Abstract
Background
Hydrophobins are proteins containing eight conserved cysteine residues that occur uniquely in mycelial fungi. Their main function is to confer hydrophobicity to fungal surfaces in contact with air or during attachment of hyphae to hydrophobic surfaces of hosts, symbiotic partners or themselves resulting in morphogenetic signals. Based on their hydropathy patterns and solubility characteristics, hydrophobins are divided into two classes (I and II), the latter being found only in ascomycetes.
Results
We have investigated the mechanisms driving the evolution of the class II hydrophobins in nine species of the mycoparasitic ascomycetous genus Trichoderma/Hypocrea, using three draft sequenced genomes (H. jecorina = T. reesei, H. atroviridis = T. atroviride; H. virens = T. virens) an additional 14,000 ESTs from six other Trichoderma spp. (T. asperellum, H. lixii = T. harzianum, T. aggressivum var. europeae, T. longibrachiatum, T. cf. viride). The former three contained six, ten and nine members, respectively. Ten is the highest number found in any ascomycete so far. All the hydrophobins we examined had the conserved four beta-strands/one helix structure, which is stabilized by four disulfide bonds. In addition, a small number of these hydrophobins (HFBs)contained an extended N-terminus rich in either proline and aspartate, or glycine-asparagine. Phylogenetic analysis reveals a mosaic of terminal clades containing duplicated genes and shows only three reasonably supported clades. Calculation of the ratio of differences in synonymous vs. non-synonymous nucleotide substitutions provides evidence for strong purifying selection (KS/Ka >> 1). A genome database search for class II HFBs from other ascomycetes retrieved a much smaller number of hydrophobins (2–4) from each species, and most were from Sordariomycetes. A combined phylogeny of these sequences with those of Trichoderma showed that the Trichoderma HFBs mostly formed their own clades, whereas those of other Sordariomycetes occurred in shared clades.
Conclusion
Our study shows that the genus Trichoderma/Hypocrea has a proliferated arsenal of class II hydrophobins which arose by birth-and-death evolution followed by purifying selection.
Background
Hydrophobins are small proteins that are unique for mycelial fungi [1-4]. Their core structure consists of four beta strands, crosslinked by four disulfide bridges [5,6], creating a structure enabling the self-assembly at a hydrophilic/hydrophobic interface between the hydrophilic cell wall and a hydrophobic environment (such as air or the hydrophobic surface of living and non-living material). They are classified on the basis of chemical properties (hydrophobicity, solubility) into class I or class II hydrophobins [1], with class I hydrophobins having been identified in both ascomycetes and basidiomycetes, and class II hydrophobins having so far been only detected in ascomycetes [4]. Hydrophobins can fulfill a plethora of biological functions ranging from the formation of aerial structures, elicitation of morphogenesis and interaction with potential hosts or symbiotic partners [2,7-10].
In the primary sequence, the most important feature common to all hydrophobins is the characteristic pattern of eight Cys-residues, which gives rise to a common disulfide network [5,11]. Besides these conserved Cys-residues and similar hydropathy patterns, however, the hydrophobins share only a few conserved residues [4]. The poor amino acid sequence conservation of hydrophobins raises the question as to the evolutionary mechanism driving the rapid differentiation of hydrophobin gene sequences. Other genes involved in the response of organisms to their immediate environment have sometimes been shown to be driven by positive selection [12-16], so called "arms races" [17]. However, concerted evolution and birth-and-death evolution under strong purifying selection have also been reported [18-24].
The fungal genus Trichoderma/Hypocrea contains a large number of mycoparasitic species [25,26], and some of them (e.g. H. lixii = T. harzianum, H. virens = T. virens, H. atroviridis = T. atroviride, T. asperellum) are also commercially used as biological fungicides [27]. Interestingly, there are only a few reports describing the occurrence of hydrophobins in mycoparasitic strains of Trichoderma/Hypocrea [28-30] and a characterization of the function or biological role for each has not been determined. The class II hydrophobin genes hfb1 and hfb2 of the weakly mycoparasitic species H. jecorina (T. reesei) have been studied [31,32] and shown to serve different functions during vegetative development. Viterbo and Chet [33] recently showed that a class I hydrophobin from T. asperellum is involved in root colonization. Interestingly, this hydrophobin is the only class I hydrophobin identified in any Trichoderma/Hypocrea sp. so far.
The assignment of a function to individual members of large gene families like the hydrophobins is complicated by the possibility that several of them may have overlapping functions [34,35], which in turn is dependent on the selective pressures acting on the organism. Understanding the evolution of such genes and identifying stable clusters within the phylogeny may therefore help illustrate members with a potentially critical function.
Results
Protein structure of the Trichoderma class II hydrophobin proteins
In order to have a representative sample of class II HFBs from Trichoderma/Hypocrea, we first screened the available genome sequences of H. jecorina, H. atroviridis and H. virens, retrieving 6, 10 and 9 genes encoding class II proteins, respectively. Second, we searched NCBI and identified one HFB from H. lixii (= T. harzianum) strain T-22 (HL_4). We also included the hydrophobin-like protein QID3 from H. lixii strain CECT 2413 [36] in the analysis. A third hydrophobin – srh1 – from "T. harzianum" [28] was also included in this study, but as this strain had been misidentified and is in fact H. atroviridis [37] it turned out to be identical to HA_2a (see Table 1). Third, we screened the TrichoEST database [38] which – besides containing ESTs of H. atroviridis and H. virens – contains transcript sequences from additional five Trichoderma/Hypocrea species (H. lixii, T. aggressivum var. europeae, T. longibrachiatum, T. stromaticum, T. asperellum and T. cf. viride), resulting in 15 further HFBs. The identity of "T. cf. viride"was rechecked on the basis of ESTs for elongation factor 1 alpha (tef1) and RNA polymerase subunit B (rpb2) and determined to be closest to T. koningiopsis, and we will therefore name this strain T. cf. koningiopsis throughout this study. Our sample consisted of 42 class II HFBs from 9 different species of Trichoderma, covering sections Longibrachiatum, Trichoderma and Pachybasium (cf. Table 1), and thus consisting of a well distributed sample.
Table 1.
Trichoderma/Hypocrea class II hydrophobin genes*
| Trichoderma Section | Trichoderma/Hypocrea spp. | abbreviation | Scaffold | accession no. |
|---|---|---|---|---|
| Longibrachiatum | H. jecorina | HFB6 | 3:1189832–1190084 | |
| H. jecorina | HFB3 | 31:136511–136957 | ||
| H. jecorina | HFB5 | 11:163081–163444 | ||
| H. jecorina | HFB1 | 3:1189832–1190084 | ||
| H. jecorina | HFB2 | 56:80872–81271 | ||
| H. jecorina | HFB4 | 5:390006–390436 | ||
| T. longibrachiatum | TL_1 | L19T52P004R01376 | [GenBank: AJ905782] | |
| Trichoderma | T. asperellum | TA_1 | L14T53P124R00732 | [GenBank: AJ903054] |
| T. asperellum | TA_2 | L14T53P129R00833 | [GenBank: AJ903147] | |
| T. asperellum | TA_3 | L14T53P116R00634 | [GenBank: AJ902899] | |
| T. asperellum | TA_4 | L14T53P135R01371 | [GenBank: AJ903666] | |
| H. atroviridis | HA_1b | 1:719649–719242 | [GenBank: EU053447] | |
| H. atroviridis | HA_1c | 1:1159027–1159456 | GenBank: EU053448] [GenBank: EU053449]; | |
| H. atroviridis | HA_2a = SRH1 | 2:2051503–2051110 | [GenBank: CAA72539] | |
| H. atroviridis | HA_2b | 2:2503511–2503091 | [GenBank: EU053450] | |
| H. atroviridis | HA_2c | 2:3431445–3432224 | [GenBank: EU053456] | |
| H. atroviridis | HA_5a | 5:343144–342721 | [GenBank: EU053451] | |
| H. atroviridis | HA_6a | 6:627631–626945 | [GenBank: EU053452] | |
| H. atroviridis | HA_6b | 6:738694–738316 | [GenBank: EU053453] | |
| H. atroviridis | HA_6c | 6:1048590–1048979 | [GenBank: EU053454] | |
| H. atroviridis | HA_22a | 22:79408–78987 | [GenBank: EU053455] | |
| T. cf. Koningiopsis | TCK_1 | L21T78P020R01908 | [GenBank: AJ909436] | |
| T. cf. Koningiopsis | TCK_2 | L21T78P014R01340 | [GenBank: EV554903] | |
| T. cf. Koningiopsis | TCK_3 | L21T78P012R01144 | [GenBank: EV554904] | |
| Pachybasium | H. virens | HV_1a | 1:2048115–2048517 | [GenBank: EU053457] |
| H. virens | HV_1b | 1:2185848–2186274 | [GenBank: EU053458] | |
| H. virens | HV_1c | 1:1718557–1718103 | [GenBank: EU053459] | |
| H. virens | HV_1d | 1:909158–909955 | [GenBank: EU053460] | |
| H. virens | HV_2a | 2:562868–563256 | [GenBank: EU053461] | |
| H. virens | HV_13a | 13:955443–955019 | [GenBank: EU053462] | |
| H. virens | HV_18a | 18:175336–174917 | [GenBank: EU053463] | |
| H. virens | HV_21a | 21:388893–389498 | [GenBank: EU053464] | |
| H. virens | HV_22a | 22:232164–232586 | [GenBank: EU053465] | |
| H. lixii | HL_1 | L03T34P016R01491 | [GenBank: AJ896766] | |
| H. lixii | HL_2 | L02T34P126R11028 | [GenBank: AJ896364] | |
| H. lixii | HL_3 | L03T34P047R04364 | [GenBank: AJ897108] | |
| H. lixii | QID3 | [GenBank: X71913.1] | ||
| H. lixii | HL_4 | [GenBank: ABN64104] | ||
| T. aggressivum var. europeae | TAE_1 | L50TH2P009R00852 | [GenBank: ES768856] | |
| T. aggressivum var. europeae | TAE_2 | L50TH2P018R01702 | [GenBank: ES768855] | |
| T. aggressivum var. europeae | TAE_3 | L50TH2P001R00008 | [GenBank: AJ904501] | |
| T. stromaticum | TS_1 | L55TSTP002R00109 | [GenBank: ES768859] |
No accession numbers are given for H. jecorina, because its genome genome is available online; genome and EST data were obtained from the following strains: H. jecorina QM6a; H. atroviridis IMI 206040; H. virens, Gv 29-8; T. asperellum T53, T. longibrachiatum T52; T. aggressivum var. europeae CBS 453.93 H. lixii CECT 2413; T. cf. koningiopsis T78; T. stromaticum CBS 101875.
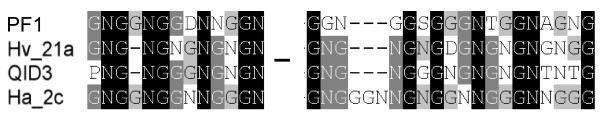
Prediction of the encoded protein sequences showed that most of the predicted HFBs had the expected structure of 90 – 110 amino acids, which includes a 15–20 aa signal peptide, the 65 aa core structure displaying the eight cysteines which are predicted to have four 4 beta-strands and a single helix. However, five of them (one from H. jecorina; HFB6) and two each from H. atroviridis (Ta_2c and Ta_6a) and H. virens (Tv_1d and Tv_21a) contained also an additional N-terminal segment of 64 – 133 amino acids which was characteristically rich in P, G and N/D, and for which no secondary structure could be predicted with certainty. This extended N-terminus is similar to the one found in the T. harzianum pseudohydrophobin QID3, in which one conserved cysteine residue is replaced by a serine [36] and indicates that it is a general feature of a small group of Trichoderma class II hydrophobins. Moreover, within a subgroup of them, a characteristic G xN repeat was found to be conserved (Fig. 1).
Figure 1.
Amino acid alignment of a portion of the extended N-terminus of QID3, Ha_2c, Hv_21a and the Passalora fulva hydrophobin PF1 (for accession number see Table 4). The gap (indicated by a "-") is of different length in the four sequences and therefore not shown. Black background indicates absolutely conserved amino acids. Grey background indicates conservation in at least three of the four proteins.
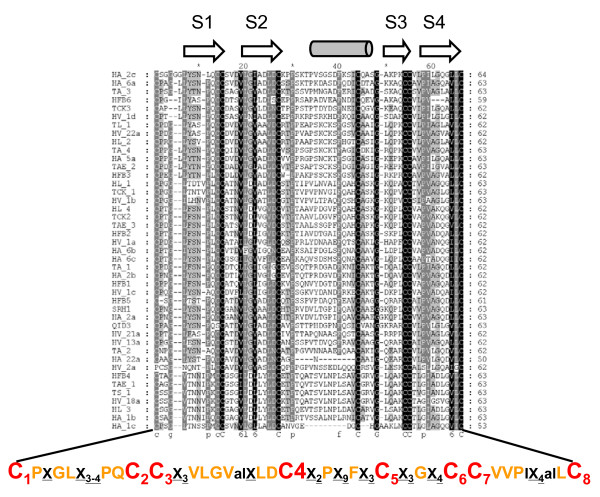
Fig. 2 shows the aa-alignment of the Trichoderma class II hydrophobins. Most of the conserved residues are located in the four β-strands and around the conserved cysteines. The aa's forming the α-helix, in contrast, showed very little conservation. Two of the HFBs, i.e. Ta_1c and Ta_22a, had part of the helical domain deleted and likely represent pseudogenes. This hypothesis is also supported by our lack of finding transcripts of these two HFBs in a total of 40.000 ESTs from different stages of H. atroviridis development (C.P. Kubicek and S.E. Baker, unpublished data). However they otherwise showed the conserved amino acid sequence pattern typical for class II HFBs, and were therefore retained in the analysis. The alignment led to the identification of 32 aa (of a total of 64 aa) that were functionally conserved (Fig. 2).
Figure 2.
Amino acid alignment of the class II hydrophobins of Trichoderma/Hypocrea used in this study. The aa sequences were trimmed to show only the area from the first to the eight cysteine. Absolutely conserved aa's are within a black background, and functionally conserved aa's highlighted in grey. The symbols and letters over the alignment show the position of the four beta-strands (S1–S4) and the single helix (indicated by a horizontal cylinder). The sequence below the alignment proposes an updated consensus sequence for the Trichoderma/Hypocrea class II HFBs, as derived from this study: therein, the cysteines are in red and numbered in order of their appearance in the sequence; X denotes any amino acid, and the subscript the number of them; "al" denotes any aliphatic, hydrophobic amino acid (A, V, L, I,)
Genomic organisation of the Trichoderma hydrophobin genes
In order to identify the mechanisms acting on the evolution of the Trichoderma hydrophobin genes, we first looked at their genomic organisation and exon structures. The six T. reesei hydrophobins are located on five different, large scaffolds, and even the two which are located on the same scaffold (hfb1 and hfb6) are separated by over 700,000 bp, and are therefore unlinked. Similar, although several of the hfb genes of H. atroviridis and H. virens were located on the same scaffold, they were separated by over 100,000 bp. In order to analyse whether any of these loci would be syntenic across Trichoderma species, we subjected each of the H. jecorina hfb genes plus 5 kb of its up- and downstream nt-sequences to a TBLAST search in the genome sequences of H. virens and H. atroviridis. The loci flanking the six hfb genes in H. jecorina did not flank any of the H. virens and H. atroviridis genes found by this analysis, although the flanking genes alone were sometimes located in the same region on a different scaffold (data not shown). Together, these data suggest that the genome regions containing hydrophobin gene loci have undergone extensive recombination during their evolution.
Intron/Exon structure of the Trichoderma hydrophobin genes
All of the chromosomal hfb genes of H. jecorina, H. virens and H. atroviridis contain two introns, which are very similar although not identical in size, and are positionally conserved. Interestingly, the length of the second exon encodes the aa sequence which folds exactly into the single α-helix of the hydrophobin and its third beta-sheet (cf. [5]) is absolutely conserved in all genes.
Phylogeny of the Trichoderma class II hydrophobin proteins
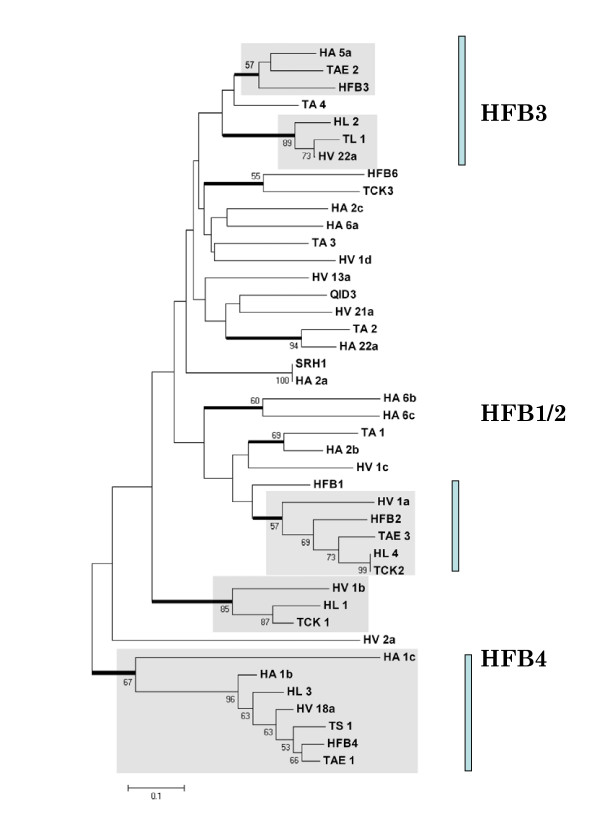
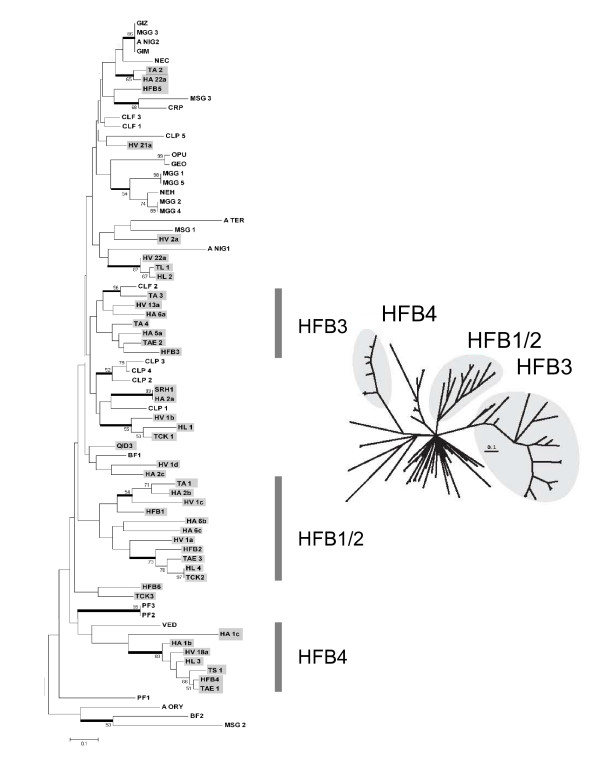
A phylogeny, based on neighbour joining of the amino acid sequence area from C1 to C8+1, is given in Fig. 3. It is conspicuous that the internal branches of the tree are essentially unresolved, and statistically supported clades only occur in terminal branches. Bayesian analysis of the same dataset produced essentially consistent results (data not shown). Five strongly supported clades contain HFBs from more than two Trichoderma species, i.e. the clades containing H. jecorina HFB1/HFB2; the clade containing TCK1; the clade containing TL1; the clade containing H. jecorina HFB3; and the clade containing H. jecorina HFB4. We considered it possible that the poor resolution of the internal tree branches could be due to the lack of conservation in the α-helix, the phylogeny was also performed on an alignment from which the aa's forming the helix had been removed. However, this did not improve clade stability (data not shown).
Figure 3.
Phylogenetic analysis of Trichoderma class II hydrophobins. The already published proteins from H. jecorina (HFB1-6) is marked in red. Branchess receiving significant support (> 50% bootstrap values) are indicated with a fat line. Significantly supported clades, which contain hydrophobins from at least 3 different species, are underplayed in grey. The vertical bars mark the clades termed HFB1/2, HFB3 and HFB4.
Evidence for gene duplications within the Trichoderma HFB proteins
One feature, which became obvious from the phylogenetic analysis and which is unaffected by the low internal branching support, is the high number of paralogous proteins. Examples for this are: Ha_1b and Ha_1c; Ha_6a and Ha_2c; Hv_21a and Hv_22a; TCK1 and TCK2; Ha_6b and Ha_6c; and HFB1 and HFB2. Most of these twins form a terminal branch, or are connected by a single node, indicating that they arose by gene duplication. The Trichoderma class II hydrophobins thus display a significant pattern of gene duplications in their evolutionary history.
Nucleotide Sequence Divergence of the hydrophobin genes
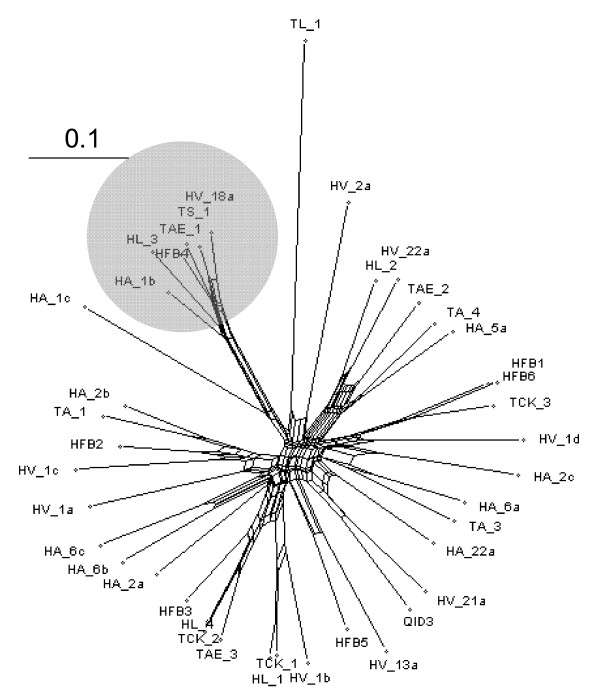
In order to obtain an insight in the mechanisms driving the evolution of the Trichoderma hfb genes, we investigated their nucleotide sequences. Introns were thereby excluded. Bayesian analysis, based on an alignment of the nucleotide sequences starting from the triplet encoding the first cysteine (C1) and ending with that of the eighth cysteine (C8), produced a phylogenetic tree which basically showed the same clade structure as the tree based on aa alignment, only with poorer support of some of the interior branches (data not shown). We investigated the nucleotide phylogeny split decomposition [39,40], a method depicting the shortest pathway by linking sequences, rather than forcing them into a bifurcating tree. The resulting tree is shown in Fig. 4, demonstrating indeed a dense network in the interior branches. The highest probability for a tree like structure was obtained with branch leading to the "Hfb4" clade (cf. Fig. 3). Since such networks may be the consequence of recombination, we applied the Phi-test, implemented in SplitsTree. However, the results of the phi-test favour the rejection of the null hypothesis of recombination (p = 0.888). Consistent results were obtained by using a sliding window approach in TOPALi (data not shown). We therefore conclude that the interior network in the tree revealed by the split decomposition method is due to a loss of genes (and thus branching information) and not recombination.
Figure 4.
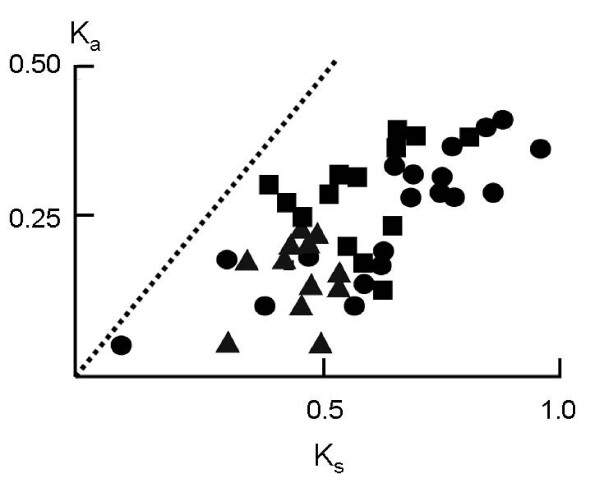
Plot of one-by-one comparisons of Ka vs. Ks for individual hydrophobin genes within clade "HFB1/2" (●), "HFB3" (■) and "HFB4" (▲) (for explanation of clades see Fig. 6). The dotted line indicates the position of Ka/Ks = 1.
The lack of recombination within the hfb genes, together with the observation of gene duplications suggests that the Trichoderma hydrophobins undergo purifying selection without concerted evolution. In such a case, the member genes would evolve independently and display a birth-and-death evolution [18]. This model of evolution assumes that new genes are created by repeated gene duplication and that some of the duplicate genes are maintained in the genome for a long time whereas others are deleted or become non-functional [18-20]. Thus nucleotide sequence differences between genes will primarily occur at synonymous sites, thus resulting in KS >> Ka.
We therefore separately tested the total number of synonymous and nonsynonymous sites, as well as the number of nonsynonymous substitutions per nonsynonymous site (Ka), and the number of synonymous -or silent-substitutions per synonymous -or silent-site (Ks) [41], both within the total gene exon sequence as well as within the two major exons separately (Table 2). In total, the number of differences at nonsynonymous sites strongly exceeded those at synonymous sites. However, the number of nucleotide substitutions at synonymous sites (KS) was significantly higher than the number of substitutions in nonsynonymous sites (Ka) in the complete gene and in exon 1, and slightly higher in exon 2. Plotting KS vs. Ka for members of selected clades, which had obtained support in the aa-phylogeny (cf. Fig. 3) showed that the KS values for some gene-to-gene comparisons are very high (up to 1.0) and have apparently reached the saturation level [21]. Interestingly, different clades showed different maximal Ka values, the "HFB4" clade thereby displaying the lowest numbers (Fig. 5).
Table 2.
Nucleotide diversity of the Trichoderma/Hypocrea class II hydrophobin genes
| all three exons | exon 1 | exon 2 | |
|---|---|---|---|
| no of sites | 234 | 101 | 35 |
| variable sites | 95 | 52 | 30 |
| no syn sites | 21.39 | 13.65 | 8.52 |
| no nonsyn sites | 65.61 | 40.35 | 24.48 |
| Eta, no of mutations | 218 | 123 | 76 |
| GC content | 0.585 | 0.598 | 0.567 |
| Pi nt diversity | 0.4259 | 0.38773 ± 0.0164 | 0.42532 ± 0.0139 |
| Theta per site (from Eta) | 0.5 | 0.49 | 0.49 |
| Tajima's D | -0.51296 | -0.74828 | -0.44716 |
| KS | 0.613 | 0.608 | 0.594 |
| KA | 0.367 | 0.312 | 0.376 |
Figure 5.
Phylogenetic tree of the nucleotide sequences of Trichoderma/Hypocrea class II hydrophobin genes by the split decomposition method. The "HFB4" clade, whose branch shows the least reticulate network, is highlightened in grey.
The Trichoderma HFB proteins form unique clades within the ascomycetous hydrophobins
The findings of gene duplication and apparently non-functional genes raised the question as whether a similar pattern of paralogous genes would be found also in other ascomycetes, and whether their members would help to stabilize the clades formed by the Trichoderma class II HFBs. To this end, we screened the available genome databases of other ascomycetes by TBLAST, using members of each of the Trichoderma HFB clades as a query (see Materials and Methods). In addition, we searched the NCBI database for previously described class II HFBs. The result was interesting in so far as most other ascomycetes for which a draft genome sequence is available contain a much smaller number of hydrophobin genes than Trichoderma, M. grisea being richest with 5 proteins (Table 3). While we cannot absolutely rule out that some HFB-encoding genes slipped through our analysis because of low similarity to query sequences, this would at the same time imply that they do not belong to any of the clades established for Trichoderma, and thus not affect the purpose of this study (but see also the Discussion below). The respective 26 sequences were aligned with those of Trichoderma and used for a phylogenetic analysis. The result, shown in Fig. 6, illustrates two points: first, most of the proteins from Trichoderma formed their own clades. Second, most of hydrophobins from the other 26 fungi formed clades which received only poor support, which is best seen by the star phylogeny obtained by Bayesian analysis (inset in Fig. 6). Third, gene duplications were evident for those fungi, for which more than two genes had been found, e.g. M. grisea MGG1 and5, and MGG2 and 4; for P. fulva PF2 and 3.
Table 3.
Class II hydrophobin genes from other ascomycetes used in this study
| subphyllum | family | species | Protein name | Accession number * |
|---|---|---|---|---|
| Leotiomycetes | Sclerotiniaceae | Botryotinia fuckeliana | BF1 | [B. fuckeliana genome database: BC1G_03994.19] |
| Botryotinia fuckeliana | BF2 | [B. fuckeliana genome database: BC1G_01012.1 ] | ||
| Eurotiomycetes | Trichocomaceae | Aspergillus oryzae | A_ORY | [GenBank: AAO16870.1] |
| Aspergillus terreus | A_TER | [GenBank: XM_001213908] | ||
| Aspergillus niger | A_NIG1 | [GenBank: XM_001394993] | ||
| Aspergillus niger | A_NIG2 | [GenBank: AAN76355.1] | ||
| Dothiodiomycetes | Mycosphaerellaceae | Mycosphaerella graminicola | MSG3 | [M. graminicola genome database: FGENESH2_PG.C_SCAFFOLD_8000534] |
| Mycosphaerella graminicola | MSG2 | [M. graminicola genome database: FGENESH2_PG.C_SCAFFOLD_2000556] | ||
| Mycosphaerella graminicola | MSG1 | [M. graminicola genome database: FGENESH2_PG.C_SCAFFOLD_11000390] | ||
| Passalora fulva | PF3 | [GenBank: CAC27408.1] | ||
| Passalora fulva | PF1 | [GenBank: CAC27407.1] | ||
| Passalora fulva | PF2 | [GenBank: CAB39312.1] | ||
| Sordariomycetes | Nectriaceae | Gibberella moniliformis | GIM | [GenBank: AY158024] |
| Gibberella zeae | GIZ | [GenBank: FG01831.1] | ||
| Nectria haematococca | NEH | [N. haematococca genome database: e_gw.1.52.181.1] | ||
| Phyllachorales | Verticillium dahliae | VED | [GenBank: AAY89101] | |
| Cryphonectriaceae | Cryphonectria parasitica | CRP | [GenBank: L09559] | |
| Clavicipitaceae | Claviceps fusiformis | CLF | [GenBank: CAB61236.1] | |
| Claviceps purpurea | CLP | [GenBank: CAD10781.1] | ||
| Ophiostomataceae | Ophiostoma ulmi | OPU | [GenBank: Z800849 | |
| Magnaporthaceae | Magnaporthe grisea | MGG4 | [GenBank: XM 364289] | |
| Magnaporthe grisea | MGG1 | [GenBank: AF126872] | ||
| Magnaporthe grisea | MGG2 | [GenBank: XM_001522792] | ||
| Magnaporthe grisea | MGG3 | [GenBank: XM_382007] | ||
| Magnaporthe grisea | MGG5 | [GenBank: XM_364289] | ||
| Sordariaceae | Neurospora crassa | NEC | [GenBank: XM_954189] |
Figure 6.
NJ analysis of amino acid sequences of class II hydrophobins from Trichoderma and other ascomycetes. Conditions and design of figure are similar as for Fig. 3. Accession numbers and/or genome database entries for the non-Trichoderma sequences are provided in Table 2. The inset on the right bottom shows the topology of an unrooted Bayesian tree (the three Trichoderma clades being highlightened in grey).
Expression analysis of the "HFB4"-clade hydrophobin genes from H. atroviridis
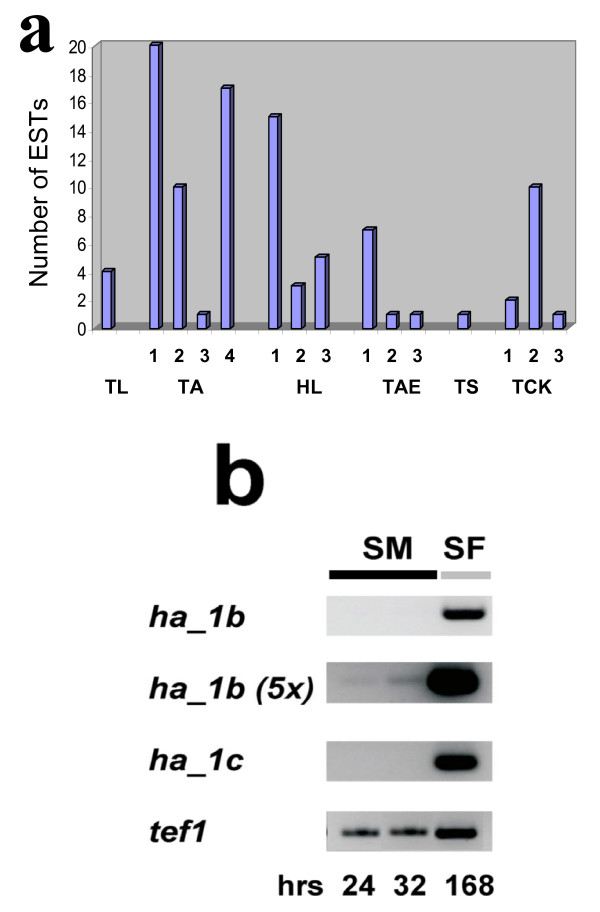
In order to obtain an estimate of the relative expression of the various Trichoderma/Hypocrea hydrophobin genes, we first compared the numbers of ESTs in the TrichoEST database (Fig. 7a). The transcripts, which were most abundant, were from HFBs clustering in different clades, indicating that there is no cluster or group which is preferentially strongly expressed. In order to investigate the expression of members of the largest supported clade (the "HFB4" clade), we grew H. atroviridis in submerged and surface culture and examined the expression of the Ha_1b- and Ha_1c-encoding genes by RT_PCR (Fig. 7b). The results show that both genes are indeed expressed, and thus both duplicated copies are probably still functional, but are found only during growth on solid and not in submerged medium.
Figure 7.
Gene expression of the Trichoderma/Hypocrea hydrophobins. (A) Number of ESTs found for the respective hfb genes during screening of the TrichoEST database. Species are abbreviated as follows: TL, T. longibrachiatum; TA, T. asperellum, HL, H. lixii; TAE, T. aggressivum var. europeae; TS, T. stromaticum; TCK, T. cf. koningiopsis. Strain numbers are given in the legend to Table 2. Individual hydrophobin genes are indicated by their respective numbers (cf. Table 1), and no number is given for species in which only a single hydrophobin gene has been detected. (B) Expression of the H. atroviridis gene members of the "HFB4" clade. SM, submerged cultivation; SF, surface cultivation. "5×" indicates that the 5-fold amount of PCR product had been loaded onto the gel.
Discussion
In this paper, we have investigated the evolutionary processes which give rise to the biodiversity of the fungal class II hydrophobin genes and proteins. This class of hydrophobins has so far been reported to be restricted to Ascomycetes only. Results from this study, however, suggest that the distribution of these genes may be even more restricted, i.e. the majority of the members of this class was actually found in the Sordariomycetes, and only few were found in Leotiomycetes and Eurotiomycetes. This picture may however be biased by the fact that Sordariomycetes are overrepresented sequenced genomes, and the six genes which we retrieved from the two species of Dothidiomycetes (Passalora, Mycosphaerella) suggest that this subphylum may also be rich in class II hydrophobins. We cannot completely rule out that the low number retrieved for Leotio- and Eurotiomycetes could be due to a failure to identify these genes by BLAST search. However, our approach also identified several class I hydrophobin genes from all these fungi (data not shown), and we would therefore assume that our screening was broad enough to identify all class II genes. Also, our results are in agreement with the results of manual annotation of several fungal genomes (Aspergillus spp., M. grisea, N. crassa, G. zeae, N. haematococcae). Therefore, while it is possible that a potential HFB encoding gene has been overlooked, our data indicate that while most species contained only 1 or 2 genes (e.g. Gibberella, Nectria, Botryotinia, Aspergillus spp.), species of Trichoderma/Hypocrea clearly exceed this with their gene number (i.e. 6 genes in H. jecorina, 9 in H. virens and 10 in H. atroviridis). The reason for this remains obscure: neither the morphology of the hyphae, the conidia or of the perithecium of Hypocrea/Trichoderma show microscopic differences which may necessitate new or multiple hydrophobins to support these structures. What differentiates this fungal genus from others, however, is its mycoparasitic and necrotrophic lifestyle [25]. While completely speculative at this moment, it is nevertheless possible that a versatile arsenal of class II hydrophobins may help the fungus to attach to the hyphae of a broad range of asco- and basidiomycetes. An amplified spectrum of genes has also been found for the chitinases of H. jecorina, which undoubtedly also aid to its mycoparasitic abilities [42]. With the availability of the hydrophobin gene sequences now in hand for two strongly mycoparasitic species – H. atroviridis and H. virens – this work lays a strong phylogenetic foundation to investigate this possibility by means of respective knock-out strains and expression analysis.
The results from this paper show that the class II hydrophobin genes of Trichoderm/Hypocrea contain a high number of duplicated genes, and at least two cases of pseudogenes. This suggests that the class II hydrophobins evolve by a death-and-birth mechanism [22], a term which has been created for a process in which genes undergo gene duplications, resulting in the maintenance of some of the copies for a considerable period of time whereas other copies are rapidly lost or converted to pseudogenes. Our data render the operation of concerted evolution unlikely, because of the high sequence divergence and also by the absence of recombination at the hydrophobin loci. (in concerted evolution, member genes evolve together as a unit by mechanisms such as gene conversion or unequal crossing-over). The fact that most of the duplicated genes occupy terminal branches in phylogenetic trees and that only few obvious pseudogenes were found, indicates that the rate of evolution of the class II hydrophobins in Trichoderma is relatively fast. This rapid evolution, and equally rapid loss of some genes is also reflected in the findings that the clades leading to the hfb genes in Trichoderma seldom contain members of other fungi, and their evolution thus took place after formation of the genus Trichoderma.
In addition, the numbers of synonymous differences of nucleotide sequences between genes from the same species are very large and frequently close to the saturation level. This high level of synonymous differences further supports the claim of a birth-and-death evolution at the DNA level, and supports the long time persistence of these genes in the genome. On the other hand, genes from different species (e.g. H. atroviridis and H. virens) but belonging to the same phylogenetic clade are highly similar (cf. Fig. 3). Such a long-term conservation of amino acid sequence is best explained by strong purifying selection. Interestingly, and in contrast to Rajashekar et al. [43], we found only a few individual cases where the Ka/Ks ratio was >1 and would reveal a history of accelerated evolution. If such a period of accelerated evolution occurred, most of the gene duplicates from this time apparently have not been maintained and the Trichoderma/Hypocrea hydrophobin genes characterized in this study are therefore mostly of recent origin.
The present study also expands our knowledge on the structure of class II hydrophobins. While most of them are small, compact proteins, which contain little other structures than the four beta-sheets and the single helix [5,6], we have detected several proteins which display a long extended N-terminus (ENT). With respect to class II HFBs, such structures have so far only been found in H. jecorina HFB6 [44], and in the pseudohydrophobin QID3 [36]. Interestingly, an ENT was recently also identified in the class I HFB Hum3 from Ustilago mayidis [45]. Lora et al. [46] hypothesized that the ENT of QID3 mediates cell wall binding because it resembles a module which is also present in plant bimolecular proteins [47-49]. Interestingly, our work reveals that there are at least two types of these ENTs: a major one, typified by H. virens HV_21a, H. atroviridis HA_2c, H. lixii QID3, and also in P. fulva HCF6, and in the spacers between the hydrophobin units in the multipartite genes of C. paspali and C. fusii, which are characterized by a conserved repeat of glycine and asparagines; and second type, shown by e.g. H. atroviridis HA_6a, H. virens HV_1d, and H. jecorina HFB6, in which the repeated motif is replaced by several PG/PD repeats, a P-rich stretch or a D-rich stretch, respectively. These proteins did not cluster together, indicating that these proteins do not show a common ancestry of the cysteine-containing core domain. Among the proteins with this terminus, one (HV_13a) is intriguing as its ENT is very short, which gives rise to the speculation that this extended N-terminus may arise by segment duplication. Support for this hypothesis would also come from the multipartite hydrophobins found in Claviceps spp. [50,51], wherein paralogous hydrophobin gene copies are connected by P, G and N-rich loops, and which may have been trapped in the stage of gene duplications at the extended N-terminus before recombining individual copies into new loci. It is thereby intriguing to observe the similarity of the nucleotide sequence of the "GN" repeat (GGTAAT) to that found to act as a recombination hot-spot in Penicillium chrysogenum (TGTAA [A/T]; [52]). Therefore, the occurrence of the Claviceps multipartite hydrophobins would be due to multiplication of some of the class II hydrophobins by tandem duplication [53,54], for which these sequences could act as recombination targets.
Nevertheless, it may still be likely that these ENTs are not only evolutionary artefacts: the [GN] repeats are reminiscent of S. cerevisiae Ure2p, a regulator of nitrogen catabolism, which can become transformed into a prion form by polymerization into filaments [55]. These filaments have an amyloid fibril backbone formed by an N-rich sequence which form a parallel superpleated beta-structure. The prion domain is thereby divided into nine seven-residue segments, each with a four-residue strand and a three-residue turn, that zig-zag in a planar serpentine arrangement, the interior of the filament being stabilized by H-bond networks generated by the stacking of N side chains. Interestingly, hydrophobins themselves are known to form amyloid-like structures [56-58], and we consider it therefore possible that the ENTs form defined structures which additionally contribute to the structural rigidity of the hydrophobins.
During phylogenetic analysis of the amino acid sequence, most hydrophobins from Trichoderma/Hypocrea did not group into strongly supported clades. However, a few exceptions were noted, notably the clades containing H. jecorina HFB1, HFB2 and HFB4, respectively. Clade "HFB4" is intriguing as its members – in contrast to HFB1 and HFB2 [31] were not expressed in submerged culture but only found in surface cultivation. This clade may thus contain hydrophobins relevant for hyphal growth. Unfortunately, the differences in aa-sequence with that of the other Trichoderma hydrophobins do not provide a clear clue as to the understanding of its function. One notable change is the substitution of the phenylalanine residue in the middle of the single helix, which is otherwise conserved in the class II hydrophobins from all other fungi (with the exception of A. niger and V. lecanii which contain an L and M, respectively), by a leucine, which may give rise to weaker hydrophobic interaction within the protein (hydrophobicity index F = 2.24; L = 1.99). However, Linder et al. [3] speculated that the aromatic ring of F39 is inserted between two Pro-residues (P11 and P50) from the two β hairpin structures into the protein and may serve to stabilize the fold through hydrogen bonds. Interestingly, members of the "HFB 4 clade" consistently have P11 replaced by an A which may hydrophobically interact with this L. In addition, the first beta-strand contains a conserved motif of two asparagines which provide it with a positive charge. Hydropathy plots show that members of the "HFB4 clade" have almost no hydrophobicity in the area between aa20 and aa40, and their helix is in contrast positively charged. While the consequence of these changes on the structure and function is however unclear our phylogenetic analysis sets the stage for future functional studies that may include transcript and gene deletion analysis.
Conclusion
Summarizing, this study offers a model of evolution which gave rise to the biodiversity of class II hydrophobins. The more than 70 members identified in this study enabled us to delineate the consensus for both essential aa sequence parts as well as for the tolerance to aa modifications to these small compact proteins. Our phylogenetic analysis will inform future functional genomic studies aimed at determination of more specific functions for each of the Trichoderma/Hypocrea class II hydrophobins. In view of the strong potential of the hydrophobins in "white" biotechnology, this information may offer their further improvement by molecular evolution [59-61].
Methods
Conditions of fungal growth
Hypocrea atroviridis (anamorph: Trichoderma atroviride) strain P1 (ATCC 74058) was maintained on potato dextrose agar (PDA) (Difco, Franklin Lakes, NJ, USA). The following medium was used for its cultivation (g·l-1; [41]): D-glucose, 10; peptone, 0.35; Tween 80, 0.175; KH2PO4, 0.68; K2HPO4, 0.87; (NH4)2SO4, 1.7; KCl, 0.2; CaCl2, 0.2; MgSO4·7H2O, 0.2; FeSO4·7H2O, 0.02; ZnSO4·7H2O, 0.02; MnSO4·7H2O, 0.02. The fungus was grown either in 1 L shake-flasks containing 200 ml of the medium at 25°C (250 RPM) or on plates (in this case the medium was solidified by the addition of 15 g·l-1 agar).
Analysis of hydrophobin gene expression
Mycelia were withdrawn at selected time points as indicated and total RNA isolated by the method of Chomzynski and Sacchi [62]. The RNA extract was treated with DNAse I (Fermentas, St. Leon-Rot, Germany) and purified using the RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany). The purified RNA was reverse transcribed using the RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas) with the oligo(dT)18 primer supplied by the manufacturer.
Appropriate aliquots of the cDNA were used for amplification by PCR utilising the GoTaq™ system (Promega, Madison, WI, USA). The assays contained 2.5 mM MgCl2 and 0.4 μM of the forward and reverse primer each (Table 4). The primers were designed in such a way that they aligned to different exons of the hfb genes to detect a possible contamination with genomic DNA. The amplification protocol consisted of an initial 1 min denaturation step at 95°C, followed by 28 cycles of denaturation (1 min at 95°C), annealing (1 min, see Tab. 1 for temperatures) and elongation (1 min at 72°C) and a final elongation step (7 min at 72°C). 40 μl of each assay were separated on 2% agarose containing 0.5 μg·ml-1 ethidium bromide. Expression of the elongation factor 1-alpha (tef1) gene [63] served as a loading control. For negative controls, the DNAse digestion, cDNA synthesis and the PCR were repeated without addition of the reverse transcriptase, in which case no amplicons were detected, thus confirming that the detected bands indeed result from cDNA synthesised from RNA (data not shown).
Table 4.
Primers used for RT-PCR
| Target | Primer name | 5' -> 3' sequence | T2 [°C] | Size [bp] |
|---|---|---|---|---|
| ha_1 b | hfb4RTfw | CTGCTTCTGAGGTCGTCGAG | 59.0 | 244 |
| hfb4RTrv | GGAAGAGCATCCTGGCAC | |||
| ha-1c | hfb5RTfw | CTCTTTACATTGGGCCTCG | 55.5 | 208 |
| hfb5RTrv | CAAGAGTGCAGCAATTGAGC | |||
| tef1 | tef1RTFw | GTACTGGTGAGTTCGAGGCTG | 59 | 350 |
| tef1RTRv | GGGCTCGATGGAGTCGATG |
In silico screening for hydrophobin sequences from Trichoderma and other fungi
To obtain the Trichoderma class II hydrophobins, we used the sequences of the six class II hydrophobins of H. jecorina [44] as a tool to retrieve genes encoding proteins with similarity from the genome sequence databases of Hypocrea virens and Hypocrea atroviridis and the TrichoEST EST database which includes EST sequences from 9 different Trichoderma spp. [38] using TBLAST (protein vs translated nucleotide). In the case of duplicates, the genomic sequence was given preference. Table 1 summarizes the genes, proteins, accession numbers or locations of the sequences thereby retrieved.
To obtain class II hydrophobin genes from other ascomycetes, the genes compiled by Linder et al. [3] were used as a starting point for a TBLAST search of the NCBI data base, using the filtering option turned off, and sequences which had not yet been included by Linder et al. [3] were retrieved. Apart of genes deposited from specific research, this database contains genome sequences from Neurospora crassa, Magnaporthe grisea, Gibberella zeae, and several Aspergillus spp. In addition, the same procedure was used to mine the genome databases of Nectria cinnabarina [64], Mycophaerella graminicola [65] and Botryotinia fuckeliana (= Botrytis cinerea, [66]), fungi whose sequences have not yet been included in the NCBI database. Genes thereby retrieved were then themselves used as a query in BLAST search as described above, and the procedure repeated until no new gene/protein was detected. The amino acid sequences of the retrieved proteins were aligned, and class I hydrophobins (identified according to the criteria described by Linder et al. [3]; e.g. by the difference in the number of amino acids between the conserved cysteins and their hydropathy profile) removed.
Phylogenetic analysis
DNA and protein sequences were visually aligned using Genedoc 2.6 [67]. Phylogenetic trees were constructed by the neighbor-joining (NJ) method [68], using the computer program MEGA, Version 4.0 [69], and by Bayesian analysis (MrBayes v3.0B4 program). The model of evolution and prior settings for individual loci was GTR + I + Γ. Metropolis-coupled Markov chain Monte Carlo (MCMCMC) sampling was performed with four incrementally heated chains that were simultaneously run for 1 and 3 millions of generations. To check for potentially poor mixing of MCMCMC, each analysis was repeated three times. The convergence of MCMCMC was monitored by examining the value of the marginal likelihood through generations. Convergence of substitution rate and rate heterogeneity model parameters was also checked. Bayesian posterior probabilities (PP) were obtained from the 50% majority rule consensus of trees sampled every 100 generations after removing the 500 first trees using the "burn" command. PP values lower then 0.95 were not considered significant.
Test for recombination
Two different procedures were applied to detect recombination by comparing adjacent sequence windows and to detect significant departures from a single phylogenetic history within the same alignment. First, we used difference of sums of squares (DSS, [70]) to compare the fit of genetic distance matrices for two adjacent windows to the same tree topology to produce the DSS statistic, the significance of which was determined through parametric bootstrapping in TOPALi [71].
In addition we tested whether recombination would be apparent from the phylogeny of hydrophobin genes. This was done by the split decomposition method in SplitsTree, version 2.4 [40], using pairwise distances under the Kimura 3-ST model [72]. This method visualizes recombination events by depicting the shortest pathway linking sequences, rather than forcing them into a bifurcating tree [39,40].
Tests for evolutionary mechanisms
To test the fit of the sequences to the model of neutral evolution, the D test statistic proposed by Tajima and Nei [73] was computed with the DnaSP program [74]. To this end, the Genedoc alignment was exported as a PHYLIP interleaved format. Only coding sequences, after removal of the preprosequence-encoding nt areas, were used for the analyses. Introns were removed from chromosomal nt-sequences by comparison with available cDNA (EST) sequences, or by relying on prediction of consensus splicing sites [75,76]. In the case of the multipartite hydrophobins from Claviceps spp. [50,51], each hydrophobin-encoding nt-area was treated as a separate entity. The extent of nucleotide divergence was estimated by using the uncorrected p distance [77]. The proportions of synonymous (were calculated pS) and nonsynonymous (pN) differences per site by the modified Nei-Gojobori method implemented in DNASp [12].
Competing interests
The author(s) declares that there are no competing interests.
Authors' contributions
CPK designed the study, performed the sequence alignments and evolutionary analyses and drafted the manuscript. SB coordinated the H. atroviridis genome sequence analysis and annotated the H. atroviridis and H. virens sequences. CG carried out the expression studies. CMK coordinate the H. virens genome sequence analysis and provided the H. virens sequence. ISD designed and performed the phylogenetic analyses. All authors read and approved the final manuscript.
Contributor Information
Christian P Kubicek, Email: ckubicek@mail.zserv.tuwien.ac.at.
Scott Baker, Email: scott.baker@pnl.gov.
Christian Gamauf, Email: gamauf@mail.zserv.tuwien.ac.at.
Charles M Kenerley, Email: c-kenerley@neo.tamu.edu.
Irina S Druzhinina, Email: druzhini@mail.zserv.tuwien.ac.at.
Acknowledgements
The authors are grateful to Heather Wilkinson for discussions on the manuscript. This work was supported by the EC-funded project TrichoEST (QLK3-2002-02032). CPK wants to acknowledge the important contribution of their colleagues from the TrichoEST consortium to the generation of the EST database. The genomic sequences for H. atroviridis and H. virens were provided by the DOE Joint Genome Institute, Walnut Creek, CA. This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program and the by the University of California, Lawrence Livermore National Laboratory under Contract No. W-7405-Eng-48, Lawrence Berkeley National Laboratory under contract No. DE-AC03-76SF00098 and Los Alamos National Laboratory under contract No. W-7405-ENG-36.
References
- Wessels JG. Fungi in their own right. Fungal Genet Biol. 1999;27:134–145. doi: 10.1006/fgbi.1999.1125. [DOI] [PubMed] [Google Scholar]
- Talbot NJ. Fungal biology. Coming up for air and sporulation. Nature. 1999;398:295–296. doi: 10.1038/18575. [DOI] [PubMed] [Google Scholar]
- Linder MB, Szilvay GR, Nakari-Setala T, Penttila ME. Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol Rev. 2005;29:877–896. doi: 10.1016/j.femsre.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Wösten HA. Hydrophobins: multipurpose proteins. Annu Rev Microbiol. 2001;55:625–646. doi: 10.1146/annurev.micro.55.1.625. [DOI] [PubMed] [Google Scholar]
- Hakanpaa J, Paananen A, Askolin S, Nakari-Setala T, Parkkinen T, Penttila ME, Linder MB, Rouvinen J. Atomic resolution structure of the HFBII hydrophobin, a self-assembling amphiphile. J Biol Chem. 2004;279:534–539. doi: 10.1074/jbc.M309650200. [DOI] [PubMed] [Google Scholar]
- Hakanpaa J, Szilvay GR, Kaljunen H, Maksimainen M, Linder M, Rouvinen J. Two crystal structures of Trichoderma reesei hydrophobin HFBI – the structure of a protein amphiphile with and without detergent interaction. Protein Sci. 2006;15:2129–2140. doi: 10.1110/ps.062326706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot MA, Talbot NJ. Building filaments in the air: aerial morphogenesis in bacteria and fungi. Curr Opin Microbiol. 2004;7:594–601. doi: 10.1016/j.mib.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Tucker S, Talbot NJ. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu Rev Phytopathol. 2001;39:385–417. doi: 10.1146/annurev.phyto.39.1.385. [DOI] [PubMed] [Google Scholar]
- Klimes A, Dobinson KF. A hydrophobin gene, VDH1, is involved in microsclerotial development and spore viability in the plant pathogen Verticillium dahliae. Fungal Genet Biol. 2006;43:283–294. doi: 10.1016/j.fgb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y, Ando A, Tamai Y, Yajima T. Pileus differentiation and pileus-specific protein expression in Flammulina velutipes. Fungal Genet Biol. 2007;44:14–24. doi: 10.1016/j.fgb.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Kwan AH, Winefield RD, Sunde M, Matthews JM, Haverkamp RG, Templeton MD, Mackay JP. Structural basis for rodlet assembly in fungal hydrophobins. Proc Natl Acad Sci USA. 2004;103:3621–3626. doi: 10.1073/pnas.0505704103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber E, Köbnik R. Positive selection of the Hrp pilin HrpE of the plant pathogen Xanthomonas. J Bacteriol. 2006;188:1405–1410. doi: 10.1128/JB.188.4.1405-1410.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benderoth M, Textor AJ, Windsor T, Mitchell-Olds J, Gershenzon J, Kroymann J. Positive selection driving diversification in plant secondary metabolism. Annu Rev Phytopathol. 2006;44:469–487. doi: 10.1146/annurev.phyto.44.032905.091147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NF, Wickham ME, Coombes BK, Finlay BB. Crossing the line: selection and evolution of virulence traits. PLoS Patho. 2006;2:e42. doi: 10.1371/journal.ppat.0020042. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mita S, Santoni S, Hochu I, Ronfort J, Bataillon T. Molecular evolution and positive selection of the symbiotic gene NORK in Medicago truncatula. J Mol Evol. 2006;62:234–244. doi: 10.1007/s00239-004-0367-2. [DOI] [PubMed] [Google Scholar]
- Haraguchi Y, Sasaki A. Host-parasite arms race in mutation modifications: indefinite escalation despite a heavy load? J Theor Biol. 1996;183:121–137. doi: 10.1006/jtbi.1996.9999. [DOI] [PubMed] [Google Scholar]
- Nei M, Hughes AL. In: HLA 1991. Proc 11th Histocompatibility Workshop and Conference. Tsuji K, Aizawa M, Sasazuli T, editor. Vol. 2. Oxford University Press; Balanced polymorphism and evolution by the birth-and-death process in the MHC loci; pp. 27–38. [Google Scholar]
- Ota T, Nei M. Divergent evolution and evolution by the birth-and-death process in the immunoglobulin VH gene family. Mol Biol Evol. 1994;15:469–482. doi: 10.1093/oxfordjournals.molbev.a040127. [DOI] [PubMed] [Google Scholar]
- Nei M, Gu X, Sitnikova T. Evolution by a birth-and-death process in multigene families of the vertebrate immune system. Proc Natl Acad Sci USA. 1997;94:7799–7806. doi: 10.1073/pnas.94.15.7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Rogozin IB, Piontkivska H. Purifying selection and birth-and-death evolution in the ubiquitin gene family. Proc Natl Acad Sci USA. 2000;97:10866–10871. doi: 10.1073/pnas.97.20.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang RH, Tyler BM, Whisson SC, Hardham AR, Govers F. Ancient origin of elicitin gene clusters in Phytophthora genomes. Mol Biol Evol. 2006;23:338–351. doi: 10.1093/molbev/msj039. [DOI] [PubMed] [Google Scholar]
- Loria R, Kers J, Joshi M. Evolution of plant pathogenicity in Streptomyces. Proc Natl Acad Sci USA. 2006;103:9118–9123. doi: 10.1073/pnas.0601738103. [DOI] [PubMed] [Google Scholar]
- Klein D, Eveleigh DE. In: Trichoderma and Gliocladium. Basic biology, taxonomy and genetics. Kubicek CP, Harman GE, editor. Vol. 1. Taylor & Francis Ltd, London, U; 1998. Ecology of Trichoderma; pp. 57–74. [Google Scholar]
- Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species – opportunistic, avirulent plant symbionts. Nature Rev Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- Hjeljord L, Tronsmo A. In: Trichoderma and Gliocladium. Enzymes, Biological Control and Commercial Applications. Harman GE, Kubicek CP, editor. Vol. 2. Taylor and Francis, London; 1998. Trichoderma and Gliocladium in biological control: an overview; pp. 131–151. [Google Scholar]
- Munoz G, Nakari-Setala T, Agosin E, Penttila ME. Hydrophobin gene srh1, expressed during sporulation of the biocontrol agent Trichoderma harzianum. Curr Genet. 1997;32:225–230. doi: 10.1007/s002940050270. [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Gonzalez JF, Suarez MB, Redondo J, Heinrich J, Delgado-Jarana J, Hermosa R, Gutierrez S, Monte E, Llobell A, Rey M. Generation, annotation and analysis of ESTs from Trichoderma harzianum CECT 2413. BMC Genomics. 2006;27;7:193. doi: 10.1186/1471-2164-7-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino JA, Redondo J, Suarez MB, Cardoza RE, Hermosa R, Gonzalez FJ, Rey M, Monte E. Generation, annotation, and analysis of ESTs from four different Trichoderma strains grown under conditions related to biocontrol. Appl Microbiol Biotechnol. 2007;75:853–862. doi: 10.1007/s00253-007-0885-0. [DOI] [PubMed] [Google Scholar]
- Askolin S, Penttila ME, Wösten HA, Nakari-Setala T. The Trichoderma reesei hydrophobin genes hfb1 and hfb2 have diverse functions in fungal development. FEMS Microbiol Letts. 2005;253:281–288. doi: 10.1016/j.femsle.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Askolin S, Linder M, Scholtmeijer K, Tenkanen M, Penttila M, de Vocht ML, Wosten HA. Interaction and comparison of a class I hydrophobin from Schizophyllum commune and class II hydrophobins from Trichoderma reesei. Biomacromolecules. 2006;7:1295–1301. doi: 10.1021/bm050676s. [DOI] [PubMed] [Google Scholar]
- Viterbo A, Chet I. TasHyd1, anew hydrophobin gene from the biocontrol agent Trichoderma asperellum, is involved in plant root colonization. Mol Plant Pathol. 2006;7:249–258. doi: 10.1111/j.1364-3703.2006.00335.x. [DOI] [PubMed] [Google Scholar]
- van Wetter MA, Wosten HA, Wessels JG. SC3 and SC4 hydrophobins have distinct roles in formation of aerial structures in dikaryons of Schizophyllum commune. Mol Microbiol. 2000;36:201–210. doi: 10.1046/j.1365-2958.2000.01848.x. [DOI] [PubMed] [Google Scholar]
- Kershaw MJ, Wakley G, Talbot NJ. Complementation of the mpg1 mutant phenotype in Magnaporthe grisea reveals functional relationships between fungal hydrophobins. EMBO J. 1998;17:3838–3849. doi: 10.1093/emboj/17.14.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lora JM, de la Cruz J, Benitez T, Llobell A, Pintor-Toro JH. A putative carbon catabolite repressed cell wall protein from the mycoparasitic fungus Trichoderma harzianum. Mol Gen Genet. 1994;242:461–466. doi: 10.1007/BF00281797. [DOI] [PubMed] [Google Scholar]
- Kullnig C, Krupica T, Woo SL, Mach RL, Rey M, Benitez T, Lorito M, Kubicek CP. Confusion abounds over identities of Trichoderma biocontrol isolates. Mycol Res. 2001;105:770–772. doi: 10.1017/S0953756201229967. [DOI] [Google Scholar]
- The TrichoEST database. http://www.trichoderma.org
- Bandelt HJ, Dress AW. Split decomposition: a new and useful approach to phylogenetic analysis of distance data. Mol Phylogenet Evol. 1992;1:242–252. doi: 10.1016/1055-7903(92)90021-8. [DOI] [PubMed] [Google Scholar]
- Huson DH. SplitsTree: a program for analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Seidl V, Huemer B, Seiboth B, Kubicek CP. A complete survey of Trichoderma chitinases reveals a new family 18 subgroup. The FEBS J. 2005;272:5923–5939. doi: 10.1111/j.1742-4658.2005.04994.x. [DOI] [PubMed] [Google Scholar]
- Rajashekar B, Samson P, Johansson T, Tunlid A. Evolution of nucleotide sequences and expression patterns of hydrophobin genes in the ectomycorrhizal fungus Paxillus involutus. New Phytol. 2007;174:399–411. doi: 10.1111/j.1469-8137.2007.02022.x. [DOI] [PubMed] [Google Scholar]
- Neuhof T, Dieckmann R, Druzhinina IS, Kubicek CP, Nakari-Setala T, Penttilä M, von Döhren H. Direct identification of hydrophobins and their processing in Trichoderma using intact-cell MALDI-TOF MS. FEBS J. 2007;274:841–852. doi: 10.1111/j.1742-4658.2007.05636.x. [DOI] [PubMed] [Google Scholar]
- Teertstra WR, Deelstra HJ, Vranes M, Bohlmann R, Kahmann R, Kämper J, Wösten HA. Repellents have functionally replaced hydrophobins in mediating attachment to a hydrophobic surface and in formation of hydrophobic aerial hyphae in Ustilago maydis. Microbiology. 2006;152:3607–12. doi: 10.1099/mic.0.29034-0. [DOI] [PubMed] [Google Scholar]
- Lora JM, Pintor-Toro JA, Benitez T, Romero LC. Qid3 protein links plant bimodular proteins with fungal hydrophobins. Mol Microbiol. 1995;18:380–382. doi: 10.1111/j.1365-2958.1995.mmi_18020380.x. [DOI] [PubMed] [Google Scholar]
- Coupe SA, Taylor JE, Isaac PG, Roberts JA. Identification and characterization of a proline-rich mRNA that accumulates during pod development in oilseed rape (Brassica napus L.) Plant Mol Biol. 1993;23:1223–1232. doi: 10.1007/BF00042355. [DOI] [PubMed] [Google Scholar]
- Castonguay Y, Laberge S, Nadeau P, Vezina LP. A cold-induced gene from Medicago sativa encodes a bimodular protein similar to developmentally regulated proteins. Plant Mol Biol. 1994;24:799–804. doi: 10.1007/BF00029861. [DOI] [PubMed] [Google Scholar]
- He CY, Zhang JS, Chen SY. A soybean gene encoding a proline-rich protein is regulated by salicylic acid, an endogenous circadian rhythm and by various stresses. Theor Appl Genet. 2002;104:1125–1131. doi: 10.1007/s00122-001-0853-5. [DOI] [PubMed] [Google Scholar]
- Arntz C, Tudzynski P. Identification of genes induced in alkaloid-producing cultures of Claviceps sp. Curr Genet. 1997;31:357–360. doi: 10.1007/s002940050216. [DOI] [PubMed] [Google Scholar]
- De Vries OM, Moore S, Arntz C, Wessels JG, Tudzynski P. Identification and characterization of a tri-partite hydrophobin from Claviceps fusiformis. A novel type of class II hydrophobin. Eur J Biochem. 1999;262:377–385. doi: 10.1046/j.1432-1327.1999.00387.x. [DOI] [PubMed] [Google Scholar]
- Fierro F, Garcia-Estrada C, Castillo NI, Rodriguez R, Velasco-Conde T, Martin JF. Transcriptional and bioinformatic analysis of the 56.8 kb DNA region amplified in tandem repeats containing the penicillin gene cluster in Penicillium chrysogenum. Fungal Genet Biol. 2006;43:618–29. doi: 10.1016/j.fgb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Kong H, Landherr LL, Fröhlich MW, Leebens-Mack J, Ma H, Depamphilis CW. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. in press 2007, Apr 23. [DOI] [PubMed]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2001;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Kajava AV, Baxa U, Wickner RB, Steven AC. A model for Ure2p prion filaments and other amyloids: the parallel superpleated beta-structure. Proc Natl Acad Sci USA. 2004;101:7885–7890. doi: 10.1073/pnas.0402427101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten HA, de Vocht ML. Hydrophobins, the fungal coat unravelled. Biochim Biophys Acta. 2000;1469:79–86. doi: 10.1016/s0304-4157(00)00002-2. [DOI] [PubMed] [Google Scholar]
- Butko P, Buford JP, Goodwin JS, Stroud PA, McCormick CL, Cannon GC. Spectroscopic evidence for amyloid-like interfacial self-assembly of hydrophobin Sc3. Biochem Biophys Res Comm. 2001;280:212–215. doi: 10.1006/bbrc.2000.4098. [DOI] [PubMed] [Google Scholar]
- Mackay JP, Matthews JM, Winefield RD, Mackay LG, Haverkamp RG, Templeton MD. The hydrophobin EAS is largely unstructured in solution and functions by forming amyloid-like structures. Structure. 2001;9:83–91. doi: 10.1016/S0969-2126(00)00559-1. [DOI] [PubMed] [Google Scholar]
- Hektor HJ, Scholtmeijer K. Hydrophobins: proteins with potential. Curr Opin Biotechnol. 2005;16:434–439. doi: 10.1016/j.copbio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Scholtmeijer K, Janssen MI, Gerssen B, de Vocht ML, van Leeuwen BM, van Kooten TG, Wösten HA, Wessels JG. Surface modifications created by using engineered hydrophobins. Appl Environ Microbiol. 2002;68:1367–1373. doi: 10.1128/AEM.68.3.1367-1373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtmeijer K, Janssen MI, van Leeuwen MB, van Kooten TG, Hektor H, Wösten HA. The use of hydrophobins to functionalize surfaces. Biomed Mater Engin. 2004;14:447–454. [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Nakari T, Alatalo E, Penttila ME. Isolation of Trichoderma reesei genes highly expressed on glucose-containing media: characterization of the tef1 gene encoding translation elongation factor 1 alpha. Gene. 1993;136:313–318. doi: 10.1016/0378-1119(93)90486-M. [DOI] [PubMed] [Google Scholar]
- DOE Joint Genome Institute. The Nectria cinnabarina genome database v 1.0. 2006. http://genome.jgi-psf.org/Necha2/Necha2.home.html
- DOE Joint genome Institute. The Mycophaerella graminicola genome database v 1.0. 2007. http://genome.jgi-psf.org/Mycgr1/Mycgr1.home.html
- Broad Institute and Syngenta AG. The Botrytis cinerea assembly. 2006. http://www.broad.mit.edu/annotation/genome/botrytis_cinerea/Home.html
- Nicholas KB, Nicholas HB. , JrGenedoc: a tool for editing and annotating multiple sequence alignments. 1997. http://www.psc.edu/biomed/genedoc
- Saitou N, Saitou M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- McGuire G, Wright F. TOPAL 2.0: improved detection of mosaic sequences within multiple alignments. Bioinformatics. 2000;16:130–134. doi: 10.1093/bioinformatics/16.2.130. [DOI] [PubMed] [Google Scholar]
- Milne I, Wright F, Rowe G, Marshall DF, Husmeier D, McGuire G. TOPALi: software for automatic identification of recombinant sequences within DNA multiple alignments. Bioinformatics. 2004;20:1806–1807. doi: 10.1093/bioinformatics/bth155. [DOI] [PubMed] [Google Scholar]
- Kimura M. Estimation of evolutionary distances between homologous nucleotide distances. Proc Natl Acad Sci USA. 1981;78:454–458. doi: 10.1073/pnas.78.1.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–285. doi: 10.1093/oxfordjournals.molbev.a040317. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Kupfer DM, Drabenstot SD, Buchanan KL, Lai H, Zhu H, Dyer DW, Roe BA, Murphy JW. Introns and splicing elements of five diverse fungi. Eukaryot Cell. 2004;3:1088–1100. doi: 10.1128/EC.3.5.1088-1100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Duyvesteijn RG, Gale L, Usgaard T, Cornelissen BJ, Ma LJ, Ward TJ. The presence of GC-AG introns in Neurospora crassa and other euascomycetes determined from analyses of complete genomes: implications for automated gene prediction. Genomics. 2006;87:338–347. doi: 10.1016/j.ygeno.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford University Press, New York; 2000. [Google Scholar]