Abstract
RNA replication and systemic trafficking play significant roles in developmental regulation and host–pathogen interactions. Viroids are the simplest noncoding eukaryotic RNA pathogens and genetic units that are capable of autonomous replication and systemic trafficking and offer excellent models to investigate the role of RNA structures in these processes. Like other RNAs, the predicted secondary structure of a viroid RNA contains many loops and bulges flanked by double-stranded helices, the biological functions of which are mostly unknown. Using Potato spindle tuber viroid infection of Nicotiana benthamiana as the experimental system, we tested the hypothesis that these loops/bulges are functional motifs that regulate replication in single cells or trafficking in a plant. Through a genome-wide mutational analysis, we identified multiple loops/bulges essential or important for each of these biological processes. Our results led to a genomic map of viroid RNA motifs that mediate single-cell replication and systemic trafficking, respectively. This map provides a framework to enable high-throughput studies on the tertiary structures and functional mechanisms of RNA motifs that regulate viroid replication and trafficking. Our model and approach should also be valuable for comprehensive investigations of the replication and trafficking motifs in other RNAs.
INTRODUCTION
RNA replication not only represents a crucial milestone in the evolution of life based on the RNA World scenario (Gilbert, 1986; Joyce, 2002) but also impacts modern life as exemplified by the amplification of infectious RNAs and RNA silencing signals (Baulcombe, 2004; Ding and Voinnet, 2007). Upon synthesis, some plant RNAs traffic into neighboring cells or even to distant organs to regulate global gene expression and profoundly influence development and defense (Lough and Lucas, 2006; Ding and Itaya, 2007; Ding and Voinnet, 2007). RNA trafficking, an integral part of viral systemic infection, often involves the active role of viral-encoded proteins (Lucas, 2006).
Structural elements in viral RNAs that regulate replication have been extensively studied (Hull, 2002; Flint et al., 2004; Miller and White, 2006). By contrast, the molecular mechanisms that control RNA cell-to-cell trafficking remain poorly understood. The idea that an RNA contains specific structural motifs to mediate trafficking (Citovsky and Zambryski, 2000; Lucas et al., 2001) is gaining increasing experimental support. Two viroid motifs that mediate trafficking between specific cells have been identified (Qi et al., 2004; Zhong et al., 2007). A cis-element in the 5′ untranslated region of potexviral RNA, which plays a role in replication (Miller et al., 1998), mediates cell-to-cell trafficking of a fused reporter RNA (Lough et al., 2006). The recent demonstration that Brome mosaic virus RNAs can traffic long distance in the absence of replication suggests that these RNAs have structural elements directly recognized by cellular factors for trafficking (Gopinath and Kao, 2007). The untranslated regions of potato (Solanum tuberosum) BEL5 mRNA appear to be important for long-distance trafficking in regulating tuber formation (Banerjee et al., 2006). Structural motifs yet to be identified also control the trafficking of other cellular RNAs (Haywood et al., 2005).
We use Potato spindle tuber viroid (PSTVd) as a model to investigate RNA motifs that mediate replication and systemic trafficking, given its outstanding biological and structural features. Viroids are the smallest and simplest eukaryotic RNA pathogens and genetic units that are capable of autonomous replication and systemic trafficking. Without protein-coding capacity, encapsidation, and helper viruses, the circular viroid genomic RNAs express all biological functions directly. To initiate replication in single cells, the (+)-strand circular genomic RNA is imported into the nucleus (for family Pospiviroidae) or chloroplast (for family Avsunviroidae). This is followed by transcription into (−)-strands that serve as the replication intermediates to synthesize the (+)-strands. This rolling circle of replication also includes cleavage of concatemeric transcription products and ligation of the cleaved products into circular molecules. Upon export out of the organelle (i.e., nucleus or chloroplast), the viroid RNAs embark on systemic trafficking that includes movement across different cellular boundaries in an inoculated leaf, entry into the phloem, long-distance movement within the phloem, and exit out of the phloem and into another leaf or other organ (Flores et al., 2005; Ding and Itaya, 2007; Owens, 2007). Thus, a viroid RNA presents a simple model to investigate many RNA-based biological processes.
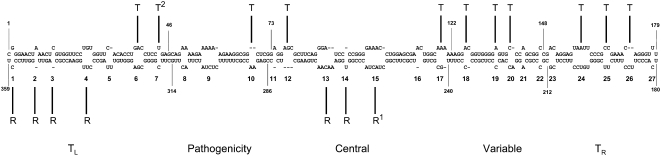
The secondary structure of PSTVd is one the best understood among RNAs, making it a highly tractable system to dissect RNA structural features that mediate specific biological functions. The 359-nucleotide PSTVd genome was predicted to form a thermodynamically favorable rod-like secondary structure in its native in vitro state (Gross et al., 1978). Based on sequence and structural comparisons among members of the family Pospiviroidae, this structure can be divided into five domains: the left and right terminal, pathogenicity, central, and variable (Figure 1; Keese and Symons, 1985). This rod-like structural model is well supported by microscopic and biophysical studies (Riesner et al., 1979), chemical/enzymatic mapping (Gast et al., 1996), and nuclear magnetic resonance spectroscopic studies (left terminal domain; Dingley et al., 2003). Mutational studies support the importance of the rod-like secondary structure of PSTVd for infection (Hammond and Owens, 1987; Hammond, 1994; Wassenegger et al., 1994; Hu et al., 1997; Owens and Thompson, 2005). Biophysical/chemical studies identified several premelting regions of low thermostability in the native structure of PSTVd, and denaturation of this structure initiated at these regions and subsequent refolding led to the formation of metastable hairpin (HP) structures via pairing of distant complementary nucleotide sequences (Riesner, 1987; Riesner et al., 1979).
Figure 1.
Functional Analysis of PSTVd Structural Motifs.
The in vitro native rod-like secondary structure of PSTVd showing the five structural domains is based on Keese and Symons (1985). The 27 loops in this secondary structure are numbered from left to right. The arrows indicate introduced mutations in the helical regions. The bars show the positions of nucleotides that are proposed to base pair to form metastable HPI and HPII structures as shown in Figure 3. TL, left terminal domain; TR, right terminal domain.
Earlier studies showed that many mutations rendered PSTVd noninfectious in inoculated tomato (Solanum lycopersicum) plants (Hammond and Owens, 1987; Loss et al., 1991; Owens et al., 1991, 1995; Qu et al., 1993; Hu et al., 1996). Such results were often interpreted as inhibited replication. However, because viroid accumulation was analyzed in either locally inoculated or systemic leaves, it was not feasible to conclude whether noninfection for a given mutant is attributed to defects in replication in single cells or to defects in systemic trafficking. For example, the lower titer of PSTVdNT compared with PSTVdNB in systemic leaves of tobacco (Nicotiana tabacum) did not result from its lower replication capacity but resulted from its confinement within the vascular tissue (Qi et al., 2004). Therefore, the specific functional defects of those noninfectious mutants remain to be resolved.
Transcription of the (+)-strand PSTVd RNA probably initiates at U359 or C1 at the left terminal loop based on in vitro studies as well as reversion of mutated nucleotides in infected tomato (Kolonko et al., 2006). HPII has long been thought to play a role in replication, particularly transcription of the (−)-strand RNA, based on the reversion of HPII-disrupting nucleotides to the wild type in infected tomato plants (Loss et al., 1991; Owens et al., 1991; Qu et al., 1993; Candresse et al., 2001). Consistent with this hypothesis, HPII could be detected in vitro and in vivo (Schröder and Riesner, 2002). HPI also appears to be important for infectivity based on the findings that mutations predicted to disrupt HPI abolish infectivity in tomato plant (Hammond and Owens, 1987). Direct experimental data, including loss-of-function genetic evidence, are necessary to further test the role of these structures in replication in single cells.
There is compelling evidence for the role of some loop structures in PSTVd infection. The loop E (Figure 1) in the central region was first shown to contain tertiary structure by in vitro UV cross-linking (Branch et al., 1985) and further supported by chemical/enzymatic mapping (Gast et al., 1996). It has been shown to play a role in in vitro processing (Baumstark et al., 1997; Schrader et al., 2003) and host adaptation (Wassenegger et al., 1996; Qi and Ding, 2002; Zhu et al., 2002). Recent studies led to a precise tertiary structural model of PSTVd loop E and loss-of-function genetic evidence for its role in replication (Zhong et al., 2006) and demonstration of its existence in vivo (Eiras et al., 2007; Wang et al., 2007). A bipartite motif mediates PSTVd trafficking from bundle sheath to mesophyll in young tobacco leaves (Qi et al., 2004). The U43/C318 loop forms a tertiary structure that is necessary for PSTVd to traffic from the bundle sheath into the phloem in Nicotiana benthamiana leaves (Zhong et al., 2007).
A major obstacle in advancing mechanistic studies of viroid RNA structures in relation to specific biological functions has been a lack of comprehensive understanding of the biological significance of the numerous secondary structural features, such as the loops and bulges. For all RNAs, loops and bulges have generally been thought to consist of nucleotides that are unpaired with functions elusive. However, rapidly accumulating evidence from x-ray crystal and nuclear magnetic resonance structural studies demonstrate that most RNA loops and bulges are highly structured three-dimensional motifs, formed via non-Watson-Crick base pairing, base stacking, and other base interactions, that serve as the major sites for RNA–RNA, RNA–protein, and RNA–small ligand interactions (Leontis et al., 2002, 2006; Holbrook, 2005; Noller, 2005). Based on these advances in our understanding of the RNA loop/bulge structures and functions, we hypothesize that most, if not all, of the loops/bulges in the rod-shaped secondary structure of PSTVd represent functional motifs that have distinct functions in replication and/or trafficking.
To test this hypothesis, we conducted a genome-wide mutational analysis of the role of these loops/bulges during single-cell replication and systemic trafficking in N. benthamiana. PSTVd replicates efficiently in cultured cell protoplasts (Qi and Ding, 2002) and easily develops systemic infection in this plant (Hu et al., 1997; Zhu et al., 2001). Through this analysis, we identified multiple loops/bulges as functional motifs that are critical for replication or trafficking. In addition, we found that several PSTVd mutants previously shown to be noninfectious in tomato were capable of replication in N. benthamiana single cells but were defective in systemic trafficking. Finally, we obtained evidence that HPI and HPII are not essential for PSTVd replication in N. benthamiana. These results support the hypothesis that the loops/bulges in the rod-shaped secondary structure of PSTVd serve as major functional motifs that likely interact with cellular factors to accomplish various aspects of replication and systemic trafficking during infection. We present a genomic map of the most critical motifs, which provides a framework to enable high-throughput studies on the tertiary structures and functional mechanisms of RNA motifs that regulate viroid replication and trafficking. Our model and approach should also be valuable for comprehensive investigations of the replication and trafficking motifs in other RNAs.
RESULTS
Generation of PSTVd Mutants with Disrupted Loops
We used the intermediate strain of PSTVd (wild type; Gross et al., 1978) as our experimental model. For simplicity of description, the term “loop” is used in this report to broadly include both loops and nucleotide bulges. Numbered from left to right, there are a total of 27 loops in the PSTVd secondary structure (Figure 1). We previously showed loop 15 (loop E) to be essential for replication (Zhong et al., 2006) and loop 7 (U43/C318 motif) to be critical for systemic trafficking (Zhong et al., 2007). Therefore, they were not included for detailed analyses in this study. However, we did include A99C, a loop E–disruptive mutant that is defective in replication (Zhong et al., 2006), as a negative control in addition to mock inoculation in all replication experiments.
To examine the function of the other 25 loops in PSTVd replication and systemic trafficking, we generated a series of mutants with each containing one disrupted loop. Specifically, we designed mutations to close each internal loop by introducing canonical Watson-Crick base pairs or by deleting bulged nucleotides. The two terminal loops cannot simply be closed due to structural constraints. Therefore, we generated mutations to enlarge each of them based on previous studies (Hammond and Owens, 1987). None of the mutations affected the overall PSTVd secondary structure as predicted by mfold (Zuker, 2003; Figure 2).
Figure 2.
Thermodynamic Prediction of Secondary Structures of Loop Mutants by mfold (Zuker, 2003).
Only partial sequences are shown. Red letters denote introduced mutations.
We also investigated the effects of helical mutations on PSTVd infection (see arrows in Figure 1 for specific mutations). These mutations were shown in previous studies to affect PSTVd infection (Hammond and Owens, 1987; Owens et al., 1991). These included G46C (Hammond and Owens, 1987), G26C, and G65U (Owens et al., 1991) that completely abolished infectivity in tomato plants. Whether they inhibited replication in single cells or systemic trafficking was unknown.
Finally, we also generated mutants with disrupted HPII and HPI to test the function of these metastable structures in replication and systemic trafficking. Based on the HPII structural model and previous analyses (Loss et al., 1991; Qu et al., 1993), each of the substitutions C229U, C231A, and G324U is predicted to disrupt HPII structure (see arrows in Figure 3). Each of the substitutions G80A and C109U is predicted to disrupt HPI (Hammond and Owens, 1987; see arrows in Figure 3). Double mutations C231A/G324U and G80A/C109U are predicted to restore formation of HPII and HPI, respectively (Qu et al., 1993; Hammond and Owens, 1987).
Figure 3.
The Nucleotide Sequences and Secondary Structures of HPI and HPII.
The arrows indicate the nucleotide substitutions used in this study.
For all mutants, we tested their replication in single cells and systemic trafficking in a whole plant of N. benthamiana. Specific results are presented below.
Genomic Locations of Loops That Are Most Critical for Replication in Single Cells
To determine the replication capacity of each mutant, we used the accumulation level of its circular genomic RNA in inoculated protoplasts of N. benthamiana cultured cells as an indicator (Qi and Ding, 2002). The protoplast assay avoids complications of cell-to-cell movement associated with assays using inoculated or systemic leaves because the accumulation level of a mutant in a leaf is determined by its replication capacity in single cells and the extent of its movement between cells. Three days after inoculation of protoplasts, RNA was extracted and analyzed by RNA gel blot hybridization using PSTVd-specific riboprobes. The accumulation levels of circular PSTVd RNA were quantified, based on results from three biological replicates for each mutant, and the data are presented as the percentage of the wild-type level. As shown in Figure 4, in negative control experiments, mutant A99C (which has nucleotide A99 substituted by C to disrupt loop E) accumulated to <5% of the wild-type level, consistent with previous results (Zhong et al., 2006). The other loop mutants exhibited different levels of accumulation compared with the wild type, ranging from ∼10 to >110% (Figure 4). Representative RNA gel blots showing wild-type and mutant PSTVd accumulation levels are presented in Figure 5A.
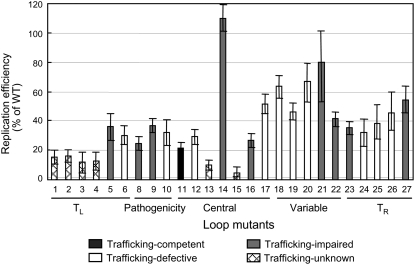
Figure 4.
Summary of Replication and Systemic Trafficking Functions of Loop Mutants.
The replication level of a mutant is presented as the percentage of the wild-type level. The systemic trafficking function of a loop mutant is indicated by column shading, with black denoting trafficking-competent mutants, white denoting trafficking-defective mutants, gray denoting trafficking-impaired mutants, and hatched denoting trafficking-unknown mutants.
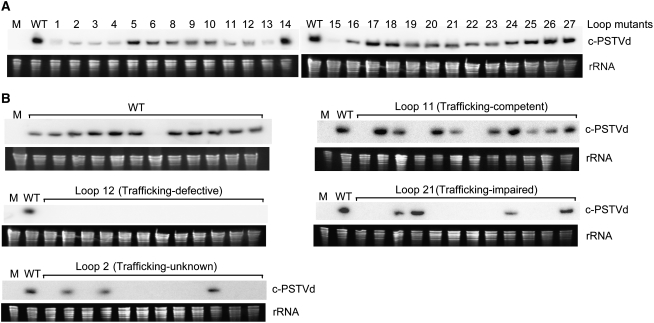
Figure 5.
RNA Gel Blot Analyses of the Replication and Systemic Trafficking Functions of PSTVd Mutants.
(A) Representative RNA gel blots showing accumulation of the circular molecules of the wild type and mutants of PSTVd (c-PSTVd) in N. benthamiana protoplasts.
(B) Representative RNA gel blots showing presence or absence of the circular molecules of the wild type, trafficking-competent (loop 11), trafficking-defective (loop 12), trafficking-impaired (loop 21), and trafficking-unknown (loop 2) mutants of PSTVd, respectively, in 12 of the N. benthamiana plants inoculated in each case. M, mock inoculation.
Examination of the overall patterns reveals that the loops most critical for replication in single cells reside in the distal end of the left terminal domain and the central region. Specifically, enlarging loop 1, deleting loops 2 and 3, and closing loop 4 in the left terminal domain reduced replication to below 16%, and disruption of loop 15 (loop E) and loop 13 in the central region reduced replication to 5 to 10% of the wild-type level. These low levels of replication indicate that presence of each of these loops is critical for replication.
The loops in the proximal end of the left terminal (loops 5 and 6), variable (loops 18 to 22), pathogenicity (loops 8 to 10), and right terminal domains (loops 23 to 27) are also important, but less critical, for replication. Disruption of loops in these domains reduced replication to 25 to 80% of the wild-type levels. Among these, the variable domain appears to be the least important for replication, given the overall higher replication levels of mutants in this domain compared with those of mutants in the other domains (Figure 4).
Further analyses reveal a complex pattern of loop mutant behavior in the central region. While disruption of loop 15 (loop E) and loop 13 reduced replication to 5 to 10% of the wild-type level as mentioned above, deletion of loop 14 surprisingly enhanced replication to >110% of the wild-type level. Disruption of the other loops reduced replication to 20 to 50% of the wild-type levels. Strikingly, as discussed below, while disruption of loop 11 reduced replication to ∼21%, it is the only mutant that maintained normal systemic trafficking.
PSTVd Loops That Are Critical for Systemic Trafficking
To identify PSTVd loops that might be important for systemic trafficking, we mechanically inoculated each mutant onto the first two true leaves of 2-week-old N. benthamiana seedlings. At 4 weeks after inoculation, the presence or absence of PSTVd in systemic leaves was determined by dot and RNA gel blots. Twelve plants, from three biological replicates each involving four plants, were inoculated with each mutant and the wild type. The results are pooled and presented in Tables 1 to 3. To determine the trafficking function of a mutant, several factors need to be considered: (1) the replication level of the mutant in protoplasts, (2) the number of inoculated plants that showed systemic infection, and (3) the maintenance of introduced mutations in the RNA progeny. Because reduced replication levels could influence the trafficking capacity of PSTVd as a population, it is important to determine the minimal replication level, relative to the wild type, that is still sufficient to sustain systemic infection. As shown in Figure 4 (black column), deletion of loop 11 reduced PSTVd accumulation to slightly >20% of the wild-type level in protoplasts. However, this mutant systemically infected nine out of the 12 inoculated plants (Figure 5B; Table 5). Sequencing of viroid progeny from four of the systemically infected plants showed maintenance of the deletion in two plants and additional mutations in the other two plants (Table 5). Although the biological effects of additional mutations remain to be understood, it is clear that 20% of the wild-type replication level is sufficient for systemic trafficking. Based on these considerations, we provisionally classified the trafficking functions of loop mutants into four categories (Figure 4, Tables 1 to 3; for representative RNA gel blot data, see Figure 5B): (1) trafficking competent, (2) trafficking defective, (3) trafficking impaired, and (4) trafficking unknown. Each is described in detail below.
Table 1.
Trafficking-Defective Mutants
| Loop | Mutations | Domaina | Replicationb | Traffickingc |
|---|---|---|---|---|
| Wild type | 100% | 11/12 | ||
| 6 | G36U/A37C/C38G | TL | 30% | 0/12 |
| 10 | A290Δ/A291Δ | P | 32% | 0/12 |
| 12 | A77Δ/G78Δ/C79Δ | CR | 29% | 0/12 |
| 17 | A118G/A119C/A120Δ | CR | 52% | 0/12 |
| 18 | A126G/C127Δ | V | 63% | 0/12 |
| 19 | C227U | V | 46% | 0/12 |
| 20 | A222G/C223Δ | V | 73% | 0/12 |
| 24 | U157G/A158G/U161A | TR | 25% | 0/12 |
| 25 | C166A/C167A | TR | 29% | 0/12 |
| 26 | C172GGAd | TR | 46% | 0/12 |
Structural domains in native secondary structure. TL, left terminal; P, pathogenicity; CR, central region; V, variable; TR, right terminal.
Replication efficiency of mutants expressed as a percentage of wild-type PSTVd.
Trafficking function is expressed as number of plants showing systemic infection, determined by dot blot, over total number of plants inoculated.
Insertion of GA between residues 172 and 173.
Table 2.
Trafficking-Impaired Mutants
| Loop | Mutations | Domain | Replication | Trafficking | Genetic Stability |
|---|---|---|---|---|---|
| Wild type | 100% | 11/12 | S | ||
| 5 | U332Δ/U333G | TL | 36% | 2/12 | M/R |
| 8 | A50G/A51U | P | 24% | 1/12 | R |
| 9 | C301Δ/C303U/A305U | P | 37% | 2/12 | M/R |
| 14 | U267Δ | CR | 110% | 4/12 | M |
| 16 | A112Δ/A113Δ | CR | 27% | 2/12 | M |
| 21 | A142U | V | 82% | 4/12 | M |
| 22 | C147G | V | 41% | 5/12 | M/R |
| 23 | A150C/C151G | TR | 35% | 4/12 | M |
| 27 | U177A/U178A | TR | 53% | 1/12 | M |
See Table 1 for legend. S, stable; R, reversion to the wild type; M, additional mutations.
Table 3.
Trafficking-Unknown Mutants
| Loop | Mutations | Domain | Replication | Trafficking | Genetic Stability |
|---|---|---|---|---|---|
| Wild type | 100% | 11/12 | S | ||
| 1 | G2U/A4C/C6G | TL | 16% | 1/12 | R |
| 2 | A8Δ | TL | 16% | 3/12 | M |
| 3 | C13Δ | TL | 12% | 2/12 | M |
| 4 | U22A/U24G | TL | 13% | 0/12 | |
| 13 | A271Δ/A272Δ/C273U/A274C/A275C | CR | 10% | 1/12 | M |
See Table 1 for legend. S, stable; R, reversion to the wild type; M, additional mutations.
Table 5.
Sequencing of Viroid Progeny from a Fraction of Systemically Infected Plants
| Loop | Original Mutations | Trafficking | No. of Plants Sequenced | Progeny Sequences (No. of Plants) |
|---|---|---|---|---|
| 11 | A74Δ | 9/12 | 4 | A74Δ(2) |
| A74G/U312C(1) | ||||
| U257A(1) | ||||
| 5 | U332Δ/U333G | 2/12 | 1 | WT+C42U/U355C(1)a |
| 9 | C301Δ/C303U/A305U | 2/12 | 1 | WT+U257A(1)a |
| 14 | U267Δ | 4/12 | 2 | U267Δ/C216U(1) |
| U267Δ/U316C(1) | ||||
| 16 | A112Δ/A113Δ | 2/12 | 2 | A112Δ/A113Δ/G243U(1) |
| A112Δ/A113Δ/U247A(1) | ||||
| 21 | A142U | 4/12 | 4 | A142U/G221U(3) |
| A142U/A219U(1) | ||||
| 22 | C147G | 5/12 | 3 | WT(1) |
| C231A(1) | ||||
| C167A(1) | ||||
| 23 | A150C/C151G | 4/12 | 2 | WT(1) |
| A150C/C151G/A135G(1) | ||||
| 27 | U177A/U178A | 1/12 | 1 | U177A/U178A/A182U(1) |
| 2 | A8Δ | 3/12 | 2 | A8Δ/A171G(2) |
| 3 | C13Δ | 2/12 | 2 | C13Δ/U316C(2) |
| 13 | A271Δ/A272Δ/C273U/A274C/A275C | 1/12 | 1 | C273U/A274C/A275C/U316C(1) |
| HPII | C229U | 3/12 | 3 | WT(2) |
| C229U/C216U(1) | ||||
| HPII | C231A | 9/12 | 5 | C231A(2) |
| WT(1) | ||||
| C231A/C216U(2) | ||||
| HPII | G324U | 8/12 | 4 | G324U(2) |
| WT(1) | ||||
| G324U/U329G(1) |
WT, reversion to wild-type sequences. Letters in bold indicate new mutations.
Mixture of wild-type and additional mutations.
Trafficking-Competent Mutant
As discussed above, the loop 11 mutant is the only one in this category.
Trafficking-Defective Mutants
These mutants accumulated to >20% of the wild-type levels in the protoplasts but failed to accumulate in systemic leaves in all of the 12 inoculated plants. These mutants included loops 6, 10, 12, 17, 18, 19, 20, 24, 25, and 26 (Table 1, white columns in Figure 4). These loops are mostly clustered in the variable and right terminal domains and at the junction between the pathogenicity domain and central region.
Trafficking-Impaired Mutants
These mutants accumulated to >20% of the wild-type levels in the protoplasts. However, they showed systemic infection in <50% of the inoculated plants. As shown in Table 2 and gray columns in Figure 4, loop mutants 5, 8, 9, 14, 16, 21, 22, 23, and 27 belong to this group. We sequenced viroid progeny from a fraction of the systemically infected plants to determine whether the trafficking mutants maintained only the original mutations, acquired new mutations, or reverted to the wild type. In all cases, we detected reversions or additional mutations (Table 5). Because the effects of these spontaneous mutations on PSTVd systemic trafficking are not clear, the conservative designation of “trafficking impaired” suggests that the original mutations did not abolish, but inhibited to some extent, the trafficking function.
Trafficking-Unknown Mutants
Loop mutants 1, 2, 3, 4, and 13 accumulated to <20% of the wild-type level in protoplasts (Table 3, hatched columns in Figure 4). Systemic accumulation of the mutants in this group was absent in most of the inoculated plants (Table 3). Sequencing of RNA progeny from a fraction of the systemically infected plants detected additional mutations (Table 5). How these additional mutations affect trafficking and replication is unclear at this stage. Because 20% is below the currently known replication level sufficient to support trafficking, we do not know whether compromised systemic accumulation of these mutants is due to the low replication levels or specific defects in systemic trafficking.
Mutations in Helical Regions alter Neighboring Loop Structures and Inhibit PSTVd Systemic Trafficking
As discussed above, mutations G46C (Hammond and Owens, 1987), G26C, and G65U (Owens et al., 1991) completely abolished PSTVd infectivity in tomato plants, but whether they inhibited replication or trafficking is unclear. As summarized in Table 4, our analyses showed that these mutants replicated to 23 to 46% of the wild-type, levels but all failed to accumulate in systemic leaves of N. benthamiana plants. Sequencing of viroid progeny from the only plant systemically infected by G65U showed reversion to the wild-type sequence.
Table 4.
Replication and Trafficking of Helical, HPI, and HPII Mutants
| Mutation | Metastable Structure | Domain | Replication | Trafficking | Genetic Stability | Tomato Infectivity |
|---|---|---|---|---|---|---|
| Wild type | 100% | 11/12 | S | S | ||
| G26C | TL | 23% | 0/12 | N1 | ||
| G46C | TL | 42% | 0/12 | N1 | ||
| G65U | P | 46% | 1/12 | R | N2 | |
| C229U | HPII-Dis | V | 54% | 3/12 | R/M | R3 |
| C231A | HPII-Dis | V | 95% | 9/12 | S/R/M | N3 |
| G324U | HPII-Dis | TL | 82% | 8/12 | S/R/M | N3 |
| C231A/G324U | HPII-Res | TL+V | 86% | 8/12 | S | N3 |
| G80A | HPI-Dis | CR | 53% | 0/12 | N1 | |
| C109U | HPI-Dis | CR | 72% | 0/12 | N1 | |
| G80A/C109U | HPI-Res | CR | 66% | 0/12 | N1 |
See Table 1 for legend. S, stable; R, reversion to the wild type; M, additional mutations; N, noninfectious; Dis, predicted to disrupt HPI or HPII structure; Res, predicted to restore HPI or HPII structure. 1, Hammond and Owens (1987); 2, Owens et al. (1991); 3, Qu et al. (1993).
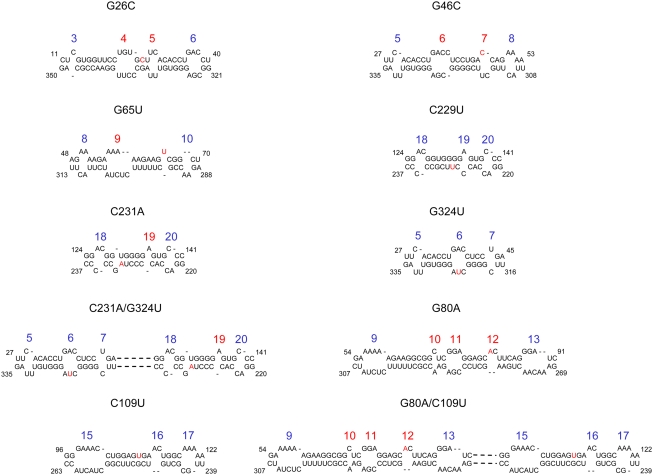
Secondary structural prediction by mfold (Zuker, 2003) showed that all mutations not only disrupted local helices but also affected the adjacent loops (Figure 6). For example, G26C enlarges loops 4 and 5, and G46C enlarges loops 6 and 7. G65U alters loop 9 and creates a small bulge between loops 9 and 10. These data suggest that defects in systemic trafficking may result from alteration of the loops, which were shown independently to be important for trafficking by the above loop-disruption mutations. However, we cannot exclude the role of helical structures at this stage.
Figure 6.
Thermodynamic Prediction of Secondary Structures of Helical, HPII, and HPI Mutants by mfold (Zuker, 2003).
Only partial sequences are shown. Red letters denote introduced mutations. The blue numbers indicate no change, and the red numbers indicate structural changes in the loops. The dashed lines represent unchanged structures.
Disruption of HPII and HPI Formation Does Not Inhibit PSTVd Replication in Single Cells
Previous studies showed that mutations predicted to disrupt the HPII core structure always reverted to wild type in infected tomato plants, suggesting the importance of HPII for replication (Loss et al., 1991; Owens et al., 1991; Qu et al., 1993). However, reversion is not direct evidence of HPII functions. To directly test the potential role of HPII in replication, we analyzed the replication and trafficking functions of selected HPII mutants. As shown in Table 4, C229U, C231A, and G324U mutations, each predicted to disrupt the HPII core structure, maintained replication functions in N. benthamiana protoplasts. While C231A, G324U, and C231A/G324U also trafficked systemically, C229U showed impairment in systemic trafficking. Sequencing confirmed maintenance of the introduced mutations in plants inoculated by C231A, G324U, and C231A/G324U. We also detected several additional mutations in the viroid progeny from plants inoculated by C231A and G324U but not by C231A/G324U (Table 5). These mutations are not expected to affect the HPII structure, but their biological effect is unknown. These data showed that HPII formation is not essential for PSTVd replication in N. benthamiana cells. Analysis by mfold showed no effect of C229U substitution on the local secondary structure of PSTVd (Figure 6). The inhibitory effect of this mutation on systemic trafficking suggests that either HPII plays a role in trafficking or more likely the specific nucleotides in the short helix between loops 18 and 19 play a role, together with these loops, in trafficking.
HPI is also suggested to be important for infectivity in tomato plants (Hammond and Owens, 1987). As shown in Table 4, the two HPI-disrupting single mutations, G80A and C109U, as well as the HPI-restoring double mutant G80A/C109U (Hammond and Owens, 1987) replicated to 53 to 72% of the wild-type levels, but all failed to establish systemic infection in N. benthamiana. Therefore, HPI formation is not necessary for PSTVd replication in this plant. Whether it is involved in systemic trafficking is an interesting issue to be pursued further (see Discussion).
DISCUSSION
Despite more than three decades of extensive studies, how a viroid RNA functions to accomplish single-cell replication and systemic trafficking remains largely unknown. More specifically, although the PSTVd secondary structural model, including the structural domains, has been well studied by many means, there is very limited knowledge of how this model is related to various specific biological functions necessary to establish systemic infection. One approach to break this bottleneck is to obtain a genomic map of the functional motifs that will enable high-throughput mechanistic studies. To this end, we conducted a genome-wide mutational analysis to determine the roles of various PSTVd loops in single-cell replication or systemic trafficking. We found that disruption of nearly every loop had an impact on either replication or systemic trafficking of PSTVd. There are two plausible explanations for our observations. First, the closing of some loops may lead to enhanced stability of the native structure of PSTVd, preventing its denaturation to form alternative structures necessary for certain functions in replication or trafficking. Second, many, if not all, of the loops in the PSTVd secondary structure serve as functional motifs that likely interact with cellular factors to regulate distinct biological processes necessary to establish systemic infection. It is important to stress that these are not mutually exclusive possibilities. The nucleotide sequences of a loop may be critical for the formation of metastable structures, whereas its tertiary structure can serve as a functional motif.
For the structural stability model, an example is the A135G substitution that closes the A135/C227 loop (loop 19 in our numbering scheme), which stabilizes a “premelting” region that may lead to inhibition of HPII formation (Owens et al., 1991). The A135G substitution rendered PSTVd noninfectious in tomato (Owens et al., 1991). We closed loop 19 by C227U substitution (Figure 2), which could have similar stabilizing effects as A135G substitution does. This mutation reduced PSTVd replication to 46% of the wild-type level and abolished its systemic trafficking capacity in N. benthamiana (Figure 4, Table 1).
In a more direct approach to test the biological functions of putative metastable structures, we analyzed the effects of HPII and HPI mutations on replication in single cells or systemic trafficking. HPII has long been thought to play an important role in PSTVd replication (Loss et al., 1991; Owens et al., 1991; Qu et al., 1993; Candresse et al., 2001). In this study, we showed that none of the tested HPII-disruptive mutations abolished PSTVd replication in N. benthamiana protoplasts. In fact, the HPII mutants replicated to 54 to 95% of the wild-type levels. More significantly, we recovered HPII-disrupting mutants from systemically infected leaves for the first time. Taken together, these data provide compelling evidence suggesting that HPII is not essential for replication in N. benthamiana. At this stage, however, we cannot rule out the possibility that HPII functions in a host-specific manner. Furthermore, there remains a possibility that PSTVd can use HPII and the stable secondary structure as alternative platforms to regulate transcription, based on in vitro studies (Repsilber et al., 1999). Further experiments are clearly needed to test critically these possibilities. We also showed that HPI-disrupting mutants maintained 53 to 72% of the replication functions in N. benthamiana, suggesting that HPI is not essential for replication in N. benthamiana cells. The observations that the two HPI-disrupting mutants and the HPI-restoring mutant failed to traffic systemically raise the intriguing question whether HPI plays a role in trafficking. On one hand, the inhibitory effect of G80A on trafficking can be explained in the HPI context. On the other hand, G80A mutation distorts loops 10 and 12, which are essential for trafficking (Figure 6, Table 4). The C109U mutation does not alter the helix or neighboring loops. Its trafficking-inhibitory effect suggests that either the HPI structure or specific nucleotides in the helix plays a role in trafficking. The double mutant G80A/C109U is predicted to restore HPI but not the native secondary structure (Hammond and Owens, 1987). Therefore, its inhibition of PSTVd trafficking appears to argue against a role of HPI in trafficking. However, we showed previously that certain compensatory mutations can predictably restore an RNA motif but cannot restore fully the motif function (Zhong et al., 2006). Thus, further experiments will be necessary to address the potential role of HPI in systemic trafficking in N. benthamiana and to determine whether it functions in a host-specific manner.
Our functional motif model is consistent with the demonstration that most RNA loops and bulges are highly structured three-dimensional motifs that serve as the major sites for RNA–RNA, RNA–protein, and RNA–small ligand interactions (Leontis et al., 2002, 2006; Holbrook, 2005; Noller, 2005). This interpretation is also supported by the findings that the tertiary structure of loop E (loop 15) and U43/C318 loop (loop 7) is critical for replication (Zhong et al., 2006) and phloem entry (Zhong et al., 2007), respectively. Figure 7 presents a PSTVd genomic map of loop motifs that are most critical for replication or systemic trafficking in N. benthamiana. To be conservative, we include only loops the disruption of that (1) reduced PSTVd replication to below 16% of the wild-type level, (2) enhanced the replication above the wild-type level, and (3) rendered PSTVd defective in systemic trafficking. We also combined findings from our previous work on the requirement of loop E (loop 15) for replication (Zhong et al., 2006) and U43/C318 motif (loop 7) for vascular entry (Zhong et al., 2007). This map shows that, in the PSTVd secondary structure, the motifs most critical for replication are clustered in the distal end of the left terminal domain and central region, whereas the trafficking motifs are mostly clustered in the variable and right terminal domains, the proximal end of the left terminal domain, and at the junction of pathogenicity/central domains. This map represents a major advance over previous mutational studies by (1) its genome-wide identification of functional motifs and by (2) its distinction of motif functions for replication at the cellular level and trafficking at the whole-plant level. It integrates biological functions with the well-established structural domains at the whole genomic level and establishes a new framework for whole-genome approaches to elucidate the infection mechanisms of a viroid RNA. To determine whether these motifs are present in other members of the genus Pospiviroid, we examined the equivalent positions of other viroid sequences in the genus that are predicted to fold into similar secondary structures as PSTVd. Many loops are indeed present in Chrysanthemum stunt viroid, Citrus exocortis viroid, Columnea latent viroid, Mexican papita viroid, Tomato apical stunt viroid, Tomato chlorotic dwarf viroid, and Tomato planta macho viroid (see Supplemental Figure 1 online; Singh et al., 2003; Zhong et al., 2007; Subviral RNA Database, http://subviral.med.uottawa.ca/cgi-bin/home.cgi). Such secondary structural conservation lends further support to the functional importance of these loops. However, RNA motifs are often more conserved at the tertiary structural rather than sequence/secondary structural levels (Leontis et al., 2006). Therefore, a future focus of studies will be the determination of the tertiary structures of all the motifs, as recently reported for loop 7 (Zhong et al., 2007) and loop 15 (loop E; Zhong et al., 2006) to understand fully structural conservation in relation to function.
Figure 7.
A Genomic Map of PSTVd Loop Motifs That Are Essential/Critical for Replication and Systemic Trafficking.
1Data from Zhong et al. (2006); 2 data from Zhong et al. (2007). R, replication; T, trafficking.
Our results provide loss-of-function evidence to further establish the role of the left terminal loop and the central region in replication. In particular, the importance of loop 1 for replication is consistent with the in vitro mapping of transcription initiation sites at the left terminal loop (Kolonko et al., 2006). The finding also provides clear evidence that failed replication at the cellular level can account for the noninfection of mutant PSTVd-P, based on which loop 1 mutations were designed, in N. benthamiana (Hu et al., 1997) and likely also in tomato (Hammond and Owens, 1987). It will be of great interest to determine whether loops 1 to 4 in the left terminal domain function at the same or distinct steps of replication. The importance of the central region in replication has long been suggested (Keese and Symons, 1985). It is clearly involved in processing in vitro (Baumstark et al., 1997; Schrader et al., 2003). We previously provided genetic evidence for the requirement of loop E (loop 15) in this region for replication in vivo (Zhong et al., 2006). As additional genetic evidence to support the essential role of the central region in replication, we identified loop 13 as another motif in this region that is essential for replication. Furthermore, deletion of loop 14 (bulged U) consistently led to enhanced replication, albeit at a modest level. The significance of this striking observation remains to be understood. It does, however, raise the intriguing question of whether positive and negative regulatory elements exist in PSTVd to maintain an optimal level of replication beneficial to the viroid population. With regard to mechanisms, defects in intracellular trafficking such as nuclear import, RNA stability (including resistance to general nuclease activities and RNA silencing), transcription, subcellular localization, or in vivo processing would affect accumulation of a mutant in the protoplasts. Thus, our collection of PSTVd mutants provides valuable materials for future studies to probe the specific roles of the PSTVd loops in these processes and identification of the cellular factors that interact with these loops for function.
We previously showed that a bipartite motif was necessary and sufficient to mediate PSTVd trafficking from the bundle sheath to mesophyll in young tobacco leaves (Qi et al., 2004) and that loop 7 (U/C motif) is required for trafficking from the bundle sheath into the phloem in N. benthamiana (Zhong et al., 2007). Whether these motifs function alone or synergistically with additional motifs to mediate trafficking in these cases remains an open issue. Furthermore, there is no information about the motifs that mediate trafficking across other cellular boundaries. Our identification of many motifs that are essential or important for systemic trafficking should make it possible to address these issues. It should be pointed out that the distinction between trafficking-defective and trafficking-impaired mutants is quite arbitrary at this stage, with the reason why systemic infection occurred in a small fraction of the plants inoculated by the latter group of mutants remaining to be understood. Nonetheless, further investigations using a combination of cellular, molecular, and structural approaches (Qi et al., 2004; Zhong et al., 2007) should provide important insights into the specific role of each of the identified loops in trafficking across various cellular boundaries. Here, two observations are particularly interesting. The identification of loops 24, 25, and 26 at the right terminal domain as essential for trafficking is consistent with previous findings that mutations in the right terminal loop inhibited PSTVd systemic infection when inoculated onto tomato (Hammond, 1994). This is also the region that has been shown to interact with VIRP1 (Gozmanova et al., 2003; Maniataki et al., 2003), a bromodomain-containing protein from tomato recently shown to be important for PSTVd infection (Kalantidis et al., 2007). Thus, the right terminal region can be a focus for further studies on the RNA–protein interactions that direct trafficking across some cellular boundaries yet to be identified. A potential implication of our findings is that multiple motifs may also be involved in regulating systemic trafficking of viral and cellular RNAs, an important issue that remains to be addressed.
In summary, our results provide a foundation for genome-wide mechanistic studies of the PSTVd structural elements in replication and systemic trafficking. These include more extensive studies on the relative contributions of the native versus metastable structures in replication and trafficking using a combination of biophysical, structural, genetic, and molecular approaches. Furthermore, our results provide a guide for targeted studies on the tertiary structures of various motifs and how each of them functions in a specific step of replication and trafficking. Finally, the pool of mutants with known functional defects will facilitate investigation of the cognate cellular factors. These findings and their implications support the notion that PSTVd can be a productive model to investigate comprehensively how distinct RNA motifs interact with cellular factors to regulate a wide range of cellular processes of general biological significance. Our model and approaches should be useful to investigate comprehensively the structure-function relationships of other RNAs.
METHODS
Plant Materials and Growth Conditions
Nicotiana benthamiana plants were grown in a growth chamber controlled at 14-h-light (27°C)/10-h-dark (24°C) cycles. Cultured cells of N. benthamiana were maintained in Murashige and Skoog medium (MS salts; Life Technologies) supplemented with 30 g/L of sucrose, 256 mg/L of KH2PO4, 100 mg/L of myo-inositol, 1 mg/L of thiamine, and 1 mg/L of 2,4-D with a final pH of 5.5. The detailed protocols are described by Zhong et al. (2005).
PSTVd cDNA Construction
Plasmid pRZ6-2 containing cDNAs of PSTVdInt was constructed by Hu et al. (1997) and was a gift from Robert Owens. All PSTVd-derived mutants were generated by site-directed mutagenesis using the Quickchange site-directed mutagenesis kit (Stratagene) using pRZ:PSTVdInt as the template. The introduced mutations were verified by sequencing. Construction of pInter(–) was described by Qi and Ding (2002).
In Vitro Transcription
To prepare PSTVd inocula, HindIII-linearized plasmid pRZ6-2 containing PSTVd cDNA was used as the template for in vitro transcription with T7 MEGAscript (Ambion). To prepare riboprobes for RNA gel blotting, [α-32P]UTP-labeled antisense riboprobes were prepared by in vitro transcription using T7 MAXIscript kit (Ambion) using SpeI-linearized pInter(–) as the template. After in vitro transcription, the DNA templates were removed by digestion with RNase-free DNase I. The RNA transcripts were purified with MEGAClear kit (Ambion). Nonradioactive and radioactive RNA transcripts were quantified by UV spectrometry or scintillation counting, respectively.
Plant and Protoplast Infection
The in vitro transcripts of PSTVd variants were used to inoculate the carborundum-dusted first two true leaves of 2-week-old N. benthamiana plants (300 ng/plant). DEPC-H2O was used for mock inoculation. Four weeks after inoculation, total RNAs were extracted from systemic leaves for RNA gel blot analysis. N. benthamiana protoplasts were prepared and transfected with PSTVd transcripts by electroporation as described by Zhong et al. (2005). At 3 d after inoculation, transfected protoplasts were collected for RNA extraction and gel blot analysis.
RNA Extraction and RNA Gel Blots
Total RNA from infected plants was isolated using Trizol reagent (Invitrogen), and total RNA from protoplasts was extracted using the RNeasy plant mini kit (Qiagen) according to the manufacturer's instructions. Five micrograms of total RNA was separated on a 5% polyacrylamide/8 M urea gel. After electrophoresis, the RNA was transferred to a Hybond-XL nylon membrane (Amersham Biosciences) using a vacuum blotting system (Amersham) and immobilized by UV cross-linking. Hybridization with [α-32P]UTP-labeled riboprobes was performed at 65°C using ULTRAhyb reagent (Ambion). After overnight hybridization, the membranes were washed twice in 2× SSC/0.1% SDS for 15 min and twice in 0.2× SSC/0.1% SDS for 15 min at 65°C and exposed to a Storage Phosphor Screen (Kodak). Hybridization signals were quantified with the Molecular Imager FX using Quantity One-4.1.1 software (Bio-Rad).
Sequencing of RNA Progeny
The protocols for preparing cDNAs of the PSTVd progeny isolated from plants were essentially as described by Qi and Ding (2002). Briefly, cDNAs of PSTVd RNA were RT-PCR amplified and sequenced in both directions using the ABI377 DNA sequencer (Perkin-Elmer) at the DNA Sequencing Facility at Ohio State University.
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure 1. Secondary Structural Conservation of Some PSTVd Loops in Other Species of Genus Pospiviroid, Including Chrysanthemum stunt viroid, Citrus exocortis viroid, Columnea latent viroid, Mexican papita viroid, Tomato apical stunt viroid, Tomato chlorotic dwarf viroid, and Tomato planta macho viroid.
Supplementary Material
Acknowledgments
Xiaorong Tao generated some of the mutants for this study. We are indebted to Robert Owens and Asuka Itaya for insightful discussions. We thank Asuka Itaya, Ying Wang, and Ryuta Takeda for critical reading of the manuscript. This work was supported by grants from the National Science Foundation (IBN-0238412 and IOB-0620143).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Biao Ding (ding.35@osu.edu).
Online version contains Web-only data.
References
- Banerjee, A.K., Chatterjee, M., Yu, Y., Suh, S.G., Miller, W.A., and Hannapel, D.J. (2006). Dynamics of a mobile RNA of potato involved in a long-distance signaling pathway. Plant Cell 18 3443–3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D. (2004). RNA silencing in plants. Nature 431 356–363. [DOI] [PubMed] [Google Scholar]
- Baumstark, T., Schroder, A.R., and Riesner, D. (1997). Viroid processing: Switch from cleavage to ligation is driven by a change from a tetraloop to a loop E conformation. EMBO J. 16 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch, A.D., Benenfeld, B.J., and Robertson, H.D. (1985). Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc. Natl. Acad. Sci. USA 82 6590–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candresse, T., Gora-Sochacka, A., and Zagorski, W. (2001). Restoration of secondary hairpin II is associated with restoration of infectivity of a non-viable recombinant viroid. Virus Res. 75 29–34. [DOI] [PubMed] [Google Scholar]
- Citovsky, V., and Zambryski, P. (2000). Systemic transport of RNA in plants. Trends Plant Sci. 5 52–54. [DOI] [PubMed] [Google Scholar]
- Ding, B., and Itaya, A. (2007). Control of directional macromolecular trafficking across specific cellular boundaries: A key to integrative plant biology. J. Integr. Plant Biol. 49 1227–1234. [Google Scholar]
- Ding, S.W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingley, A.J., Steger, G., Esters, B., Riesner, D., and Grzesiek, S. (2003). Structural characterization of the 69 nucleotide potato spindle tuber viroid left-terminal domain by NMR and thermodynamic analysis. J. Mol. Biol. 334 751–767. [DOI] [PubMed] [Google Scholar]
- Eiras, M., Kitajima, E.W., Flores, R., and Daros, J.A. (2007). Existence in vivo of the loop E motif in potato spindle tuber viroid RNA. Arch. Virol. 152 1389–1393. [DOI] [PubMed] [Google Scholar]
- Flint, S.J., Enquist, L.W., Racaniello, V.R., and Skalka, A.M. (2004). Principles of Virology. (Washington, DC: ASM Press).
- Flores, R., Hernandez, C., Martinez de Alba, A.E., Daros, J.A., and Di Serio, F. (2005). Viroids and viroid-host interactions. Annu. Rev. Phytopathol. 43 117–139. [DOI] [PubMed] [Google Scholar]
- Gast, F.U., Kempe, D., Spieker, R.L., and Sanger, H.L. (1996). Secondary structure probing of potato spindle tuber viroid (PSTVd) and sequence comparison with other small pathogenic RNA replicons provides evidence for central non-canonical base-pairs, large A-rich loops, and a terminal branch. J. Mol. Biol. 262 652–670. [DOI] [PubMed] [Google Scholar]
- Gilbert, W. (1986). The RNA world. Nature 319 618. [Google Scholar]
- Gopinath, K., and Kao, C.C. (2007). Replication-independent long-distance trafficking by viral RNAs in Nicotiana benthamiana. Plant Cell 19 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozmanova, M., Denti, M.A., Minkov, I.N., Tsagris, M., and Tabler, M. (2003). Characterization of the RNA motif responsible for the specific interaction of potato spindle tuber viroid RNA (PSTVd) and the tomato protein Virp1. Nucleic Acids Res. 31 5534–5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, H.J., Domdey, H., Lossow, C., Jank, P., Raba, M., Alberty, H., and Sänger, H.L. (1978). Nucleotide sequence and secondary structure of potato spindle tuber viroid. Nature 273 203–208. [DOI] [PubMed] [Google Scholar]
- Hammond, R.W. (1994). Agrobacterium-mediated inoculation of PSTVd cDNAs onto tomato reveals the biological effect of apparently lethal mutations. Virology 201 36–45. [DOI] [PubMed] [Google Scholar]
- Hammond, R.W., and Owens, R.A. (1987). Mutational analysis of potato spindle tuber viroid reveals complex relationships between structure and infectivity. Proc. Natl. Acad. Sci. USA 84 3967–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haywood, V., Yu, T.S., Huang, N.C., and Lucas, W.J. (2005). Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant J. 42 49–68. [DOI] [PubMed] [Google Scholar]
- Holbrook, S.R. (2005). RNA structure: The long and the short of it. Curr. Opin. Struct. Biol. 15 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y., Feldstein, P.A., Bottino, P.J., and Owens, R.A. (1996). Role of the variable domain in modulating potato spindle tuber viroid replication. Virology 219 45–56. [DOI] [PubMed] [Google Scholar]
- Hu, Y., Feldstein, P.A., Hammond, J., Hammond, R.W., Bottino, P.J., and Owens, R.A. (1997). Destabilization of potato spindle tuber viroid by mutations in the left terminal loop. J. Gen. Virol. 78 1199–1206. [DOI] [PubMed] [Google Scholar]
- Hull, R. (2002). Matthew's Plant Virology. (San Diego, CA: Academic Press).
- Joyce, G.F. (2002). The antiquity of RNA-based evolution. Nature 418 214–221. [DOI] [PubMed] [Google Scholar]
- Kalantidis, K., Denti, M.A., Tzortzakaki, S., Marinou, E., Tabler, M., and Tsagris, M. (2007). Virp1 is a host protein with a major role in Potato spindle tuber viroid infection in Nicotiana plants. J. Virol. 81 12872–12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keese, P., and Symons, R.H. (1985). Domains in viroids: Evidence of intermolecular RNA rearrangements and their contribution to viroid evolution. Proc. Natl. Acad. Sci. USA 82 4582–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolonko, N., Bannach, O., Aschermann, K., Hu, K.H., Moors, M., Schmitz, M., Steger, G., and Riesner, D. (2006). Transcription of potato spindle tuber viroid by RNA polymerase II starts in the left terminal loop. Virology 347 392–404. [DOI] [PubMed] [Google Scholar]
- Leontis, N.B., Lescoute, A., and Westhof, E. (2006). The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 16 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis, N.B., Stombaugh, J., and Westhof, E. (2002). Motif prediction in ribosomal RNAs Lessons and prospects for automated motif prediction in homologous RNA molecules. Biochimie 84 961–973. [DOI] [PubMed] [Google Scholar]
- Loss, P., Schmitz, M., Steger, G., and Riesner, D. (1991). Formation of a thermodynamically metastable structure containing hairpin II is critical for infectivity of potato spindle tuber viroid RNA. EMBO J. 10 719–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough, T.J., Lee, R.H., Emerson, S.J., Forster, R.L., and Lucas, W.J. (2006). Functional analysis of the 5′ untranslated region of potexvirus RNA reveals a role in viral replication and cell-to-cell movement. Virology 351 455–465. [DOI] [PubMed] [Google Scholar]
- Lough, T.J., and Lucas, W.J. (2006). Integrative plant biology: Role of phloem long-distance macromolecular trafficking. Annu. Rev. Plant Biol. 57 203–232. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J. (2006). Plant viral movement proteins: Agents for cell-to-cell trafficking of viral genomes. Virology 344 169–184. [DOI] [PubMed] [Google Scholar]
- Lucas, W.J., Yoo, B.C., and Kragler, F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2 849–857. [DOI] [PubMed] [Google Scholar]
- Maniataki, E., Martinez de Alba, A.E., Gesser, R.S., Tabler, M., and Tsagris, M. (2003). Viroid RNA systemic spread may depend on the interaction of a 71-nucleotide bulged hairpin with the host protein VirP1. RNA 9 346–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, E.D., Plante, C.A., Kim, K.H., Brown, J.W., and Hemenway, C. (1998). Stem-loop structure in the 5′ region of potato virus X genome required for plus-strand RNA accumulation. J. Mol. Biol. 284 591–608. [DOI] [PubMed] [Google Scholar]
- Miller, W.A., and White, K.A. (2006). Long-distance RNA-RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 44 447–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller, H.F. (2005). RNA structure: reading the ribosome. Science 309 1508–1514. [DOI] [PubMed] [Google Scholar]
- Owens, R. (2007). Potato spindle tuber viroid: The simplicity paradox resolved? Mol. Plant Pathol. 8 549–560. [DOI] [PubMed] [Google Scholar]
- Owens, R.A., Chen, W., Hu, Y., and Hsu, Y.H. (1995). Suppression of potato spindle tuber viroid replication and symptom expression by mutations which stabilize the pathogenicity domain. Virology 208 554–564. [DOI] [PubMed] [Google Scholar]
- Owens, R.A., and Thompson, S.M. (2005). Mutational analysis does not support the existence of a putative tertiary structural element in the left terminal domain of Potato spindle tuber viroid. J. Gen. Virol. 86 1835–1839. [DOI] [PubMed] [Google Scholar]
- Owens, R.A., Thompson, S.M., and Steger, G. (1991). Effects of random mutagenesis upon potato spindle tuber viroid replication and symptom expression. Virology 185 18–31. [DOI] [PubMed] [Google Scholar]
- Qi, Y., and Ding, B. (2002). Replication of Potato spindle tuber viroid in cultured cells of tobacco and Nicotiana benthamiana: The role of specific nucleotides in determining replication levels for host adaptation. Virology 302 445–456. [DOI] [PubMed] [Google Scholar]
- Qi, Y., Pélissier, T., Itaya, A., Hunt, E., Wassenegger, M., and Ding, B. (2004). Direct role of a viroid RNA motif in mediating directional RNA trafficking across a specific cellular boundary. Plant Cell 16 1741–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, F., Heinrich, C., Loss, P., Steger, G., Tien, P., and Riesner, D. (1993). Multiple pathways of reversion in viroids for conservation of structural elements. EMBO J. 12 2129–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repsilber, D., Wiese, S., Rachen, M., Schröder, A.W., Riesner, D., and Steger, G. (1999). Formation of metastable RNA structures by sequential folding during transcription: Time-resolved structural analysis of potato spindle tuber viroid (-)-stranded RNA by temperature-gradient gel electrophoresis. RNA 5 574–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesner, D. (1987). Structure formation. In The Viroids, T.O. Diener, ed (New York: Plenum), pp. 63–98.
- Riesner, D., Henco, K., Rokohl, U., Klotz, G., Kleinschmidt, A.K., Domdey, H., Jank, P., Gross, H.J., and Sänger, H.L. (1979). Structure and structure formation of viroids. J. Mol. Biol. 133 85–115. [DOI] [PubMed] [Google Scholar]
- Schrader, O., Baumstark, T., and Riesner, D. (2003). A mini-RNA containing the tetraloop, wobble-pair and loop E motifs of the central conserved region of potato spindle tuber viroid is processed into a minicircle. Nucleic Acids Res. 31 988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder, A.R., and Riesner, D. (2002). Detection and analysis of hairpin II, an essential metastable structural element in viroid replication intermediates. Nucleic Acids Res. 30 3349–3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R.P., Ready, K.F.M., and Nie, X. (2003). Viroids of solanaceous species. In Viroids, A. Hadidi, R. Flores, J.W. Randles, and J.S. Semancik, eds (Collingwood, Australia: CSIRO), pp. 125–133.
- Wang, Y., Zhong, X., Itaya, A., and Ding, B. (2007). Evidence for the existence of the loop E motif of Potato spindle tuber viroid in vivo. J. Virol. 81 2074–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M., Heimes, S., and Sänger, H.L. (1994). An infectious viroid RNA replicon evolved from an in vitro-generated non-infectious viroid deletion mutant via a complementary deletion in vivo. EMBO J. 13 6172–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M., Spieker, R.L., Thalmeir, S., Gast, F.U., Riedel, L., and Sänger, H.L. (1996). A single nucleotide substitution converts potato spindle tuber viroid (PSTVd) from a noninfectious to an infectious RNA for Nicotiana tabacum. Virology 226 191–197. [DOI] [PubMed] [Google Scholar]
- Zhong, X., Itaya, A., and Ding, B. (2005). Transfecting protoplasts by electroporation to study viroid replication. In Cuurent Protocols in Microbiology, R. Coico, T. Kowalik, J.M. Quarles, B. Stevenson, and R.K. Taylor, eds (New York: John Wiley & Sons), pp. 16D.14.11–16D.14.11. [DOI] [PubMed]
- Zhong, X., Leontis, N.B., Qian, S., Itaya, A., Qi, Y., Boris-Lawrie, K., and Ding, B. (2006). Tertiary structural and functional analyses of Loop E motif in viroid RNA reveal its essential role in RNA-templated RNA replication by the nuclear transcription machinery. J. Virol. 80 8566–8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, X., Tao, X., Stombaugh, J., Leontis, N., and Ding, B. (2007). Tertiary structure and function of an RNA motif required for plant vascular entry to initiate systemic trafficking. EMBO J. 26 3836–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Green, L., Woo, Y.M., Owens, R., and Ding, B. (2001). Cellular basis of potato spindle tuber viroid systemic movement. Virology 279 69–77. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., Qi, Y., Xun, Y., Owens, R., and Ding, B. (2002). Movement of potato spindle tuber viroid reveals regulatory points of phloem-mediated RNA traffic. Plant Physiol. 130 138–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.