Abstract
The grass family comprises the most important cereal crops and is a good system for studying, with comparative genomics, mechanisms of evolution, speciation, and domestication. Here, we identified and characterized the evolution of shared duplications in the rice (Oryza sativa) and wheat (Triticum aestivum) genomes by comparing 42,654 rice gene sequences with 6426 mapped wheat ESTs using improved sequence alignment criteria and statistical analysis. Intraspecific comparisons identified 29 interchromosomal duplications covering 72% of the rice genome and 10 duplication blocks covering 67.5% of the wheat genome. Using the same methodology, we assessed orthologous relationships between the two genomes and detected 13 blocks of colinearity that represent 83.1 and 90.4% of the rice and wheat genomes, respectively. Integration of the intraspecific duplications data with colinearity relationships revealed seven duplicated segments conserved at orthologous positions. A detailed analysis of the length, composition, and divergence time of these duplications and comparisons with sorghum (Sorghum bicolor) and maize (Zea mays) indicated common and lineage-specific patterns of conservation between the different genomes. This allowed us to propose a model in which the grass genomes have evolved from a common ancestor with a basic number of five chromosomes through a series of whole genome and segmental duplications, chromosome fusions, and translocations.
INTRODUCTION
The grass family is the fourth largest among flowering plants and comprises some of the agronomically most important crop species, such as wheat (Triticum ssp), maize (Zea mays), and rice (Oryza sativa). Grass genomes differ greatly in size, ploidy level, and chromosome number. Bread wheat (Triticum aestivum; 2n = 42) belongs to the Pooideae family and has a hexaploid genome (AABBDD) of 17 Gb that originated through two polyploidization events (Feldman et al., 1995; Blake et al., 1999; Huang et al., 2002). Rice (2n = 24) is diploid and belongs to the Ehrhartoideae family. With a size of 0.4 Gb, its genome is 40 times smaller than that of bread wheat. Fossil data and phylogenetic studies estimated that the different grass families diverged from a common ancestor 50 to 70 million years ago (MYA) (for reviews, see Kellogg, 2001; Gaut, 2002).
Comparative studies between grasses, mostly cereals such as barley (Hordeum vulgare), wheat, maize, rice, and sorghum (Sorghum bicolor), have been the focus of intense research in the past decade (for a recent review, see Salse and Feuillet, 2007). Early comparative studies relied on cross–restriction fragment length polymorphism (RFLP) mapping analyses of closely related species. They revealed significant macrocolinearity between the cereal genomes and led to the construction of a consensus grass map based on 25 rice linkage blocks (reviewed in Devos and Gale, 2000; Feuillet and Keller, 2002; Devos, 2005). These results, however, were obtained from low-resolution genetic maps with an average of one marker every 10 centimorgan that allowed the detection of only large rearrangements. Moreover, the maps were constructed with low-copy RFLP markers that were selected for their ability to provide a signal in cross-hybridizations, thereby limiting the detection of whole or partial genome duplication events. It also has been difficult to assess orthologous and paralogous relationships in gene families, since comparative mapping by RFLP often identified paralagous rather than orthologous sequences, leading to an underestimation of colinearity.
In the past 5 years, international initiatives have led to the development of additional genomic resources that allow comparative genomic studies between the grass genomes at a higher level of resolution (microcolinearity). The International Triticeae EST Cooperative (http://wheat.pw.usda.gov/genome/) efforts have resulted in the production of >1 million wheat ESTs (http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html, 09/28/07 release), 7107 of which, defining 16,099 loci, have been cytogenetically mapped in deletion bins (Qi et al., 2004). In rice, the International Rice Genome Sequencing Project (IRGSP) recently completed the sequence of the O. sativa ssp japonica cv Nipponbare (International Rice Genome Sequencing Project, 2005). Twelve pseudomolecules corresponding to 372,077,801 bp of finished sequence (The Institute for Genome Research [TIGR] version 4; 381,150,945 bp in IRGSP version 4) were assembled and characterized through several rounds of annotation. This resulted in estimates for the rice gene number ranging from ∼32,000 (Rice Annotation Project, 2007) for the most recent annotation of the international consortium to 42,654 genes in the TIGR version 4 (http://www.tigr.org/tigr-scripts/osa1_web/gbrowse/rice/; Yuan et al., 2003). These resources have been used to perform large-scale intraspecific and interspecific sequence comparisons between the two genomes and have helped to refine our understanding of colinearity between their chromosomes. Sorrells et al. (2003), Sorrells (2004), and Singh et al. (2007) have compared the sequences of 4485 and 3792 cytogenetically mapped wheat ESTs against the rice genome sequence. Studies focusing on single chromosome groups or regions have been performed recently as well for rice chromosome 3 compared with wheat and maize ESTs (Buell et al., 2005; Rice Chromosome 3 Sequencing Consortium, 2005) and for rice chromosome 11 compared with wheat ESTs (Singh et al., 2004). These studies increased the resolution of comparative mapping between the two species by 25- to 30-fold, revealing more rearrangements than previously observed at the genetic map level.
In addition to the assessment of colinearity between the genomes, comparative analyses can reveal ancestral genome duplications. Early studies with the first generation of molecular markers indicated the presence of duplicated loci on the genetic maps in different cereals, suggesting ancestral genome duplications and polyploidization events in the history of species that are now considered diploids. RFLP and isozyme studies in the early 1990s had already suggested that maize chromosomes share duplicated segments (Wendel et al., 1989; Ahn and Tanksley, 1993). Whole duplication of the maize genome through allotetraploidization was identified and characterized further through the evolutionary analysis of duplicated genes (Gaut and Doebley, 1997) and by interspecific comparisons between orthologous loci in rice, sorghum, and maize (Swigonova et al., 2004). In rice, early RFLP mapping studies suggested that chromosomes 1 and 5 (Kishimoto et al., 1994) as well as chromosomes 11 and 12 (Nagamura et al., 1995) contain ancient duplicated regions. The release of the genome sequence drafts from japonica and indica rice subspecies allowed whole genome sequence comparisons and further characterization of duplications in rice (Yu et al., 2002, 2005; Paterson et al., 2003, 2004; Vandepoele et al., 2003; Guyot et al., 2004; International Rice Genome Sequencing Project, 2005; Wang et al., 2005). The most recently published analysis (Yu et al., 2005) revealed a whole genome duplication (WGD) that occurred between 53 and 94 MYA (i.e., before the divergence of the cereal genomes), a recent segmental duplication between chromosomes 11 and 12, and numerous individual gene duplications. Together, these duplications cover an estimated 65.7% of the rice genome. Rice genome duplications have been studied also by TIGR using its latest genome annotation (41,046 nontransposable element–related rice protein sequences; http://www.tigr.org/tdb/e2k1/osa1/segmental_dup/index.shtml). The results indicate a large number of segmental duplications, but the low stringency of the analysis did not allow clear conclusions to be drawn on the exact nature of the rice duplications. Finally, comparative analyses between rice and other grasses have been helpful in revealing duplications in the rice genome. For example, by comparing 2600 maize mapped sequence markers with the rice sequence, Salse et al. (2004) identified six duplications between chromosomes 8-12, 2-6, 6-10, 1-5, and 3-6 that had not been detected previously. More recently, Wei et al. (2007) built a high-resolution comparative physical map between the rice and maize genomes that helped to refine maize and rice duplications. Thus, a number of studies have suggested that the grass genomes were subjected to different rounds of whole genome and segmental duplications during their evolution from a common ancestor 50 to 70 MYA. While the mechanisms by which the genomes have evolved have become more evident over the past decades, the basic chromosome number of the grass ancestor and the evolutionary path that led to the large variety of basic chromosome number found in today's grass genomes remain uncertain. Different authors have proposed basic numbers ranging from 5 to 12 chromosomes (for a review, see Gaut, 2002; Wei et al., 2007), but to date no model that would reconcile all of the data has been proposed for the structure and evolution of the ancestral grass genome.
Because it is difficult to infer orthologous (derived from a common ancestor by speciation) and paralogous (derived by duplication within one genome) relationships from sequence comparisons, stringent alignment criteria and statistical validation are essential to evaluate accurately whether the association between two or more genes found in the same order on two chromosomal segments in different genomes occurs by chance or reflects true colinearity. Several recently developed software programs, such as LineUP (Hampson et al., 2003), ADHoRE (Automatic Detection of Homologous Regions; Vandepoele et al., 2002), FISH (Fast Identification of Segmental Homology; Calabrese et al., 2003), and CloseUp (Hampson et al., 2005), help to address these questions. A number of other programs, such as Cmap (Fang et al., 2003), and websites, such as Gramene (Jaiswal et al., 2006) or the TIGR synteny projects (http://www.tigr.org/tdb/synteny/wheat/description.shtml), have been established to visualize sequence-based colinearity data obtained from comparative sequence analyses between the grass genomes. While these websites provide user-friendly graphical displays of macrocolinearity, they rely on data obtained with low-stringency alignment criteria and without statistical validation. In addition, they do not take into account the density and location of conserved genes to identify precisely paralogous and orthologous regions; therefore, they generally overestimate colinearity between different segments of the genomes.
In this study, we applied new and stringent alignment criteria and performed statistical tests systematically to redefine interchromosomal duplications in rice, identify wheat genome duplications, and reassess colinearity relationships between the two genomes. This allowed us to detect and characterize shared duplicated regions between rice and wheat, to compare them with maize and sorghum data, and to establish a model for the evolution of the grass genomes from a common ancestor with n = 5 chromosomes.
RESULTS
Improved Sequence Alignment Criteria to Infer Significant Orthologous and Paralogous Relationships
When two sequences are aligned, BLASTN (Altschul et al., 1990, 1997) produces high-scoring pairs (HSPs) that consist of two sequence fragments of arbitrary but equal length, the alignment of which is locally maximal and for which the alignment score meets or exceeds a threshold or cutoff score. HSPs are based on statistical criteria such as the e-value, score, and percentage of identity. Detecting conserved regions is limited with sequence alignments obtained by BLAST with these default parameters. To increase the significance of intraspecific and interspecific sequence alignments for inferring evolutionary relationships within and between genomes, we defined three new parameters for BLAST analysis: AL for aligned length, CIP for cumulative identity percentage, and CALP for cumulative alignment length percentage (see Methods for definitions). With these parameters, BLAST produces the highest cumulative percentage of identity over the longest cumulative length, thereby increasing the stringency in defining conservation between sequences. Publicly available rice and wheat EST sequences were reanalyzed by BLAST using these parameters, followed by a statistical test with the CloseUp software (Hampson et al., 2005) that validates nonrandom associations between groups of sequences. We applied different levels of stringencies to the intraspecific and interspecific sequence comparisons in the CloseUp analysis to take into account differences between the numbers of sequences in each data set. Since the whole rice genome is available (42,654 annotated genes), we applied higher stringency alignment criteria and statistical validation in rice than in wheat, for which only a limited subset of mapped gene sequences (6426 ESTs) is available. By combining these results with data on the chromosomal location of the sequences, we were able to distinguish orthologous and paralogous regions in both genomes with high confidence and, subsequently, to identify shared duplications between the two genomes.
Identification and Characterization of Duplicated Regions in the Rice Genome
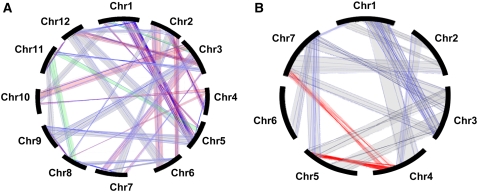
Duplicated regions in rice were identified by applying the new alignment criteria and statistical validation described above to the 42,654 non-transposable element-related genes detected in the fourth release of the TIGR rice genome annotation (http://www.tigr.org/tdb/e2k1/osa1/pseudomolecules/info.shtml). The gene sequences were aligned against themselves (BLASTN) using 70% CIP and 70% CALP. The results show that 34,903, 2410, 1121, and 4220 genes matched zero, one, two, and more than two sequences, respectively, elsewhere in the rice genome. We considered as reliable indicators of putative duplications in the genome the 2410 (5.6%) sequences that identified a single homolog (with an average value of 89.4% CIP and 91.2% CALP). They were analyzed further with CloseUp (density ratio, 2; cluster length, 40; match, 5) to distinguish between the regions corresponding to genes that result from ancient duplications and those that are associated randomly. Among the 2410 putative duplicated loci, 539 (22.4%) were statistically significant. They defined 29 duplicated regions that are distributed among the 12 rice chromosomes as follows: r1-r2/3/5/10/12, r2-r4/6/7/8/12, r3-r7/9/10/11/12, r4-r5/8/10, r5-r9/11/12, r6-r7/8/12, r7-r8, r8-r9/11, r9-r11, and r11-r12 (Figure 1A; see Supplemental Table 1 online).
Figure 1.
Intraspecific Duplications of the Rice and Wheat Genomes.
(A) Schematic representation of the 539 pairs of paralogous genes (linked by thin blue lines) defining 29 duplication blocks on the 12 rice chromosomes. The 10 duplicated regions identified previously (Yu et al., 2005) are highlighted in red, the 3 newly detected duplications are indicated in green, and the 16 duplicated regions found within the 13 segments previously identified are in gray.
(B) Schematic representation of the 10 duplicated regions and 2 translocations identified on the seven wheat chromosome groups. Duplicated segments are shown in gray, with thin blue lines representing the duplicated genes within each segment. The translocations between w4-w5 and w4-w7 are highlighted in red.
Ten of the 29 duplications (indicated by asterisks in Supplemental Table 1 online) correspond to the duplications identified previously by Yu et al. (2005) (in red in Figure 1A) and cover 47.8% of the rice genome. The 10 duplications correspond to exactly the same regions as the 18 duplicated regions reported by Yu et al. (2005), but in our analysis we have considered a single duplication event when two duplications were physically close on the same sister chromosomes. Among the 19 additional duplicated regions (between chromosomes r1-r2/3/10/12, r2-7/8/12, r3-r9/11, r4-r5, r5-r9/11/12, r6-r7/8/12, r7/r8, r8-r11, and r9/r11), 3 correspond to duplicated regions not identified previously (in green in Figure 1A). They cover 10.4% of the genome and define novel relationships between chromosomes r5 and r11, r8 and r11, and r1 and r3. The remaining 16 duplications (in gray in Figure 1A) are superimposed on the previous 13 (10 + 3) duplications. They define novel relationships between the chromosomes and represent 14.8% of the genome. Thus, in total, the 29 duplications cover 72% (267 Mb) of the rice genome, with an average density of one gene per 0.8 Mb. We conclude that the identification by our method of 10 known and 19 additional duplication blocks in rice validates the reliability of our approach for determining interchromosomal duplications and demonstrates its usefulness for further intragenomic and intergenomic comparisons.
Identification and Characterization of Duplicated Regions in the Wheat Genome
To identify duplications in the wheat genome, we first established a set of unique EST sequences with the highest possible length and for which chromosomal locations are known. The 6426 EST sequences that have been cytogenetically mapped on wheat deletion bins (Qi et al., 2004) were aligned against nonredundant EST clusters produced in the framework of Genoplante projects (available at http://urgi.versailles.inra.fr/data/banks/). Using CIP and CALP values of 95 and 85%, respectively, 90.6% (5823) of the mapped ESTs were considered to be identical (with average CIP and CALP values of 99.2 and 98.8%, respectively) to 5707 nonredundant EST contigs and were called WECs (for wheat EST contigs). The remaining 603 sequences that were not associated with a contig were called WESs (for wheat marker singletons). We then determined the number of WECs (among 5823) that are associated with a unique EST contig (among 5707) using the same CIP and CALP values. The results showed that 96.1% (5596) of the EST contigs are associated with a single WEC, whereas 3.9% (111) are associated with two (107) or three (4) WECs. These latter correspond to homoeologous WEC sequences with high sequence identity, thereby reflecting the hexaploid nature of the wheat genome. When two or three homoeologous WECs were identified, we used the sequence of the EST contig that showed the highest CIP value as a representative of the homoeologous sequence groups in the rest of the comparative analyses.
Thus, among the initial 6426 mapped EST, 98.2% (5,707 + 603 = 6310) are associated with an EST contig that represents a single locus on a group of homoeologous chromosomes. Mapping information for the 5707 WECs and 603 WESs (Qi et al., 2004) showed that 5003, 946, 224, and 137 were assigned to one, two, three, and more than three loci in wheat, respectively. The 5003 WECs and WESs mapping at a single locus were used for further analysis of the colinearity with the rice genome, whereas those mapping to two distinct loci (946) were used to study intragenomic duplications in wheat.
Among the 946 duplicated WECs and WESs, 638 were associated with a genetic position within a specific deletion bin as defined by Qi et al. (2004). We ignored the remaining 308 sequences, as the information was limited to their presence on a chromosome or a chromosome arm. Putative interchromosomal duplications were identified through the statistical analysis of the 21 possible pairwise combinations formed by duplicated WES and WEC loci among the seven wheat consensus chromosome groups (for a graphical display of all of the relationships, see Supplemental Figure 1 online). The number of putatively duplicated WEC or WES loci ranged from 13 to 94 per chromosome pair, with a total of 638. We obtained the largest numbers for the w4-w5 and w4-w7 combinations (see Supplemental Figures 1-16 and 1-18 online). Only 216 (33.9%) of the 638 putative duplications were validated using CloseUp (density ratio, 0.5; cluster length, 25; match, 5) and the information on the position of the WES and WEC sequences on consensus chromosomes (see Methods and Supplemental Table 2 online). They define 12 statistically significant duplication blocks that cover 67.5% of the genome and correspond to the following chromosome pairs: w1-w2 (16 ESTs), w1-w3 (11 ESTs), w1-w4 (5 ESTs), w1-w7 (7 ESTs), w2-w4 (6 ESTs), w2-w7 (9 ESTs), w3-w5 (14 ESTs), w3-w7 (14 ESTs), w4-w5 (69 ESTs), w4-w7 (45 ESTs), w5-w7 (10 ESTs), and w6-w7 (10 ESTs) (Figure 1B). No statistically significant duplications were detected between w1-w5, w1-w6, w2-w6, w3-w6, and w4-w6 (see Supplemental Table 2 and Supplemental Figure 1 online). We found the highest number of duplicated genes between chromosomes w4-w5 (No. 9 in Supplemental Table 2 online; 69 paralogs) and w4-w7 (No. 10 in Supplemental Table 2 online; 45 paralogs). These regions correspond to two known translocations in wheat (Mickelson-Young et al., 1995; Miftahudin et al., 2004). They appear as duplications in our analysis because we used consensus chromosomes, and when a homoeologous region has been translocated to another chromosome, it will show similarity to the remaining homoeologous region on the chromosome group of origin. To distinguish between duplications and translocations, we systematically reanalyzed every duplicated region on each of the homoeologous chromosome groups (see Supplemental Figures 1-1 to 1-21 online). The results confirmed that only the duplications identified between w4 and w5 as well as between w4 and w7 correspond to translocations.
Thus, with this genome-wide analysis, we identified 10 duplicated regions and 2 translocations on the seven wheat chromosome groups. The identification of 10 well-defined duplicated regions allowed us to further study their origin through comparisons with the rice duplications.
Identification of Orthologous Regions between Rice and Wheat
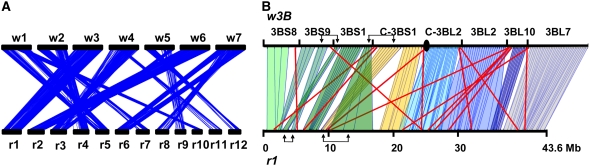
To identify orthologous regions between the rice and wheat genomes, we aligned the 5003 WEC and WES sequences that mapped at a single locus in wheat against the 42,654 rice genes. Using 60% CIP and 70% CALP for the sequence alignment (BLASTN), 36% (1805) of the wheat sequences showed similarity (average value of 83.9% CIP and 90.6% CALP) to a unique gene in rice. Subsequent CloseUp analysis (density ratio, 2; cluster length, 20; match, 5) indicated that 1108 of them can be considered true orthologs. They are present in 13 orthologous regions that cover 83.1 and 90.4% of the rice and wheat genomes, respectively, and correspond to the following chromosome pairs: w1-r5 (102 genes), w1-r10 (43 genes), w2-r4 (110 genes), w2-r7 (51 genes), w3-r1 (207 genes), w4-r3 (148 genes), w4-r11 (17 genes), w5-r3 (40 genes), w5-r9 (36 genes), w5-r12 (42 genes), w6-r2 (156 genes), w7-r6 (95 genes), and w7-r8 (61 genes) (Figure 2A; see Supplemental Table 3 online). In fact, the w5-r3 relationship does not reflect true colinearity between these chromosomes but the wheat translocation between w4 and w5 and the orthology between wheat chromosome 4 and rice chromosome 3. Thus, in total, 12 true orthologous relationships can be defined (Figure 3).
Figure 2.
Identification of 13 Orthologous Regions between Rice and Wheat.
(A) Schematic representation of the 13 orthologous regions identified between rice (r1 to r12) and wheat (w1 to w7) chromosomes. The 1108 pairs of orthologous genes are depicted as thin blue lines.
(B) Schematic representation of the 149 orthologs (vertical lines) identified between wheat chromosome 3B (w3B) and rice chromosome 1 (r1). Different colored blocks represent the colinear regions identified between r1 and w3B. Rearrangements between orthologous genes are highlighted with red lines. Two large inversions of colinear regions are indicated with arrows above and below the wheat and rice chromosomes, respectively. Wheat deletion bins are indicated above the 3B chromosome, whereas rice chromosome 1 is divided into 10-Mb segments.
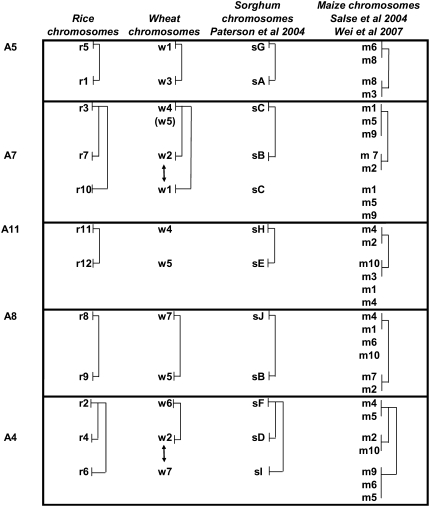
Figure 3.
Orthologous Relationships and Shared Duplications between the Rice, Maize, Wheat, and Sorghum Genomes.
Colinear wheat, rice, sorghum, and maize chromosomes are displayed on the same line in the figure. Duplications are indicated with solid lines. The five blocks of shared duplications identified in the four genomes are displayed on the right side of the ancestral chromosomes (A5, A7, A11, A8, and A4) that they define. The artefactual syntenic relationship identified between w5 and r3 that reflects the wheat w4-w5 translocation is indicated in parentheses. The two duplications shared between wheat and rice chromosomes that do not share a common ancestry (w1-r10/w2-r7 and w7-r6/w2-r4) but are found on orthologous chromosomes in both species are indicated with double arrows.
We then compared the linear order of the rice genes with the positions of the orthologous ESTs in the wheat deletion bins. The results indicate that for 27.2% of them, the identified wheat ortholog is not located in the orthologous wheat deletion bin, thereby indicating rearrangements within orthologous regions. For example, of 149 orthologs present on rice chromosome 1 and wheat chromosome 3B, 21 (14.1%) are not found in the orthologous wheat bins (Figure 2B). In addition, we observed two large inversions involving deletion bins 3BS9, 3BS1, and c-3BS1 between w3B and r1 (Figure 2B). Thus, our results show that, even if the current public set of mapped wheat EST presents some limits in comparative analysis, since the linear order of the ESTs within a wheat deletion bin is not known, rearrangements can be identified and the evaluation of colinearity between wheat and rice can be improved through an accurate assessment of the sizes and positions of the orthologous regions.
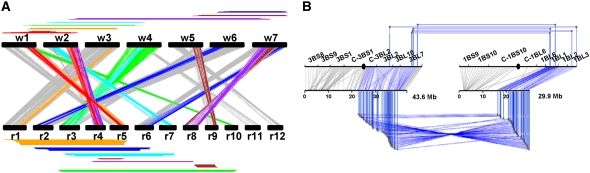
Identification of Shared Duplications between Rice and Wheat
In this study, we identified 29 duplications in rice, 10 in wheat, and 12 regions of orthology between the two genomes. Seven of the intraspecific duplications are conserved at orthologous positions between rice and wheat (Figure 4A). They are found on the following chromosome pair combinations: w1-w2/r5-r4, w1-w3/r5-r1, w1-w4/r10-r3, w2-w4/r7-r3, w2-w7/r4-r8, w5-w7/r9-r8, and w6-w7/r2-r6 (detailed in Supplemental Table 4 online). Altogether, they represent 68.3% of the rice genome and 65.9% of the wheat genome. One of the largest shared duplication (No. 2 in Supplemental Table 4 online) corresponds to a duplication between w1 and w3 that is orthologous to the r1 and r5 duplication. The conservation of duplications between the rice and wheat genomes indicates that they probably originated from an ancient duplication event that predated the divergence between the two species, 50 to 70 MYA. Not all chromosomes show remains of ancient shared duplications. No orthologous duplications were identified between rice chromosomes 11 and 12 and their wheat orthologs w4 and w5 (Figure 3). This is probably due to the fact that w4 and w5 have been involved in recent translocations, thereby disrupting the orthologous relationships.
Figure 4.
Seven Duplications Are Shared between Rice and Wheat.
(A) Schematic representation of the seven duplicated regions shared between wheat and rice. The paralogous regions are represented with the same colors in wheat (top) and rice (bottom) as well as the corresponding orthologous regions identified between the two sets of chromosomes (center). The color code is as follows: w1-w2/r5-r4 (red), w1-w3/r5-r1 (orange), w1-w4/r10-r3 (green), w2-w4/r7-r3 (light blue), w2-w7/r4-r8 (purple), w5-w7/r9-r8 (brown), and w6-w7/r2-r6 (dark blue). Rice–wheat orthologs not involved in shared duplications are indicated in gray.
(B) Schematic representation of the duplications shared between rice chromosomes 5 to 1 (42 paralogs linked by horizontal lines at bottom) and wheat chromosomes 1B to 3B (five paralogs linked by horizontal lines at top). The rice and wheat colinear regions r1-w3B and r5-w1B are linked by 149 and 66 orthologs (vertical lines), respectively.
To study the origin and features of the shared duplicated regions in more detail, we analyzed further one of the largest shared duplications that involves wheat chromosomes 3B and 1B and rice chromosomes 1 and 5. The r1-w3B orthologous regions are related through 149 genes, while 66 genes are conserved between the orthologous r5-w1B regions (Figure 4B). To identify paralogous sequences within the intraspecific duplications, we used the 246 and 115 WEC and WES sequences that map at unique positions in the duplicated regions of w3 (deletion bin, 3BL2, 3BL10, and 3BL7 for w3B [Figure 4B; see Supplemental Figure 1-2 online]) and w1 (1BL1, 1BL2, and 1BL3 for w1B [Figure 4B; see Supplemental Figure 1-2 online]) to perform a BLASTN alignment with 70% CIP and 70% CALP values. This identified eight putative paralogous genes within the w1-w3 duplicated region. In rice, the orthologous regions on chromosomes 1 and 5 contain 42 paralogs (Figure 4B). We then used the pattern of nucleotide substitution within the shared duplicated regions to estimate the duplication time in the ancestral rice and wheat genomes. The 8 wheat and 42 rice sequences were subjected to a synonymous nucleotide substitution analysis (see Supplemental Table 5 online). To validate the results, we also analyzed 20 paralogous sequences randomly selected from the r11-r12 duplication. Using a mutation rate of 6.5 × 10−9 substitutions per synonymous site per year (Gaut et al., 1996), the results indicate that the r11-r12 duplication event in rice occurred between 14 and 27.3 MYA, which is consistent with the 21 MYA suggested by Yu et al. (2005). The duplication between r1 and r5 is estimated to have occurred 53.2 to 76.3 MYA in rice; the wheat w3-w1 duplication time is estimated as 89.9 to 128.3 MYA. Thus, our estimates are in agreement with published divergence times for rice and wheat from their common ancestor (50 to 70 MYA) and support the idea that duplications occurred in the grass genome ancestor before the divergence of the different grass species.
A Model for the Structural Evolution of Rice and Wheat from a Common Cereal Ancestor
We combined the data obtained in this study on shared duplications between wheat and rice with previous comparative analyses performed between rice and maize (Salse et al., 2004; Wei et al., 2007) and between rice and sorghum (Paterson et al., 2004). The maize and sorghum chromosomes fell into the 12 groups of orthology that we had defined between rice and wheat (Figure 3). Analysis of the conservation pattern resulted in the definition of five ancestral blocks (A5, A7, A11, A8, and A4; Figure 3) containing orthologous chromosomes that exhibit shared ancestral duplications. The detailed analysis of the duplication patterns between rice, wheat, sorghum, and maize within each block (Figure 3) led us to propose a model (Figure 5) for the evolution of these four grass genomes from a common ancestor with five chromosomes that were named A4, A5, A7, A8, and A11, following the current numbering of the rice chromosomes. The first block (ancestral chromosome A5) corresponds to the r5-r1 and w1-w3 shared duplication. In sorghum, the corresponding duplication is found between the sG and sA chromosomes (Paterson et al., 2004). In maize, it is located on chromosomes m3 and m8 as well as on chromosomes m6 and m8 (Figures 3 and 5). The duplication shared between m3 and m8 and between m6 and m8 reflects the recent tetraploidization of the maize genome, whereas the conservation between m3-m8 and m6-m8 dates back to a WGD of the ancestral grass genome with five chromosomes (Figure 5, event 1). This pattern of duplications was recently confirmed by Wei et al. (2007) in a reconstruction of the maize genome evolutionary history through a comparative analysis between a high-resolution integrated physical map of maize and the rice and sorghum genomes.
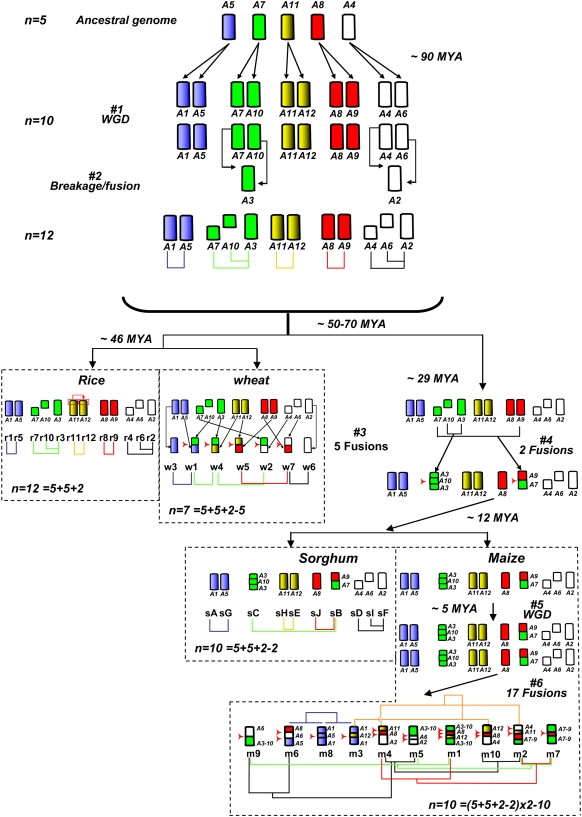
Figure 5.
Model for the Structural Evolution of the Rice, Wheat, Sorghum, and Maize Genomes from a Common Ancestor with n = 5 Chromosomes.
Chromosomes are represented with color codes to illuminate the evolution of segments from a common ancestor with five chromosomes. The five chromosomes are named according to the rice nomenclature. Different events that have shaped the structure of the different grass genomes during their evolution from the common ancestor are indicated with #. For each species, chromosome numbers are followed by a formula indicating the evolutionary origin of this number (e.g., in wheat, n = 7 = 5 [ancestor] + 5 [WGD] + 2 [aneuploid segmental duplication] – 5 [chromosome fusion]). Where possible, divergence times are indicated based on previous estimates by Gaut (2002), Swigonova et al. (2004), and Paterson et al. (2004).
The second block (ancestral chromosome A7) corresponds to the r3-r7-r10 and w4-w2-w1 shared duplications (Figures 3 and 5). Here, the r3-r7 and r3-r10 duplicated regions do not overlap and cover 52% of chromosome 3 (Figure 1A). This pattern of duplication reflects the origin of the r3 chromosome through translocations and fusions between the ancestral chromosomes A7 and A10, as suggested previously by Wang et al. (2005) (Figure 5, event 2). The same two duplications are conserved in wheat between chromosome w4 and the w2 and w1 chromosomes, suggesting that wheat chromosome 4 originates from the same ancestral event. In sorghum, a duplication was observed between the sB and sC chromosomes (Paterson et al., 2004), and it was demonstrated that sC originated from a chromosomal fusion between the ancestral r3 and r10 chromosomes (A3 and A10 in our model; Figure 5). This origin explains why only one duplication can be found in sorghum (Figure 3). The fusion between A3 and A10 occurred in the ancestral genome to maize and sorghum (Figure 5); therefore, the shared duplication found in maize between the m1-m5-m9 and m7-m2 chromosomes (Figure 3) reflects the same pattern. This was also confirmed by the maize genome analysis of Wei et al. (2007).
Block 3 (ancestral chromosome A11) corresponds to the less conserved duplication, since it is detected only between rice r11 and r12, sorghum sH and sE, and maize m2-m4 and m3-m10 (Figure 3). In wheat, this duplication cannot be detected, because of the previously mentioned translocations that have affected the orthologous w4 and w5 chromosomes.
Block 4 (ancestral chromosome A8) corresponds to the duplications shared between rice r8-r9 and wheat w7-w5 (Figure 5). These duplications were also observed by Paterson et al. (2004) on the orthologous sorghum chromosomes sJ and sB (Figure 3) and by Wei et al. (2007) between m1 and m4 and between m2 and m7 (reflecting tetraploidization) as well as between m1-m4 and m2-m7 (reflecting the ancestral duplication) (Figure 3).
Block 5 (ancestral chromosome A4) contains duplications shared between the rice r6-r2-r4, wheat w6-w2 (Figure 5A), and sorghum sI, sF, and sD (Paterson et al., 2004) orthologous chromosomes. This pattern reflects the origin of the A2 chromosome through a fusion between the A4 and A6 ancestral chromosomes (Figure 5, event 2). The duplication between w6 and w7 was not detected in wheat because of the previously mentioned translocations between w4-5 and w7. These duplications are conserved in maize (Wei et al., 2007) between chromosomes m6, m5, and m9, between chromosomes m4 and m5, and between chromosomes m2 and m10 as a result of tetraploidization of the genome. The observed duplications between m6-m5-m9, m4-m5, and m2-m10 reflect the ancestral relationships between the A2, A4, and A6 chromosomes (Figure 5). In the rice–wheat comparison, we had identified seven shared duplications, but only five of them correspond to the ancestral shared duplications that we have identified between the four grass genomes. In fact, the other two (r5-r4/w2-w1 and r4-r8/w2-w7) are found on chromosomes that do not share a common ancestry and are superimposed on regions that are already involved in WGD. They only appeared in the comparative analysis because they are located in syntenic regions.
The analysis of duplications that (1) are shared between the rice, wheat, maize, and sorghum genomes, (2) do not overlap with each other, and (3) tile together to cover the genomes almost entirely led to the model of evolution that is presented in Figure 5, in which an ancestral genome with five chromosomes (A5, A7, A11, A8, and A6) underwent a WGD (event 1) that resulted in five additional chromosomes: A1, A10, A12, A9, and A6. This tetraploidization was followed by two translocations/fusions that have resulted in two new chromosomes, A3 (=A10 + A7) and A2 (=A4 + A6) (event 2), and an intermediate ancestor with n = 12 (5 + 5 + 2) chromosomes. Subsequently, the different grass genomes would have evolved differentially from this ancestral genomic structure. In rice, additional segmental duplications occurred without modifying the basic structure of 12 chromosomes. Thus, the 29 duplication events identified in the rice genome result from three successive rounds of duplications: (1) a WGD (event 1) and two chromosome translocations (event 2); (2) 22 additional segmental duplications overlapping with the WGDs; and (3) recent duplications over ∼3 Mb at the terminal ends of chromosomes r11 and r12.
From the ancestral genome with 12 chromosomes, wheat underwent five chromosomal fusions (event 3 in Figure 5) between A5 and A10, A6 and A8, A9 and A12, A3 and A11, and A4 and A7, resulting in chromosomes w1, w7, w5, w4, and w2, respectively. Chromosomes w3 and w6 originated directly from the ancestral chromosomes A1 and A2, respectively. This resulted in an ancestral wheat genome with n = 7 chromosomes (5 + 5 + 2 − 5). Thus, the 10 duplicated regions observed here in the wheat genome reflect the ancestral WGD (event 1, for seven of the duplications) and three additional segmental duplications that have occurred since the chromosomal fusions.
For maize and sorghum, our findings and model are in complete agreement with the recent analysis of Wei et al. (2007), who showed that both have evolved from an ancestral genome with 12 chromosomes after two chromosomal fusions (between A3 and A10 and between A7 and A9; Figure 5), resulting in an intermediate ancestor with n = 10 chromosomes (5 + 5 + 2 − 2) (event 4 in Figure 5). Then, maize and sorghum evolved independently from this ancestor. While the sorghum genome structure remained similar to the ancestral genome, maize underwent a WGD resulting into an intermediate with n = 20 chromosomes (event 5 in Figure 5). This corresponds to the tetraploidization event described in previous studies (Gaut and Doebley, 1997; Swigonova et al., 2004). Following this event, numerous chromosomal fusions have led to a genome structure with 10 chromosomes (n = 10 = [5 + 5 + 2 − 2] × 2 − 10). At least 17 chromosomal fusions (event 6 in Figure 5) must have occurred to explain the relationships that can be observed today between the different maize chromosomes.
Thus, with this model that proposes a reconstruction of the rice, wheat, sorghum, and maize genomes from an ancestor with n = 5 chromosomes, it is possible to immediately identify the ancestral relationships and the origin (WGD, breakage, fusion) of the different chromosomes in each of the four genomes.
DISCUSSION
Intragenomic and Intergenomic Comparisons Using Improved Integrative Alignment Criteria Reveal Shared Duplications between Rice and Wheat
In this study, we used improved integrative sequence alignment criteria (CIP and CALP) combined with a statistical validation to reanalyze the publicly available data set of rice and wheat sequences. This allowed us to identify orthologs and paralogs with confidence and to infer new relationships compared with previous studies that were either based only on small-scale sequence comparisons (Guyot et al., 2004; Singh et al., 2004; Buell et al., 2005; Rice Chromosome 3 Consortium, 2005) or lacked statistical validation when performed at the whole genome level (Sorrells et al., 2003; La Rota and Sorrells, 2004; Singh et al., 2007). Former comparative studies were performed with a sequence alignment threshold of 80% identity over 100 bases for the best HSP (La Rota and Sorrells, 2004) or with a score of 200 for BLASTN analysis (Singh et al., 2007). We have shown previously that these values may lead to the misidentification of orthologous regions (Salse et al., 2002, 2004), as they allow the detection of members of gene families that are not truly orthologous or of homologies that are based only on conserved domains.
To validate our strategy, we first applied our stringent alignment criteria and statistical validation to the rice genome. We confirmed all of the duplications reported previously (Yu et al., 2005) and identified 19 additional regions duplicated between different rice chromosomes. Yu et al. (2005) had estimated that the duplications of the rice genome cover 65.7% of the genome. In our analysis, the same regions (10 duplications) cover 47.8% of the genome. The difference is due to the accuracy of the method used to determine the limits of the duplicated regions. Yu et al. (2005) adopted a graphical approach because of the background noise produced by BLAST. Using more stringent criteria allowed us to determine the limits of the duplicated regions with high confidence and accuracy, thereby reducing the size of the regions compared with their analysis. The accuracy and robustness of our method are supported by results obtained by the Rice Chromosomes 11 and 12 Sequencing Consortia (2005), who estimated the length of the most recent duplication on the very distal end of chromosomes 11 and 12 as 3.3 Mb. This corresponds exactly to the length that we found with our method (see Supplemental Table 1 online) and is smaller than the estimates (6.5 and 4.8 Mb on chromosomes 11 and 12, respectively) of Yu et al. (2005) for the same region. Moreover, our analysis allowed us to detect 16 novel duplications that are superimposed on regions already identified as duplicated, thereby defining additional ancestral relationships between the rice chromosomes. Interestingly, these relationships were not observed between the orthologous chromosomes in the other grass genomes. This suggests that the superimposed duplications observed here in rice are traces of very ancient duplications that predate the WGD identified in the grass ancestor. Additional grass genome sequences as well as additional tools that allow the estimation of divergence times between very ancient duplications with high accuracy are needed to support this hypothesis.
In wheat, we identified 10 duplicated regions that represent 67.5% of the genome. Previous RFLP mapping studies had suggested ancient duplications in the wheat genome. For example, Dubcovsky et al. (1996) showed that 31% of the RFLP loci in the diploid Triticum monococcum map are present more than once in the genome, and Qi et al. (2004) reported that 19% of the EST markers used for deletion bin mapping were found on nonhomoeologous sets of chromosomes. Until now, however, the extent and localization of the duplicated regions were not defined precisely, and our results provide a picture of duplications in the wheat genome. The 10 identified duplications are probably an underestimation, and it is possible that a number of duplications that were not statistically validated in this study (e.g., w2-w3, w2-w5, and w5-w6) will be confirmed in the future as additional gene-mapping information becomes available. At this time, our analyses demonstrate already that most of the genome is duplicated and strongly suggest an ancient duplication of the diploid wheat genomes before their hybridization into polyploid wheat.
Following the same approach, we reexamined the colinearity between rice and wheat and identified 13 colinear regions that cover 83.1 and 90.4% of the rice and wheat genomes, respectively. Previous comparative analyses between the rice draft sequences and wheat ESTs (Sorrells et al., 2003; Conley et al., 2004; La Rota and Sorrells, 2004; Linkiewicz et al., 2004; Munkvold et al., 2004; Randhawa et al., 2004) also found 13 orthologous segments between the two genomes. Here, we were able to identify more precisely the limits of the orthologous regions and to detect more rearrangements than previously described using a comparison between the complete rice genome sequence (International Rice Genome Sequencing Project, 2005) and a unique set of mapped wheat ESTs with the highest possible length. With this level of resolution, it is no longer possible to split wheat chromosomes into a mosaic of rice orthologous regions (Sorrells et al., 2003; Sorrells, 2004), and the colinearity between the two genomes is more fragmented that previously assumed by comparative mapping. Here, the extent of detectable rearrangements is closer to that observed through comparisons between wheat BAC sequences and orthologous rice genome sequences, in which many exceptions to microcolinearity have been observed (reviewed in Salse and Feuillet, 2007).
By combining data from the intraspecific and interspecific comparisons, we identified seven duplication events that are conserved at orthologous positions between rice and wheat. In this study, we used a combination of individual analysis of duplications within each genome and colinearity assessment between the two genomes using identical alignment criteria and statistical validation. Previous attempts to identify shared duplications between wheat and rice were only based on the analysis of regions that correspond to duplicated regions in rice showing a high proportion of gene matches with two different wheat chromosome groups. This dual synteny approach used recently by Singh et al. (2007) was based on rice genes present in 8 of the 10 regions previously known to be duplicated (Yu et al., 2005) and the identification of homologous wheat ESTs that map in deletion bins on two distinct wheat chromosomes. Using this approach on the 13 orthologous regions that were identified in our study, we found that r1 is orthologous to w3 (207 conserved genes) and to w1 (13 conserved genes) but also to w2 with the same significance (11 conserved genes). Similarly, r5 is orthologous to w1 (102 conserved genes) and to w3 (9 conserved genes) but also to w2 (8 conserved genes) (data not shown). Thus, in contrast with our strategy, it was not possible with this approach to validate statistically that the r1-r5 duplication in rice is shared between w3-w1 in wheat. Singh et al. (2007) also concluded that the region duplicated between r11 and r12 is conserved in wheat between w4-w5, suggesting that the duplication predated the rice and wheat divergence. This conflicts with our findings as well as with previous studies indicating that the r11-r12 duplication is specific to the rice lineage and relatively recent (Yu et al., 2005). These findings provide a note of caution for comparative genome-wide analyses and suggest that applying the same stringent alignment criteria and statistical validation to each of the data sets used for genome comparisons is essential to avoid misinterpretation of evolutionary patterns. Here, by basing our intragenomic and intergenomic comparisons on the highest identity over the longest sequence alignment, we used a conservative approach that excluded sequences that could lead potentially to an overestimation of sequence conservation within and between the rice and wheat genomes. Nevertheless, we were able to detect additional duplications in rice and provide a detailed assessment of genome duplications in wheat, demonstrating the robustness of this approach. Finally, the seven duplications that are shared in rice and wheat are likely underestimated, since the data set of wheat genes for which a chromosomal position is known is currently limited. Future large EST mapping or genome sequencing projects will be required to obtain a complete assessment of the duplications in the wheat genome and of their conservation with the rice genome.
Evolution of the Grass Genomes from an n = 5 Common Ancestor
Individual genome duplication analyses in rice, sorghum, and maize have already suggested an ancestral WGD predating the divergence between the different cereal genomes (Paterson et al., 2004; Yu et al., 2005; Wang et al., 2005), but to date the original basic number of chromosomes remained uncertain (Gaut, 2002). Recently, Wei et al. (2007) thoroughly studied the origin of the maize chromosomes and proposed a model for the evolution of the cereal genomes from an ancestor with n = 12 chromosomes whose structure is similar to the rice genome. Our model is in complete agreement with the proposed evolutionary path from this ancestor but provides additional evidence for the origin of the 12 ancestral chromosomes.
By integrating new data on the duplications of the wheat genome into a detailed comparative analysis of the pattern of shared duplications between rice, wheat, maize, and sorghum, we were able to propose a basic chromosome number of n = 5 for the common ancestor of the cereal genomes. We suggest that predating the divergence between the cereal genomes, 50 to 70 MYA, the ancestor with n = 5 chromosomes underwent a WGD that resulted in an n = 10 intermediate. Following this tetraploidization event, two interchromosomal translocations and fusions led to the construction of two new chromosomes, resulting in the n = 12 intermediate ancestor described previously. This scenario reconciles previous contradictory suggestions that had proposed a grass ancestor with either n = 5 or n = 12 chromosomes (Gaut, 2002), since the evolution from 5 to 12 chromosomes would have happened before the divergence between the different grass genomes. In our model, rice would have retained this original chromosome number, whereas it would have been reduced in the wheat, maize, and sorghum genomes. This is probably a common phenomenon in plant chromosome number evolution. Chromosome fusions, translocations, and inversions have been proposed recently for the evolution of the Arabidopsis thaliana genome (n = 5) from an ancestor at n = 8. Comparative analyses with the related species Arabidopsis lyrata (n = 8) and Capsella rubella (n = 8) have shown that chromosomes 1, 2, and 5 originated through the fusion of translocated and inverted segments of ancestral chromosomes (for review, see Schubert, 2007). The five fusions suggested by our model in wheat likely occurred in the ancestral genome of the Triticeae (wheat, barley, and rye [Secale cereale]) that have a basic chromosome number of n = 7. Preliminary evidence for traces of the presence of ancient duplications was recently reported in a barley/rice colinearity study (Stein et al., 2007) using the dual synteny approach employed by Singh et al. (2007).
Compared with the numerous rearrangements that resulted in a dramatic reduction of the maize chromosome number (from 20 to 10) after tetraploidization (Wei et al., 2007; our data), our model suggests that only two chromosomal fusions have occurred since tetraploidization of the ancestor with n = 5 chromosomes. This low level of rearrangements is similar to what is observed in wheat, in which even after several rounds of polyploidization there is very little disruption of colinearity between the wild diploid ancestors and the polyploid wheat genomes. Therefore, it is possible that the ancestral grass genome contained a gene that prevented homoeologous pairing and reduced the impact of a whole genome doubling, similar to the Ph1 gene in wheat (Martinez-Perez et al., 1999). Since transposable elements can also participate in large rearrangements (Bennetzen, 2005), differences in the amount of transposable elements may influence the degree of genome rearrangements as well. Thus, one can speculate that the ancestral grass genome had a lower amount of transposable elements than maize, thereby reducing their impact on genome rearrangements following the tetraploidization event.
Our model proposes an evolutionary path that reconciles all previous observations and apparent inconsistencies that have been reported in individual grass genome studies. It can serve as a basis for further analyses once the sorghum and maize genome sequences are completed. It will be interesting also to see whether our model remains compatible with the evolution of the emerging temperate grass genome model, Brachypodium distachyon (n = 5), which is at an intermediate evolutionary stage between rice and wheat (Draper et al., 2001) and for which an 8× whole genome shotgun sequence will be completed within the next year (Huo et al., 2006). Finally, to support our hypothesis of an ancestor with n = 5 chromosomes, additional comparative studies are needed with genomes from species outside of the Poales. Members of the Arecales (coconut [Cocos nucifera], palms [Palmae]) or the Zingiberales (banana [Musa spp], ginger [Zingiber officianale]) are good candidates in which to conduct such studies.
METHODS
Nucleic Acid Sequence Alignments
Three new parameters were defined to increase the stringency and significance of BLAST sequence alignment by parsing BLASTN results and rebuilding HSPs or pairwise sequence alignments. The first parameter, AL (aligned length), corresponds to the sum of all HSP lengths. The second, CIP (cumulative identity percentage), corresponds to the cumulative percentage of sequence identity obtained for all of the HSPs (CIP = [∑ ID by HSP/AL] × 100). The third parameter, CALP (cumulative alignment length percentage), represents the sum of the HSP lengths (AL) for all of the HSPs divided by the length of the query sequence (CALP = AL/query length). The CIP and CALP parameters allow the identification of the best alignment (i.e., the highest cumulative percentage of identity in the longest cumulative length), taking into account all HSPs obtained for any pairwise alignment. These parameters were applied to all of the BLAST alignments that were performed in this study. After testing different combinations of CIP and CALP values in the different analyses, stringent values (95% CIP, 85% CALP) were used for the identification of true paralogs among the wheat (Triticum aestivum) EST contigs, whereas less stringent parameters were applied for the analyses of colinearity (60% CIP, 70% CALP) and duplication (70% CIP, 70% CALP).
Wheat and Rice Sequence Databases
The 6426 wheat ESTs representing 15,569 nonredundant loci that were assigned to deletion bins by Qi et al. (2004) were downloaded from the GrainGene website (http://wheat.pw.usda.gov/). Since some ESTs may correspond to nonoverlapping 3′ and 5′ ends of the same cDNA sequence, we developed a unigene set of mapped wheat ESTs by aligning these sequences against the public EST clusters that were produced in the framework of Genoplante projects (http://urgi.versailles.inra.fr/data/banks/). To ensure that each EST is associated with a unique EST contig while maximizing the length of the alignment, very stringent CIP (95%) and CALP (85%) values were applied to the analysis. This led to the identification of WESs (i.e., ESTs that are not associated with any EST cluster) and WECs (i.e., ESTs associated with an EST contig with 95% CIP and 85% CALP). WEC and WES sequences were used for the identification of duplications within the wheat genome and for establishing orthologous relationships between rice (Oryza sativa) and wheat. Since ESTs are not ordered within deletion bins and because deletion bin size is variable between the homoeologous A, B, and D genomes, comparative analysis with the three genomes would be prone to mapping errors. To simplify the analysis and ensure the correct identification of paralogs in wheat and orthologs with rice, the analysis was performed on the seven wheat homoeologous chromosome groups. Thus, any WEC and WES that was mapped in a deletion bin on two or three of the homoeologous chromosomes of a given chromosome group was considered with a unique position on a single consensus chromosome per group. The consensus position was calculated as follows: [(BSt + ((BSi/(NG + 1)) × NGR))/CSi ] × 100, where BSt = bin start coordinate, Bsi = bin size (in percentage of the remaining chromosome [Qi et al., 2004]), NG = number of genes assigned to a bin, NGR = gene rank within a bin, and CSi = total chromosome size (200 = 100 for the long arm and 100 for the short arm according to Qi et al. [2004]). ESTs that mapped on different homoeologous chromosome arms were ignored.
The sequences of the 12 rice pseudomolecules (build 4; 372,077,801 bp) were downloaded from the TIGR website (ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_4.0/), and the 42,654 genes identified by the annotation were used for the analysis of the rice duplications. We chose to perform the analysis with the TIGR annotation rather than the more conservative annotation (∼32,000 genes) that was recently released by the Rice Annotation Project (2007) to increase the probability of finding conserved orthologs even among sequences that were no longer considered in the new annotation because they were only supported by ESTs. To establish the orthologous relationships with wheat, the gene position (coordinates in megabases) on the pseudomolecules was taken into account.
Identification of Duplicated Regions in Wheat and Rice
In wheat, duplications were identified using WESs and WECs corresponding to ESTs that were mapped on two different homoeologous consensus chromosomes for each of the seven chromosome groups. In rice, duplications were identified through genome sequence comparison using the 42,654 rice annotated genes. After alignment of the rice annotated genes using 70% CIP and 70% CALP values, the genes showing a match with another gene elsewhere in the genome were defined as putative paralogous pairs that were used for the identification of duplicated regions.
Identification of Orthologous Regions between Rice and Wheat
Nonredundant WECs and WESs were aligned against the rice annotated genes with 60% CIP and 70% CALP to identify putative orthologous sequences. An in-depth analysis of the alignments showed that the presence of unconserved 5′ or 3′ untranslated regions (UTRs) considerably decreased the overall percentage of identity and disturbed the identification of orthologous versus paralogous genes. To take this problem into account, we estimated the average length of the 5′ and 3′ UTRs in our data set. The average lengths were 250 ± 65.6 and 400 ± 112.5 bp of the 5′ and 3′ UTRs, respectively, for 85% of the WECs or WESs considered in our study. Thus, in addition to sequences showing high similarity (CIP) over the whole length (CALP), we considered as potential orthologs or paralogs all genes for which pairwise sequence alignments showed a minimum of 60% CIP and 70% CALP after systematically excluding the UTR sequences.
A gene (G) was considered rearranged between rice and wheat (i.e., not in same order compared with the flanking orthologs) when Gwp was not contained in the Awp ± sd value (with Gpw representing the EST coordinate in wheat and Awp representing the mean calculated for the 10 upstream and 10 downstream ESTs based on their positions in rice).
Statistical Analysis
Putative paralogous pairs identified during the duplication analyses in the wheat and rice genomes, as well as orthologous gene pairs identified from the colinearity analysis between rice and wheat, were validated through CloseUp analysis (http://www.igb.uci.edu/servers/cgss.html). The statistical analysis is based on a permutation test (Monte Carlo) performed through a randomization of the initial data to select conserved blocks of genes that are not obtained by chance (i.e., that are not obtained in any random distribution performed). A colinear or duplicated region was considered statically significant when the gene density within the region was validated by CloseUp permutation tests with the following parameters: density ratio/cluster length/match number of 0.5/25/5 for wheat and 2/40/5 for rice. The statistical parameters were less stringent for the duplication analysis in wheat because less sequence information (6426) is available compared with rice (42,654). For the validation of orthologous regions in the colinearity analysis, default parameters with a density ratio of 2, a cluster length of 20, and a match number of 5 were used.
Graphical Display
Duplications as well as colinearity were graphically visualized using Genome Pixelizer software (http://niblrrs.ucdavis.edu/GenomePixelizer/GenomePixelizer_Welcome.html). Three data sheets were provided to the software: marker information (name, position on chromosome, chromosome number); link information (data obtained for the statistical identification of paralogous or orthologous sequence pairs); and graphical information (number, size of chromosomes).
Nucleotide Substitution Rate Analysis of the Shared Duplication between Rice Chromosomes 1 to 5 and Wheat Chromosomes 3B to 1B
Synonymous nucleotide substitution values (Nei and Gojobori, 1986) were calculated between paralogous genes in rice (duplication between chromosomes r1-r5) and their wheat orthologs (duplication between chromosomes w1-w3) using the software package MEGA3 (Kumar et al., 2004) after sequence alignment using CLUSTAL W (Thompson et al., 1994). Dating of duplication events was based on a mutation rate of 6.5 × 10−9 substitutions per synonymous site per year (Gaut et al., 1996).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Graphical Representation of the 216 Statistically Validated Wheat Paralogs in Each of the 21 Possible Pairwise Combinations between the Seven Wheat Chromosome Groups.
Supplemental Table 1. CloseUp Analysis of Duplicated Gene Pairs Reveals 29 Duplicated Regions in the Rice Genome.
Supplemental Table 2. CloseUp Analysis of Duplicated Gene Pairs Reveals 10 Duplicated Regions in the Wheat Genome and Two Translocations.
Supplemental Table 3. Detailed Features for the 13 Orthologous Regions Identified between Rice and Wheat.
Supplemental Table 4. Detailed Features for the Seven Duplication Blocks Shared between Wheat and Rice.
Supplemental Table 5. Nucleotide Substitution Rates of the Paralogous Genes Present in the Shared Duplication between Rice Chromosomes 1 to 5 and Wheat Chromosome Groups 1 to 3.
Supplementary Material
Acknowledgments
This work was supported by grants from the Agence Nationale de la Recherche (Grant ANR-05-BLANC-0258-01) and from the Institut National de la Recherche Agronomique.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Catherine Feuillet (catherine.feuillet@clermont.inra.fr).
Online version contains Web-only data.
References
- Ahn, S., and Tanksley, S.D. (1993). Comparative linkage maps of the rice and maize genomes. Proc. Natl. Acad. Sci. USA 90 7980–7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. (2005). Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet. Dev. 15 621–627. [DOI] [PubMed] [Google Scholar]
- Blake, N.K., Lehfeldt, B.R., Lavin, M., and Talbert, L.E. (1999). Phylogenetic reconstruction based on low copy DNA sequence data in an allopolyploid: The B genome of wheat. Genome 42 351–360. [PubMed] [Google Scholar]
- Buell, C.R., et al. (2005). Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res. 15 1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese, P.P., Chakravarty, S., and Vision, T.J. (2003). Fast identification and statistical evaluation of segmental homologies in comparative maps. Bioinformatics 19 74–80. [DOI] [PubMed] [Google Scholar]
- Conley, E.J., et al. (2004). A 2600-locus chromosome bin map of wheat homeologous group 2 reveals interstitial gene rich islands and colinearity with rice. Genetics 168 625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, K.M. (2005). Updating the ‘crop circle.’ Curr. Opin. Plant Biol. 8 155–162. [DOI] [PubMed] [Google Scholar]
- Devos, K.M., and Gale, M.D. (2000). Genome relationships: The grass model in current research. Plant Cell 12 637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, J., Mur, L.A., Jenkins, G., Ghosh-Biswas, G.C., Bablak, P., Hasterok, R., and Routledge, A.P. (2001). Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol. 127 1539–1555. [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky, J., Luo, M.C., Zhong, G.Y., Bransteiter, R., Desai, A., Kilian, A., Kleinhofs, A., and Dvorak, J. (1996). Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L. Genetics 143 983–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Z., Polacco, M., Chen, S., Schroeder, S., Hancock, D., Sanchez, H., and Coe, E. (2003). cMap: The comparative genetic map viewer. Bioinformatics 19 416–417. [DOI] [PubMed] [Google Scholar]
- Feldman, M., Lupton, F.G.H., and Miller, T.E. (1995). Wheats. In Evolution of Crops, 2nd ed, J. Smartt and N.W. Simmonds, eds (London: Longman Scientific), pp. 184–192.
- Feuillet, C., and Keller, B. (2002). Comparative genomics in the grass family: Molecular characterization of grass genome structure and evolution. Ann. Bot. (Lond.) 89 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S. (2002). Evolutionary dynamics of grass genomes. New Phytol. 154 15–28. [Google Scholar]
- Gaut, B.S., and Doebley, J.F. (1997). DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., Morton, B.R., McCaig, B.C., and Clegg, M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyot, R., Yahiaoui, N., Feuillet, C., and Keller, B. (2004). In silico comparative analysis reveals a mosaic conservation of genes within a novel colinear region in wheat chromosome 1AS and rice chromosome 5S. Funct. Integr. Genomics 4 47–58. [DOI] [PubMed] [Google Scholar]
- Hampson, S., McLysaght, A., Gaut, B., and Baldi, P. (2003). LineUp: Statistical detection of chromosomal homology with application to plant comparative genomics. Genome Res. 13 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson, S.E., Gaut, B.S., and Baldi, P. (2005). Statistical detection of chromosomal homology using shared-gene density alone. Bioinformatics 21 1339–1348. [DOI] [PubMed] [Google Scholar]
- Huang, S.X., Sirikhachornkit, A., Faris, J.D., Su, X.J., Gill, B.S., Haselkorn, R., and Gornicki, P. (2002). Phylogenetic analysis of the acetyl-CoA carboxylase and 3-phosphoglycerate kinase loci in wheat and other grasses. Plant Mol. Biol. 48 805–820. [DOI] [PubMed] [Google Scholar]
- Huo, N., Gu, Y.Q., Lazo, G.R., Vogel, J.P., Coleman-Derr, D., Luo, M.C., Thilmony, R., Garvin, D.F., and Anderson, O.D. (2006). Construction and characterization of two BAC libraries from Brachypodium distachyon, a new model for grass genomics. Genome 49 1099–1108. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project (2005). The map-based sequence of the rice genome. Nature 436 793–800. [DOI] [PubMed] [Google Scholar]
- Jaiswal, P., et al. (2006). Gramene: A bird's eye view of cereal genomes. Nucleic Acids Res. 34 D717–D723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, E.A. (2001). Evolutionary history of the grasses. Plant Physiol. 125 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto, N., Higo, H., Abe, K., Arai, S., Saito, A., and Higo, K. (1994). Identification of the duplicated segments in rice chromosomes 1 and 5 by linkage analysis of cDNA markers of known functions. Theor. Appl. Genet. 88 722–726. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- La Rota, M., and Sorrells, M.E. (2004). Comparative DNA sequence analysis of mapped wheat ESTs reveals the complexity of genome relationships between rice and wheat. Funct. Integr. Genomics 4 34–46. [DOI] [PubMed] [Google Scholar]
- Linkiewicz, A.M., et al. (2004). A 2500-locus bin map of wheat homoeologous group 5 provides insights on gene distribution and colinearity with rice. Genetics 168 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez, E., Shaw, P., Reader, S., Aragon-Alcaide, L., Miller, T., and Moore, G. (1999). Homologous chromosome pairing in wheat. J. Cell Sci. 112 1761–1769. [DOI] [PubMed] [Google Scholar]
- Mickelson-Young, L., Endo, T.R., and Gill, B.S. (1995). A cytogenetic ladder-map of the wheat homoeologous group-4 chromosomes. Theor. Appl. Genet. 90 1007–1011. [DOI] [PubMed] [Google Scholar]
- Miftahudin, R.K., et al. (2004). Analysis of expressed sequence tag loci on wheat chromosome group 4. Genetics 168 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold, J.D., et al. (2004). Group 3 chromosome bin maps of wheat and their relationship to rice chromosome 1. Genetics 168 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamura, Y., et al. (1995). Conservation of duplicated segments between rice chromosomes 11 and 12. Breed. Sci. 45 373–376. [Google Scholar]
- Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3 418–426. [DOI] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., and Chapman, B.A. (2004). Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA 101 9903–9908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., Peterson, D.G., Estill, J.C., and Chapman, B.A. (2003). Structure and evolution of cereal genomes. Curr. Opin. Genet. Dev. 13 644–650. [DOI] [PubMed] [Google Scholar]
- Qi, L.L., et al. (2004). A chromosome bin map of 16,000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa, H.S., et al. (2004). Deletion mapping of homoeologous group 6-specific wheat expressed sequence tags. Genetics 168 677–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse, J., and Feuillet, C. (2007). Comparative genomics of cereals. In Genomics-Assisted Crop Improvement, Vol. 1, R. Varshney and R. Tuberosa, eds (Dordrecht, The Netherlands: Springer-Verlag), pp. 177–205.
- Salse, J., Piegu, B., Cooke, R., and Delseny, M. (2002). Synteny between Arabidopsis thaliana and rice at the genome level: A tool to identify conservation in the ongoing rice genome sequencing project. Nucleic Acids Res. 30 2316–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse, J., Piegu, B., Cooke, R., and Delseny, M. (2004). New in silico insight into the synteny between rice (Oryza sativa L.) and maize (Zea mays L.) highlights reshuffling and identifies new duplications in the rice genome. Plant J. 38 396–409. [DOI] [PubMed] [Google Scholar]
- Schubert, I. (2007). Chromosome evolution. Curr. Opin. Plant Biol. 10 109–115. [DOI] [PubMed] [Google Scholar]
- Singh, N.K., et al. (2004). Sequence analysis of the long arm of rice chromosome 11 for rice-wheat synteny. Funct. Integr. Genomics 4 102–117. [DOI] [PubMed] [Google Scholar]
- Singh, N.K., et al. (2007). Single-copy genes define a conserved order between rice and wheat for understanding differences caused by duplication, deletion, and transposition of genes. Funct. Integr. Genomics 7 17–35. [DOI] [PubMed] [Google Scholar]
- Sorrells, M. (2004). Cereal genomics research in the post-genomic era. In Cereal Genomics, P.K. Gupta and R.K. Varshney, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 559–584.
- Sorrells, M.E., et al. (2003). Comparative DNA sequence analysis of wheat and rice genomes. Genome Res. 13 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, N., et al. (2007). A 1,000-loci transcript map of the barley genome: New anchoring points for integrative grass genomics. Theor. Appl. Genet. 114 823–839. [DOI] [PubMed] [Google Scholar]
- Swigonova, Z., Lai, J.S., Ma, J.X., Ramakrishna, W., Llaca, V., Bennetzen, J.L., and Messing, J. (2004). Close split of sorghum and maize genome progenitors. Genome Res. 14 1916–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Annotation Project (2007). Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 17 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Chromosome 3 Sequencing Consortium (2005). Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res. 15 1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice Chromosomes 11 and 12 Consortia (2005). The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol. 3 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K., Saeys, Y., Simillion, C., Raes, J., and Van de Peer, Y. (2002). The automatic detection of homologous regions (ADHoRe) and its application to microcolinearity between Arabidopsis and rice. Genome Res. 12 1792–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele, K., Simillion, C., and Van de Peer, Y. (2003). Evidence that rice and other cereals are ancient aneuploids. Plant Cell 15 2192–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Shi, X., Hao, B., Ge, S., and Luo, J. (2005). Duplication and DNA segmental loss in the rice genome: Implications for diploidization. New Phytol. 165 937–946. [DOI] [PubMed] [Google Scholar]
- Wei, F., et al. (2007). Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet. 3 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel, J.F., Stuber, C.W., Goodman, M.M., and Beckett, J.B. (1989). Duplicated plastid and triplicated cytosolic isozymes of triosephosphate isomerase in maize (Zea mays L.). J. Hered. 3 218–228. [DOI] [PubMed] [Google Scholar]
- Yu, J., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92. [DOI] [PubMed] [Google Scholar]
- Yu, J., et al. (2005). The genomes of Oryza sativa: A history of duplications. PLoS Biol. 3 266–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Q., Ouyang, S., Liu, J., Suh, B., Cheung, F., Sultana, R., Lee, D., Quackenbush, J., and Buell, C.R. (2003). The TIGR rice genome annotation resource: Annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res. 31 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.