-ArgHCl. In parallel to the increasing refolding yield, we observed a slight decrease in the apparent pseudo-first-order rate of renaturation (Fig. 1C ▶), in agreement with earlier model calculations (Kiefhaber et al. 1991; Hevehan and De Bernardez Clark 1997).
Figure 1.
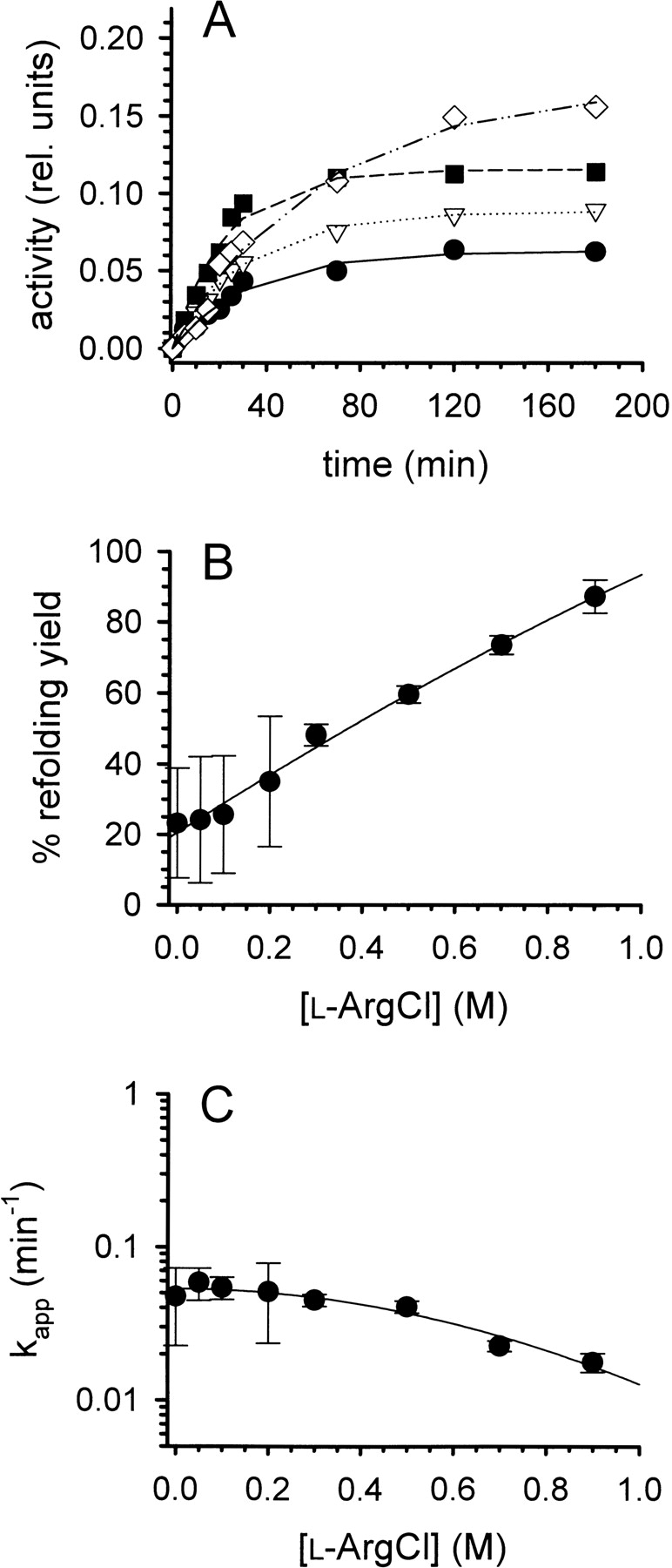
Refolding of lysozyme. (A) Denatured-reduced lysozyme was refolded at pH 8.2 and room temperature in the presence of a glutathione redox-shuffling system and increasing concentrations of l-ArgHCl as described in the text. The protein concentration was 0.25 mg mL−1. [l-ArgHCl] was 0, 0.2, 0.5, and 0.9 M (bottom to top). The refolding yield was monitored by measuring the enzymatic activity. (B) The final refolding yields were expressed as percentage of the specific activity of native lysozyme and plotted as function of [l-ArgHCl]. (C) The time course of refolding was analyzed as a pseudofirst-order reaction, yielding apparent time constants kapp. Data in B and C represent mean values ± SD from three independent experiments. Lines are meant to guide the eye.
Equilibrium unfolding
The effect of l-ArgHCl on the thermodynamic stability has been investigated for a series of proteins (Taneja and Ahmad 1994; Lin and Timasheff 1996; Arakawa and Tsumoto 2003). In these studies, no general tendency for stabilization of native protein structure by L-ArgHCl has been observed, making it unlikely that the increased renaturation yield of lysozyme in its presence is due to a thermodynamic stabilization of the native protein. To corroborate this point, we performed GuHCl-induced equilibrium unfolding experiments in the presence of increasing concentrations of l-ArgHCl (Fig. 2A ▶). No significant shift in the transition midpoint could be observed in the range of l-ArgHCl concentrations relevant to the oxidative refolding of lysozyme (up to 1.0 M). An evaluation of the data assuming a simple two-state transition and a linear GuHCl-dependency of the free energy of unfolding (ΔGU) failed to show any significant influence of l-ArgHCl on the thermodynamic stability of lysozyme (Table 1A). Equally, the stability of lysozyme against its reversible thermal denaturation, as monitored by differential scanning calorimetry (DSC), was not influenced by the presence of l-ArgHCl up to 1.0 M (Fig. 2B ▶). No significant effect on the transition temperatures Tm or the heat of unfolding ΔHTm was observed (Table 1B).
Figure 2.
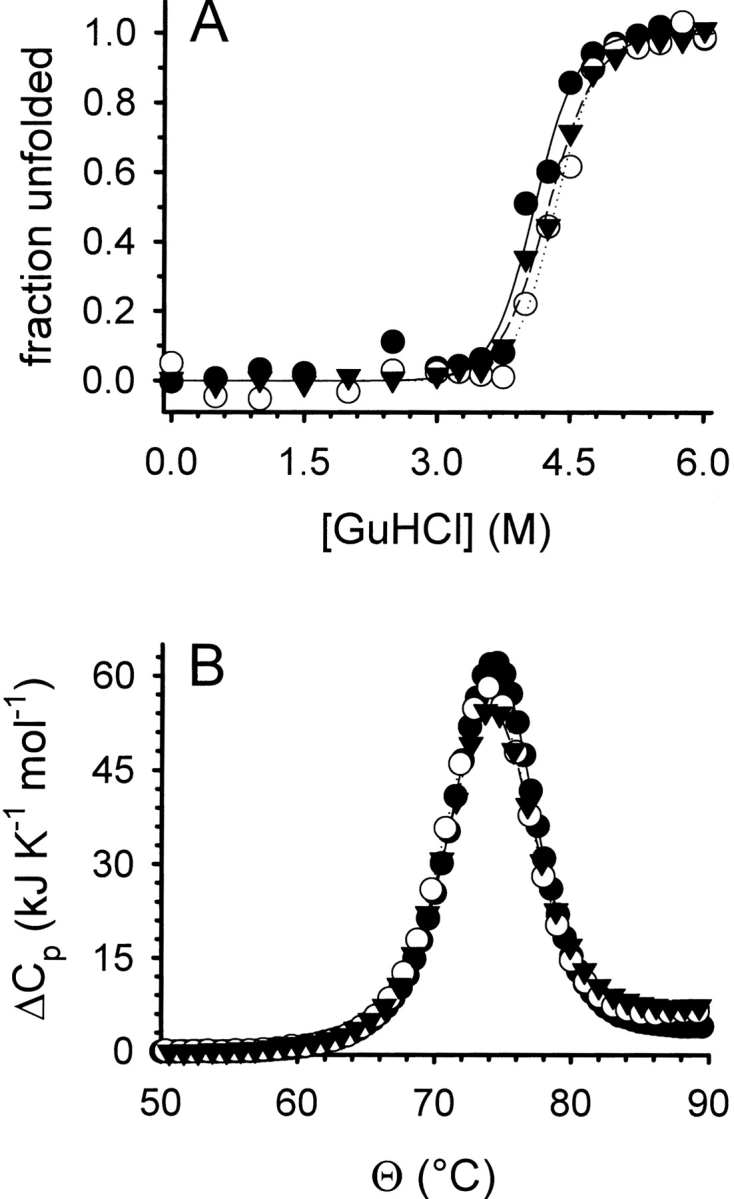
Denaturant-induced folding/unfolding (A) and thermal stability (B) of lysozyme. (A) The GuHCl-induced folding/unfolding equilibrium of lysozyme was monitored by tryptophan fluorescence at pH 7.0 without l-ArgHCl (filled circles), in the presence of 0.5 M l-ArgHCl (open circles), and of 1.0 M l-ArgHCl (filled triangles). Protein concentration was 20 μg mL−1. Data were normalized assuming a linear dependency of the intrinsic fluorescence for both the native and the denatured state. Lines represent fits to a two-state unfolding model. (B) The thermal unfolding of lysozyme was monitored by DSC at pH 6.0 without l-ArgHCl (filled circles), in the presence of 0.5 M l-ArgHCl (open circles), and of 1.0 M l-ArgHCl (filled triangles). Protein concentrations were 0.5 mg mL−1. Lines represent fits of the data to a two-state unfolding model.
Table 1.
Equilibrium stability data (A) for the guanidinium chloride-induced folding/unfolding and (B) for the thermal unfolding of lysozyme
| Aa | [l-ArgHCl] (M) | ΔGU0 (kJ mol−1) | meq (kJ mol−1 M−1) | [GuHCl]50 (M) |
| 0 | 44.2 ± 2.9 | 10.8 ± 2.4 | 4.0 ± 0.2 | |
| 0.5 | 43.2 ± 0.9 | 9.8 ± 0.8 | 4.4 ± 0.5 | |
| 1.0 | 54 ± 13 | 13 ± 3 | 4.0 ± 0.2 |
| Bb | [l-ArgHCl] (M) | Tm (°C) | ΔHTm (kJ mol−1) | |
| a Data represent error-weighted averages ± SD from four to seven independent experiments for each concentration of l-ArgHCl. | ||||
| b Data correspond to the traces shown in Figure 2B ▶. | ||||
| 0 | 74.3 | 490 | ||
| 0.5 | 73.7 | 469 | ||
| 1.0 | 73.9 | 452 | ||
Influence of l-ArgHCl on aggregation kinetics
l-ArgHCl neither caused a significant acceleration of the refolding kinetics of lysozyme nor an increase in its thermodynamic stability (see above). Therefore, the increase in the yield of the oxidative refolding of lysozyme must be due to the suppression of unproductive side reactions, namely the aggregation of folding intermediates into insoluble misfolded protein material. It has repeatedly been suggested that l-ArgHCl acts as a suppressor of aggregation in the renaturation of lysozyme (Hevehan and De Bernardez Clark 1997; Arakawa and Tsumoto 2003; Ho et al. 2003). Surprisingly, however, to our knowledge no systematic studies on the influence of l-ArgHCl on the kinetics of the aggregation of lysozyme have been published so far. As the aim of this work was to identify factors that are responsible for the effect of l-ArgHCl, we performed light-scattering measurements of the formation of aggregates under refolding conditions (Fig. 3 ▶). In the absence of l-ArgHCl, an almost instantaneous formation of microscopic aggregates was observed, as indicated by the jump in the light-scattering signal upon addition of denatured-reduced lysozyme, followed by a kinetics representing the growth of aggregates, and finally a decrease in the signal due to precipitation of macroscopic clots formed from the misfolded and aggregated protein material. These processes show a steep dependency on the protein concentration (Fig. 3A ▶). At all studied protein concentrations (up to 0.6 mg mL−1), increasing concentrations of l-ArgHCl were highly effective in suppressing the initial formation of microscopic aggregates and in delaying the formation of heavier, macroscopic clots (Fig. 3B ▶).
Figure 3.
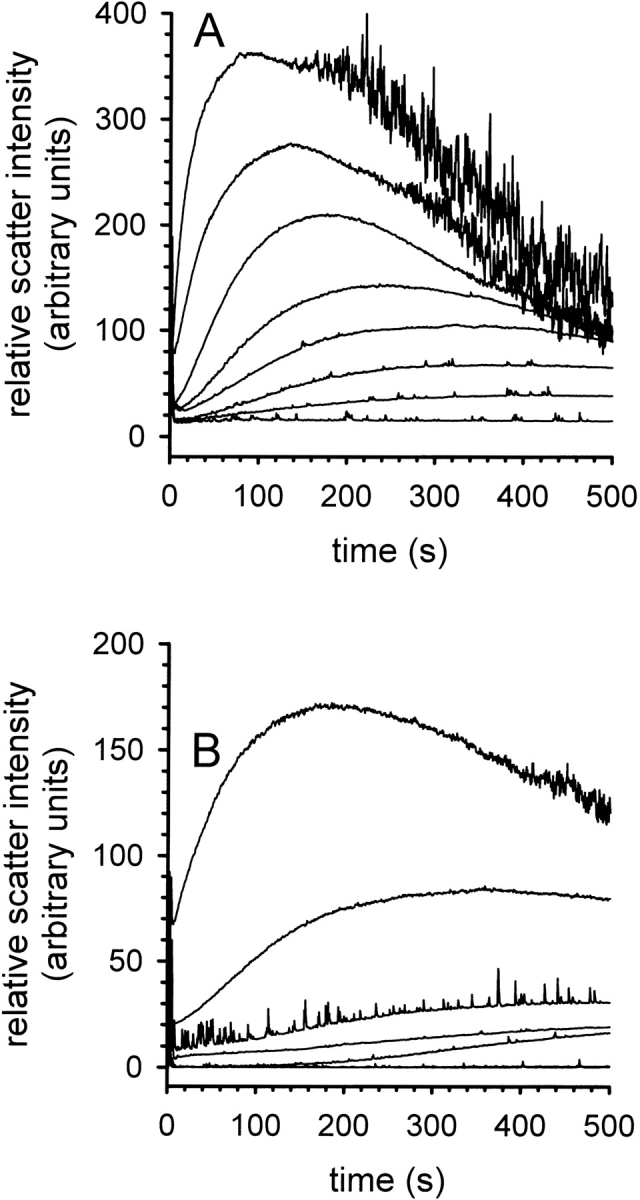
Kinetics of aggregation during the refolding of lysozyme. (A) Time-dependent formation of aggregates at increasing concentrations of denatured lysozyme in the presence of 50 mM l-ArgHCl. Lysozyme concentration was 0.1, 0.2, 0.25, 0.3, 0.35, 0.4, 0.5, and 0.6 mg mL−1 (bottom to top). (B) Time-dependent formation of aggregates in the presence of increasing concentrations of l-ArgHCl at a protein concentration of 0.3 mg mL−1. [l-ArgHCl] was 0, 0.05, 0.1, 0.15, 0.2, 0.25, and 0.4 M (top to bottom). The aggregation was monitored by light scattering at 600 nm.
Solubility of denatured lysozyme derivatives
We now wanted to address the question of whether the observed suppression of aggregation was due to purely kinetic effects or a thermodynamic stabilization of intermediates in solution. Therefore, we measured the solubility of denatured-reduced lysozyme whose cysteines had been blocked by alkylation with iodoacetamide or iodoacetic acid, to prevent refolding. The corresponding equilibrium solubilities under refolding conditions (0.1 M Tris-HCl, 1 mM EDTA [pH 8.5]) were determined as a function of the concentration of l-ArgHCl and NaCl (Fig. 4 ▶). For both forms of denatured lysozyme, the presence of NaCl at moderate to high ionic strengths resulted in solubilities below 25 μg mL−1, while l-ArgHCl had a markedly different effect. Increasing concentrations of l-ArgHCl up to 1.0 M increased the equilibrium solubility up to 0.19 ± 0.05 mg mL−1 for the iodoacetamide-modified form (Fig. 4A ▶) and to 0.8 ± 0.3 mg mL−1 for the iodoacetic acid-modified form (Fig. 4B ▶). Similar results were obtained for denatured-reduced lysozyme and for its mixed disulfide with glutathione (not shown). The observed increase in the solubility of the various denatured forms of lysozyme runs in parallel to the increase in renaturation yield in this concentration range (Fig. 1B ▶), and to the suppression of the unproductive formation of insoluble aggregated protein by l-ArgHCl (Fig. 3B ▶). An increased equilibrium solubility of unfolded species and intermediates seems to be the major cause for the effect of l-ArgHCl as a refolding enhancer.
Figure 4.
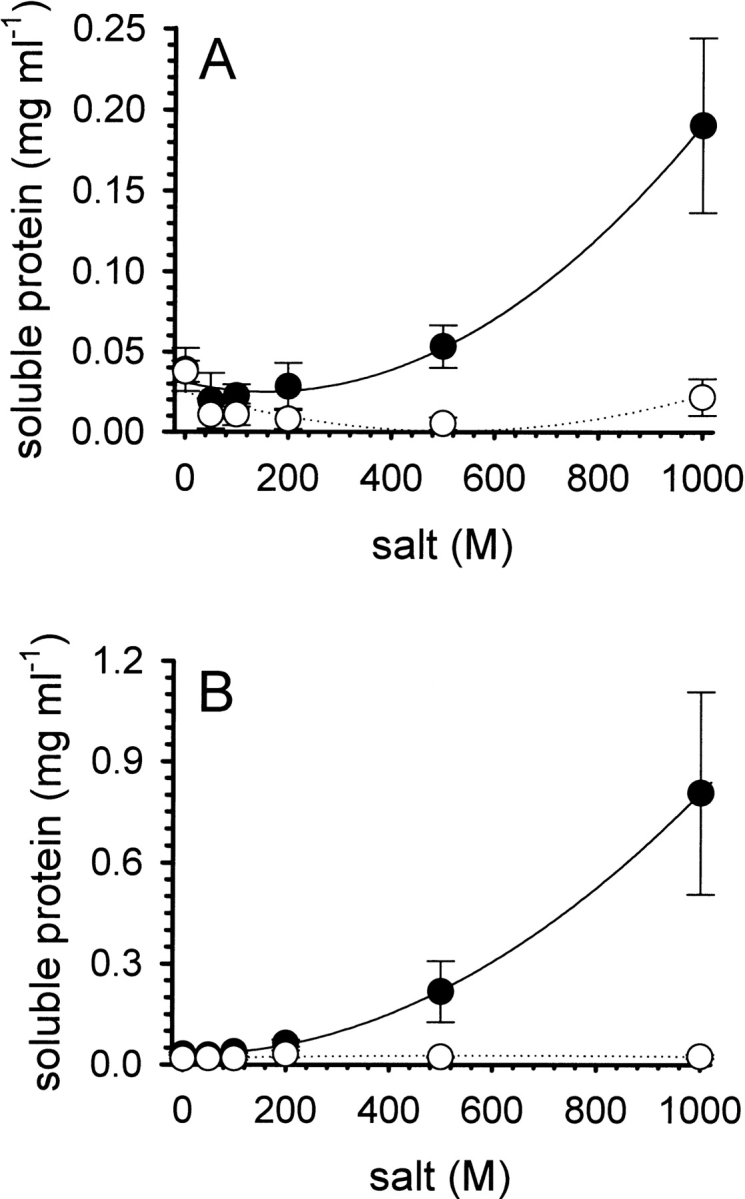
Solubility of iodoacetamide-modified (A) and iodoacetic acid-modified (B) denatured lysozyme. Preparations of the modified forms of denatured lysozyme (protein concentration ∼5 mg mL−1) were dialyzed overnight at room temperature against buffer at pH 8.5, containing increasing amounts of l-ArgHCl (filled circles) or NaCl (open circles). The concentrations of soluble protein in the supernatant were determined after centrifugation, as described in the text. Data represent mean values ± SD from four independent experiments. Lines are meant to guide the eye.
Discussion
In agreement with earlier results (Hevehan and De Bernardez Clark 1997), we observed an increase in the yield of oxidative refolding of lysozyme in the presence of increasing concentrations of l-ArgHCl. An interesting observation in this respect was that the apparent first-order rate constant for refolding was decreasing in parallel to the increasing refolding yield. This is consistent with earlier calculations (Kiefhaber et al. 1991; Hevehan and De Bernardez Clark 1997) that model the general kinetics of oxidative protein refolding as a competition between a first-order folding reaction and an unproductive aggregation process of higher order. These calculations do of course refer to the kinetics of rate-determining steps. It was shown by Goldberg et al. (1991) that the recovery of lysozyme activity closely follows the rate-determining step of refolding. Therefore, the observed decrease in the apparent refolding rate clearly implies that the effect of l-ArgHCl cannot be explained by an acceleration of the kinetics along the protein’s productive refolding pathway. Unaltered kinetics of refolding combined with a decrease in the rates of unproductive side reactions, however, provide for a satisfactory explanation of our observations.
A further finding of our study was that the difference in free energy between native and unfolded lysozyme in solution was, under the applied conditions, not significantly affected by the presence of l-ArgHCl (Fig. 2 ▶). Thus, a relative thermodynamic stabilization of the native state of lysozyme can be ruled out as major cause for the enhanced refolding yield. Differential interactions of l-ArgHCl with the native versus the unfolded protein must be balanced in such a way that their energetic contributions almost cancel out. Energetic contributions of a cosolvent toward the stability of a protein in solution arise, on the one hand, from the cohesive effect that the cosolvent exerts on water molecules (as manifest, e.g., in changes of the surface tension of the solution), and, on the other hand, from direct interactions between the cosolvent and the polypeptide chain (Kita et al. 1994; Schellman 2003; Shimizu and Smith 2004). Increasing concentrations of l-ArgHCl increase the surface tension of its aqueous solutions. Based on this effect alone, l-ArgHCl would be expected to promote the stability of the native state of proteins, similar to polyalcohols and sugars (Lee and Timasheff 1981; Xie and Timasheff 1997). Additionally however, l-ArgHCl, as opposed to other amino acid salts, shows considerable contributions from direct interactions (Kita et al. 1994). It is, in this respect, similar to the denaturing agents urea and GuHCl, and it might be assumed that its guanidino group is largely responsible for the strength of these interactions. Favorable direct interactions of a cosolvent with the functional groups of a polypeptide chain are generally destabilizing in nature, as they favor the solvent exposure of sites that are buried in the native state of a protein.
However, these interactions may be expected to play a central role for the usefulness of a cosolvent as a refolding enhancer. The aggregation of denatured protein is the main unproductive side reaction in protein renaturation, and an effective refolding enhancer as l-ArgHCl successfully suppresses this pathway (Fig. 3 ▶). Favorable interactions of lArg with specific functional groups of the polypeptide chain interactions destabilize the native form of the protein, but on the other hand, they lower the energetic cost of exposure of extended conformations of the polypeptide chain, and thus stabilize denatured conformations with respect to aggregation and precipitation. This should be manifest in an increased equilibrium solubility of denatured protein, misfolded conformations, and folding intermediates, as confirmed by the experimental results of this work (Fig. 4 ▶). Besides the ability of the guanidino group of l-Arg to act as a hydrogen bond donor, charge–charge interactions with the polypeptide chain may play an important role. The more negatively charged iodoacetic acid-modified lysozyme (Fig. 4B ▶) (as well as its mixed disulfide with glutathione) showed higher solubilities than the iodoacetamide-modified form (Fig 4A ▶).
Generally speaking, when using low molecular weight compounds as cosolvents in refolding reactions, the task is to strike a balance between, on the one hand, preserving the relative stability of the native state and, on the other hand, stabilizing denatured polypeptides and intermediates in solution in order to prevent them from following the path down to aggregation. l-Arg, with its various functional groups, seems to be very well balanced in that respect.
Materials and methods
Materials
Hen egg white lysozyme (EC 3.2.1.17, lot K23837881-809, 100,000 units mg−1) was purchased from Merck KGaA. Ultrapure guanidinium hydrochloride (GuHCl), Tris base, dithiothreitol (DTT), and electrophoresis grade EDTA were from ICN Biomedicals, Inc. l-Arginine hydrochloride (l-ArgHCl) was from Ajinomoto Co., Inc. Lyophilized cells of Micrococcus lysodeikticus were purchased from Sigma-Aldrich Chemie GmbH. All reagents were of analytical grade or of higher purity.
Protein concentrations
Lysozyme concentrations were determined photometrically using an extinction coefficient3280 of 2.63 mL mg −1 cm−1 for native and 2.37 mL mg−1 cm−1 for denatured lysozyme (Saxena and Wetlaufer 1970). The value for denatured lysozyme was also used for its derivatives. In some instances, determined protein concentrations were cross-checked by measuring tryptophan fluorescence according to Pajot (1976).
Oxidative refolding of lysozyme
Lysozyme was denatured by incubating 20 mg mL−1 of the native protein in denaturation buffer (6 M GuHCl, 100 mM DTT, 1 mM EDTA, 0.1 M Tris/HCl [pH 8.5]) for 2 h at room temperature. The solution was adjusted to pH 4.0 with 1 M HCl and dialyzed two times, for at least 4 h each, at 4°C against 100 volumes of a stirred solution of 4 M GuHCl, 1 mM EDTA, 50 mM acetic acid/NaOH (pH 4.5), to yield a stock solution of denatured-reduced lysozyme.
Oxidative refolding was initiated by rapid mixing of denatured lysozyme with 60 volumes of degassed renaturation buffer (3 mM GSH, 0.3 mM GSSG, 1 mM EDTA, 0.1 M Tris/HCl [pH 8.2]), containing 0 to 0.9 M l-arginine, under a nitrogen atmosphere at room temperature. The final concentration of lysozyme was 250 μg mL−1. Samples were drawn at times ranging from 0 to 180 min after initiation of renaturation and analyzed for lysozyme activity.
Lysozyme activity assay
Lysozyme activity was assayed by measuring the lysis of Micrococcus lysodeikticus (75 μg mL−1 lyophilized cells in 66 mM sodium phosphate buffer [pH 6.2], 25°C). Lysozyme was added to the stirred assay mixture, and activities were determined by measuring the decrease in turbidity at 450 nm over the initial 30 sec. Activities are reported as percent renaturation yield, referring to the activity of an equal concentration of native lysozyme under the same conditions.
Denaturant-dependent stability experiments
The denaturant-dependent unfolding of lysozyme was monitored by tryptophan fluorescence. Equilibrium stability experiments were performed for both unfolding and refolding by diluting concentrated native lysozyme or completely unfolded protein (in 6 M GuHCl) into buffer series containing 50 mM HEPES/NaOH (pH 7.0), and GuHCl and l-ArgHCl in the indicated concentrations. The final protein concentration was 20 μg mL−1. The samples were equilibrated for 12 h at 20°C before the fluorescence measurements were carried out. The excitation wavelength was set to 280 nm and the fraction of unfolded protein was calculated from the change in fluorescence intensity at 360 nm.
Differential scanning calorimetry
Differential scanning calorimetry (DSC) experiments were carried out with a VP-DSC instrument (Microcal, LLC). For the measurements, native lysozyme at a concentration of 2 mg mL−1 was dialyzed against buffers containing 11 mM sodium citrate (pH 6.0), 1 mM EDTA, and l-ArgHCl in the indicated concentrations. Degassed dialysis buffer was used to fill the instrument and to record the baseline. Prior to loading into the sample cell, the protein solutions were diluted with the respective dialysis buffer to a concentration of ∼0.5 mg mL−1 and degassed. Temperature scans were performed from 10°C to 100°C with a scan rate of 90 K h−1. All samples were found to show a reversible temperature transition. After transformation of the heat capacity data from time-based to temperature-based and subtracting the buffer/buffer baselines, the corrected DSC traces were fitted to a two-state model using Origin 7.0 software (OriginLab Corp.).
Aggregation kinetics
The formation of aggregates during the oxidative refolding of lysozyme was monitored by measuring the intensity of scattered light at 600 nm in an Hitachi F-4500 fluorescence spectrophotometer. In these experiments the oxidative refolding of lysozyme was carried out at 20°C in the presence of a constant final concentration of 0.3 M GuHCl carried over from the denatured protein solution.
Protein solubility
Denatured-reduced lysozyme was prepared as described above at a protein concentration of 50 mg mL−1, exhaustively dialyzed three times at 4°C against 100 volumes of 10 mM HCl and centrifuged at 15,000g for 30 min to remove any precipitate. Mixed disulfide was prepared by incubating reduced lysozyme over night at 20°C in 6 M GuHCl, buffered with 0.1 M Tris-HCl (pH 9.0) and containing a mixture of 0.1 mM GSH and 250 mM GSSG. This preparation was dialyzed against 10 mM HCl and cleared by centrifugation. Iodoacetamide-modified and iodoacetic acid-modified lysozyme were prepared by diluting denatured-reduced lysozyme into 4 volumes of 6 M GuHCl, 1 mM EDTA, 0.1 M boric acid/NaOH (pH 9.0), and 12.5 mM of iodoacetamide or iodoacetic acid. The reaction mixture was incubated at room temperature for 1 h, exhaustively dialyzed against 10 mM HCl, and cleared by centrifugation.
For the solubility experiments, ∼1 mL of the preparations were dialyzed overnight at room temperature against 100 mL of the indicated buffers containing l-ArgHCl or NaCl. The formed precipitates were pelleted by centrifugation for 30 min at 15,000g and the concentration of soluble protein in the supernatant was determined as described above, using the extinction coefficient of denatured lysozyme.
Acknowledgments
We thank Mrs. Renate Nitsch and Mr. Jörg Schildhauer for their excellent technical assistance. R.C.R. wishes to express his gratitude toward Dr. A.M. Kayastha, School of Biotechnology, Banaras Hindu University, India, for his continuous help and support. This work was supported by the Federal State (Land) of Saxony-Anhalt (grant no. 3537 A/0903 L).
Abbreviations
DTT, dithiothreitol
DSC, differential scanning calorimetry
GSH, reduced glutathione
GSSG, oxidized glutathione
GuHCl, guanidinium chloride
l-Arg, l-arginine
l-ArgHCl, l-arginine monohydrochloride
[x], concentration of compound x
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.041085005.
References
- Acharya, A.S. and Taniuchi, H. 1982. Implication of the structure and stability of disulfide intermediates of lysozyme on the mechanism of renaturation. Mol. Cell. Biochem. 44 129–148. [DOI] [PubMed] [Google Scholar]
- Arakawa, T. and Tsumoto, K. 2003. The effects of arginine on refolding of aggregated proteins: Not facilitate refolding, but suppress aggregation. Biochem. Biophys. Res. Commun. 304 148–152. [DOI] [PubMed] [Google Scholar]
- Armstrong, N., de Lencastre, A., and Gouaux, E. 1999. A new protein folding screen: Application to the ligand binding domains of a glutamate and kainate receptor and to lysozyme and carbonic anhydrase. Protein Sci. 8 1475–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano, R., Kudo, T., Makabe, K., Tsumoto, K., and Kumagai, I. 2002. Antitumor activity of interleukin-21 prepared by novel refolding procedure from inclusion bodies expressed in Escherichia coli. FEBS Lett. 528 70–76. [DOI] [PubMed] [Google Scholar]
- Brinkmann, U., Buchner, J., and Pastan, I. 1992. Independent domain folding of Pseudomonas exotoxin and single-chain immunotoxins: Influence of inter-domain connections. Proc. Natl. Acad. Sci. 89 3075–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner, J. and Rudolph, R. 1991. Renaturation, purification and characterization of recombinant Fab-fragments produced in Escherichia coli. Biotechnology 9 157–162. [DOI] [PubMed] [Google Scholar]
- De Bernardez Clark, E., Schwarz, E., and Rudolph, R. 1999. Inhibition of aggregation side reactions during in vitro protein folding. Methods Enzymol. 309 217–237. [DOI] [PubMed] [Google Scholar]
- Fahnert, B., Lilie, H., and Neubauer, P. 2004. Inclusion bodies: Formation and utilisation. Adv. Biochem. Eng. Biotechnol. 89 93–142. [DOI] [PubMed] [Google Scholar]
- Goldberg, M.E., Rudolph, R., and Jaenicke, R. 1991. A kinetic study of the competition between renaturation and aggregation during the refolding of denatured-reduced egg white lysozyme. Biochemistry 30 2790–2797. [DOI] [PubMed] [Google Scholar]
- Hevehan, D.L. and De Bernardez Clark, E. 1997. Oxidative renaturation of lysozyme at high concentrations. Biotechnol. Bioeng. 54 221–230. [DOI] [PubMed] [Google Scholar]
- Ho, J.G.S., Middelberg, A.P.J., Ramage, P., and Kocher, H.P. 2003. The likelihood of aggregation during protein renaturation can be assessed using the second virial coefficient. Protein Sci. 12 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefhaber, T., Rudolph, R., Kohler, H.-H., and Buchner, J. 1991. Protein aggregation in vivo: A quantitative model of the kinetic competition between folding and aggregation. Biotechnology 9 825–829. [DOI] [PubMed] [Google Scholar]
- Kita, Y., Arakawa, T., Lin, T.-Y., and Timasheff, S.N. 1994. Contribution of the surface free energy perturbation to protein–solvent interactions. Biochemistry 33 15178–15189. [DOI] [PubMed] [Google Scholar]
- Kudou, M., Shiraki, K., Fujiwara, S., Imanaka, T., and Takagi, M. 2003. Prevention of thermal inactivation and aggregation of lysozyme by polyamines. Eur. J. Biochem. 270 4547–4554. [DOI] [PubMed] [Google Scholar]
- Krause, M., Rudolph, R., and Schwarz, E. 2002. The non-ionic detergent Brij 58P mimics chaperone effects. FEBS Lett. 532 253–255. [DOI] [PubMed] [Google Scholar]
- Lange, C. and Rudolph, R. 2005. Production of recombinant proteins for therapy, diagnostics and industrial research by in vitro folding. In Protein folding handbook (eds. T. Kiefhaber and J. Buchner), pp. 1215–1250. Wiley-VCH, Weinheim, Germany.
- Lee, J.C. and Timasheff, S.N. 1981. The stabilization of proteins by sucrose. J. Biol. Chem. 256 7193–7201. [PubMed] [Google Scholar]
- Lin, T.-Y. and Timasheff, S.N. 1996. On the role of surface tension in the stabilization of globular proteins. Protein Sci. 5 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelis, U., Rudolph, R., Jarsch, M., Kopetzki, E., Burtscher, H., and Schumacher, G. 1995. Process for the production and renaturation of recombinant, biologically active, eukaryotic alkaline phosphatase. U.S. Patent 5,434,067.
- Oneda, H. and Inouye, K. 1999. Refolding and recovery of recombinant human matrix metalloproteinase 7 (matrilysin) from inclusion bodies expressed by Escherichia coli. J. Biochem. 126 905–911. [DOI] [PubMed] [Google Scholar]
- Orsini, G. and Goldberg, M.E. 1978. The renaturation of reduced chymotrypsinogen A in guanidine HCl. J. Biol. Chem. 253 3453–3458. [PubMed] [Google Scholar]
- Pajot, P. 1976. Fluroescence of proteins in 6-M guanidine hydrochloride. A method for the quantitative determination of tryptophan. Eur. J. Biochem. 63 263–269. [DOI] [PubMed] [Google Scholar]
- Radford, S.E., Dobson, C.M., and Evans, P.A. 1992. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature 358 302–307. [DOI] [PubMed] [Google Scholar]
- Rattenholl, A., Lilie, H., Grossmann, A., Stern, A., Schwarz, E., and Rudolph, R. 2001. The pro-sequence facilitates folding of human nerve growth factor from Escherichia coli inclusion bodies. Eur. J. Biochem. 268 3296–3303. [DOI] [PubMed] [Google Scholar]
- Roux, P., Ruoppolo, M., Chaffotte, A., and Goldberg, M.E. 1999. Comparison of kinetics of S-S bond, secondary structure, and active site formation during refolding of reduced denatured hen egg white lysozyme. Protein Sci. 8 2751–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, R. and Fischer, S. 1990. Process for obtaining renatured proteins. U.S. Patent 4,933,434.
- Samuel, D., Kumar, T.K.S., Ganesh, G., Jayaraman, G., Yang, P.-W., Chang, M.-M., Trivedi, V.D., Wang, S.-L., Hwang, K.-C., Chang, D.-K., et al. 2000. Proline inhibits aggregation during protein refolding. Protein Sci. 9 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano, H., Koumoto, Y., Ohta, K., Sasaki, Y., Segawa, S., and Tachibana, H. 1992. Efficient in vitro folding of the three-disulfide derivatives of hen lysozyme in the presence of glycerol. FEBS Lett. 303 11–14. [DOI] [PubMed] [Google Scholar]
- Saxena, V.P. and Wetlaufer, D.B. 1970. Formation of three-dimensional structure in proteins. I. Rapid nonenzymatic reactivation of reduced lysozyme. Biochemistry 9 5015–5022. [DOI] [PubMed] [Google Scholar]
- Schellman, J.A. 2003. Protein stability in mixed solvents: A balance of contact interaction and excluded volume. Biophys. J. 85 108–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, S. and Smith, D.J. 2004. Preferential hydration and the exclusion of cosolvents from protein surfaces. J. Chem. Phys. 121 1148–1154. [DOI] [PubMed] [Google Scholar]
- Shiraki, K., Kudou, M., Fujiwara, S., Imanaka, T., and Takagi, M. 2002. Biophysical effect of amino acids on the prevention of protein aggregation. J. Biochem. 132 591–595. [DOI] [PubMed] [Google Scholar]
- Suenaga, M., Ohmae, H., Tsuji, S., Itoh, T., and Nishimura, O. 1998. Renaturation of recombinant human neurotrophin-3 from inclusion bodies using a suppressor agent of aggregation. Biotechnol. Appl. Biochem. 28 119–124. [PubMed] [Google Scholar]
- Tandon, S. and Horowitz, P. 1988. The effects of lauryl maltoside on the reactivation of several enzymes after treatment with guanidinium chloride. Biochim. Biophys. Acta 955 19–25. [DOI] [PubMed] [Google Scholar]
- Taneja, S. and Ahmad, F. 1994. Increased thermal stability of proteins in the presence of amino acids. Biochem J. 303 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsumoto, K., Shinoki, K., Kondo, H., Uchikawa, M., Juji, T., and Kumagai, I. 1998. Highly efficient recovery of functional single-chain Fv fragments from inclusion bodies overexpressed in Escherichia coli by controlled introduction of oxidizing reagent—application to a human single-chain Fv fragment. J. Immunol. Methods 219 119–129. [DOI] [PubMed] [Google Scholar]
- Tsumoto, K., Ejima, D., Kumagai, I., and Arakawa, T. 2003. Practical considerations in refolding proteins from inclusion bodies. Protein Expr. Purif. 28 1–8. [DOI] [PubMed] [Google Scholar]
- Wetlaufer, D.B. and Xie, Y. 1995. Control of aggregation in protein refolding: A variety of surfactants promote renaturation of carbonic anhydrase II. Protein Sci. 4 1536–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, G. and Timasheff, S.N. 1997. The thermodynamic mechanism of protein stabilization by trehalose. Biophys Chem. 64 25–43. [DOI] [PubMed] [Google Scholar]


