Abstract
The GNOM protein plays a fundamental role in Arabidopsis thaliana development by regulating endosome–to–plasma membrane trafficking required for polar localization of the auxin efflux carrier PIN1. GNOM is a family member of large ARF guanine nucleotide exchange factors (ARF-GEFs), which regulate vesicle formation by activating ARF GTPases on specific membranes in animals, plants, and fungi. However, apart from the catalytic exchange activity of the SEC7 domain, the functional significance of other conserved domains is virtually unknown. Here, we show that a distinct N-terminal domain of GNOM mediates dimerization and in addition interacts heterotypically with two other conserved domains in vivo. In contrast with N-terminal dimerization, the heterotypic interaction is essential for GNOM function, as mutations abolishing this interaction inactivate the GNOM protein and compromise its membrane association. Our results suggest a general model of large ARF-GEF function in which regulated changes in protein conformation control membrane association of the exchange factor and, thus, activation of ARFs.
INTRODUCTION
In eukaryotic cells, specificity and integrity of membrane compartments is maintained through cargo-selective vesicle trafficking. A fundamental regulatory step in vesicle formation is the activation of small ADP-ribosylation factor (ARF) GTPases by exchanging their bound GDP for GTP, which is mediated by the catalytic SEC7 domain of ARF guanine nucleotide exchange factors (ARF-GEFs). ARF-GEFs reversibly associate with specific membranes and thus control ARF GTPase activity in space and time. However, nothing is known about the mechanism of membrane localization of large ARF-GEFs and their regulation. Pleckstrin-homology domains that bind to inositol lipids mediate membrane association of small and medium-sized ARF-GEFs in animals and yeast (Blomberg et al., 1999; Gillingham and Munro, 2007). By contrast, large ARF-GEFs, which constitute the only family conserved in all groups of eukaryotes and the only family present in plants, lack a classical membrane association domain. Instead, they comprise five noncatalytic domains, in addition to their catalytic Sec7 domain: the N-terminal dimerization and cyclophilin binding (DCB) domain, the adjacent homology upstream of SEC7 (HUS) domain, and three homology downstream of SEC7 (HDS) domains (Figure 1A) (Cox et al., 2004; Mouratou et al., 2005). However, the functional significance of the different domains is ill defined. When expressed as a transgenic protein fragment, the DCB and the HUS domain were reported to be sufficient for membrane association of the human large ARF-GEF GBF1 (Mansour et al., 1999). However, the DCB domain has been shown to mediate dimerization of large ARF-GEFs in yeast two-hybrid and in vitro experiments (Grebe et al., 2000), and the transgenic DCB-HUS fragment could therefore be passively tethered to the membrane by its interaction with endogenous full-length GBF1. Also, a mutation of a conserved motif in the HUS domain reduces membrane association of yeast Gea2p, suggesting its functional requirement (Park et al., 2005). In addition, HDS domains of Gea1p and Gea2p interact with the integral membrane protein Gmh1p, which might act as their membrane receptor. However, loss of Gmh1p does not substantially reduce the membrane association of the interacting GEFs (Chantalat et al., 2003).
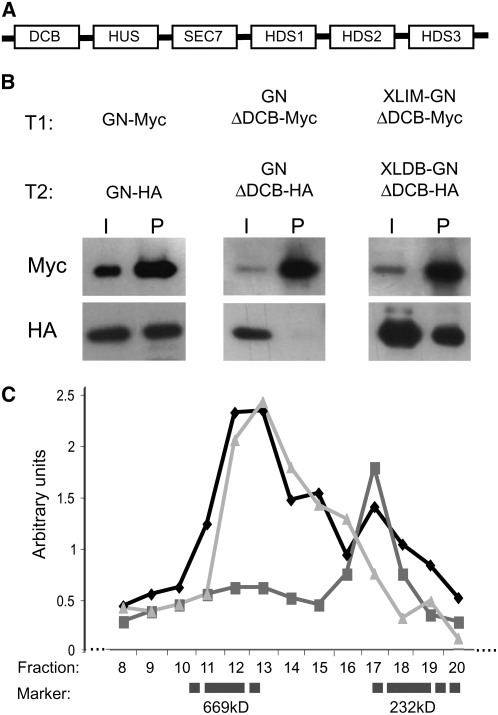
Figure 1.
In Vivo Dimerization of Large ARF-GEF GNOM.
(A) Domain organization of large ARF-GEFs (Mouratou et al., 2005).
(B) Coimmunoprecipitation of Myc- and HA-tagged GNOM variants from protein extracts of doubly transgenic (T1, transgene 1; T2, transgene 2) plants with anti-Myc beads. I, input; P, precipitate. Full-length GNOM (left panel); GNOM lacking the DCB domain (center panel); constitutively dimerized GNOMΔDCB (right panel).
(C) Chromatographic analysis of dimerization-dependent GNOM complexes. Overlays of three independent chromatographic runs each from extracts of Myc- and HA-tagged GNOM variants of doubly transgenic plants: GNOMΔDCB (squares, dark gray) and XLIM-GNOMΔDCB/XLDB-GNOMΔDCB (triangles, light gray). GNOM-Myc (diamonds, black) transgenic plant extract was added as an internal standard (overlay of six runs). Marker distribution is indicated (catalase, 232 kD; thyroglobulin, 669 kD).
Here, we demonstrate an essential role for the DCB domain of GNOM in vivo. The DCB domain had been implicated in dimerization and binding of the immunophilin At CYP19-4 (Grebe et al., 2000). Surprisingly, however, it is not the DCB-mediated dimerization that is essential, but a novel heterotypic interaction of the DCB domain with other GNOM domains that is functionally required. Mutations abolishing this interaction between GNOM domains also interfere with membrane association, which suggests an unexpected molecular mechanism for regulating membrane association and thereby determining ARF activity.
RESULTS
The DCB Domain Mediates N-Terminal Interaction between GNOM Proteins in Vivo
The amino acid sequence and secondary structure of the DCB domain is highly conserved in large ARF-GEFs across eukaryotes (Cox et al., 2004; Mouratou et al., 2005), suggesting its functional conservation. The DCB domain of the Arabidopsis thaliana large ARF-GEF GNOM has been suggested to mediate dimerization by homotypic interaction based on yeast two-hybrid and in vitro experiments, although this interaction was not shown in planta (Grebe et al., 2000). We therefore investigated whether the DCB–DCB interaction occurs in vivo by analyzing differentially epitope-tagged GNOM variants for interaction in transgenic plants. These GNOM proteins were full length (GNOM-Myc and GNOM-HA), lacked the DCB domain (GNOMΔDCB-Myc and GNOMΔDCB-HA), or had their DCB domain replaced with a heterologous dimerization module, XLIM and XLDB (XLIM-GNOMΔDCB-Myc and XLDB-GNOMΔDCB-HA) (Ung et al., 2001). GNOM-HA interacted with GNOM-Myc, whereas no in vivo interaction was observed between GNOMΔDCB variants (Figure 1B) in coimmunoprecipitation experiments, consistent with the corresponding yeast two-hybrid assay (see Supplemental Figure 1A online). Coimmunoprecipitation of XLIM-GNOMΔDCB-Myc and XLDB-GNOMΔDCB-HA indicated that the XLIM-XLDB module restores N-terminal interaction between GNOMΔDCB proteins (Figure 1B). Thus, the DCB domain of GNOM functions as a dimerization domain in vivo.
To analyze the relative abundance of GNOM dimers compared with monomers in vivo, we investigated complex formation of GNOM proteins with altered dimerization ability. Protein extracts from plants expressing epitope-tagged GNOM proteins were fractionated by size-exclusion chromatography, the fractions were analyzed by protein gel blotting, and signal density of the fractions was quantified. The vast majority of GNOM protein eluted in fractions corresponding to 570 to 670 kD (Figure 1C), which is consistent with complex sizes reported for mammalian and yeast large ARF-GEFs BIG1, BIG2, and Gea1p, respectively (Peyroche et al., 1996; Yamaji et al., 2000). Deletion of the DCB domain (GNOMΔDCB) led to a shift in protein complex size to fractions corresponding to ∼300 kD, whereas constitutive dimerization of GNOMΔDCB (XLIM-GNOMΔDCB and XLDB-GNOMΔDCB) relocated GNOMΔDCB into full-length GNOM-positive fractions (Figure 1C). Thus, the DCB–DCB domain interaction mediates the formation of GNOM dimers, rather than oligomers, as its most abundant form in vivo.
The DCB Domain Has Another Essential Function Other Than Mediating N-Terminal Dimerization
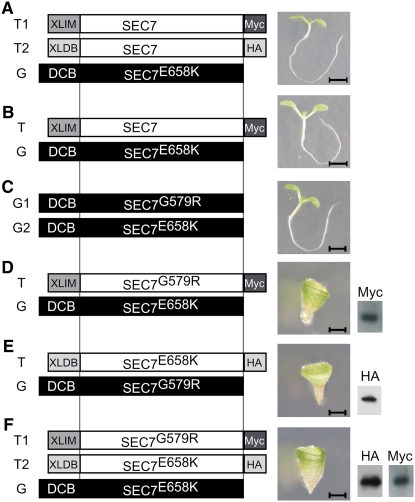
To assess the in vivo significance of dimerization, we analyzed GNOM lacking the DCB domain (GNOMΔDCB) and constitutively dimerized GNOMΔDCB (XLIM-GNOMΔDCB and XLDB-GNOMΔDCB) for their functionality in planta by assessing their ability to complement catalytically inactive gnomE658K mutants. This mutant displays early embryonic defects leading to developmental arrest of grossly abnormal seedlings (Mayer et al., 1993; Shevell et al., 1994). Constitutively dimerized GNOMΔDCB variants complemented gnomE658K, demonstrating that constitutive dimerization does not interfere with GNOM function (Figure 2A; see Supplemental Table 1 online). Surprisingly, GNOMΔDCB complemented gnomE658K, too (Figure 2B; see Supplemental Figure 2 and Supplemental Table 1 online). These data indicate that dimerization neither has to be regulated nor is it required for large ARF-GEF function. However, the in vivo significance of dimerization had been suggested by the interallelic complementation of two nonfunctional alleles, gnomG579R and gnomE658K, which encode full-length inactive proteins with single amino acid exchanges in the SEC7 domain (Figure 2C; Busch et al., 1996). To understand the molecular mechanism(s) underlying this interallelic complementation, we examined the biological role of dimerization in the context of the two complementing alleles. GNOM mutant proteins lacking the DCB domains (XLIM-GNOMG579RΔDCB or XLDB-GNOME658KΔDCB) did not complement the respective other mutant full-length protein, resulting in the gnom mutant phenotype (Figures 2D and 2E; see Supplemental Table 1 online). Interestingly, constitutive dimerization of XLIM-GNOMG579RΔDCB and XLDB-GNOME658KΔDCB did not rescue gnomE658K mutant plants either (Figure 2F; see Supplemental Table 1 online). These results indicate that the DCB domain is required for interallelic complementation and that, surprisingly, dimerization is not sufficient. This suggests that the DCB domain performs another essential function that cannot be mimicked by the XLIM–XLDB interaction module.
Figure 2.
Functional Analysis of GNOM Mutant Proteins.
(A) to (F) Interallelic complementation of endogenous (G) and transgenic (T) GNOM variants: schematic showing heterologous dimerization module XLIM and XLDB, Myc or HA tag, and amino acid exchange mutations (left panels). G1, genomic allele 1; G2, genomic allele 2; T1, transgene 1; T2, transgene 2. Phenotypes of homozygous gnom mutant seedlings harboring the respective GNOM transgene(s) (center panels). Protein gel blot analysis of transgenic expression (right panels).
(A) to (C) Complementation. Bars = 3 mm.
(D) to (F) No complementation. Bars = 0.1 mm.
The Immunophilin CYP19-4 Appears Not to Play a Role in GNOM Function
The interaction of the DCB domain with the immunophilin CYP19-4 (CYCLOPHILIN5) in Arabidopsis, suggested by yeast two-hybrid and in vitro interaction assays (Grebe et al., 2000), might represent the additional function of the DCB domain. Similarly to GNOM, the two mammalian large ARF-GEFs BIG1 and BIG2 interact with the immunophilin FKBP13 in Jurkat T cells via their conserved DCB domains (Padilla et al., 2003). Curiously, mammalian FKBP13 and Arabidopsis CYP19-4 harbor an N-terminal signal for translocation into the endoplasmic reticulum (ER) (Jin et al., 1991; Saito et al., 1999), whereas large ARF-GEFs are localized in the cytosol or to the cytosolic leaflet of target membranes (Shin and Nakayama, 2004). FKBP13, which harbors an additional ER retention signal, has been reported to localize to the ER lumen (Nigam et al., 1993). Therefore, we assessed the subcellular localization of Myc-tagged CYP19-4 expressed from the RPS5A promoter (Weijers et al., 2005). Both immunofluorescence and immunogold labeling localized CYP19-4 protein to the ER, Golgi stacks, and multivesicular bodies in seedling root tips (see Supplemental Figure 3 online). These results make it unlikely that GNOM interacts with CYP19-4 in vivo. Moreover, neither physical interaction of GNOM with CYP19-4 in planta nor alteration of the catalytic GDP/GTP exchange of GNOM in the presence of CYP19-4 in vitro nor genetic interaction of gnom and cyp19-4 mutants was detected (Geldner, 2003; Lau, 2005). Taken together, our results suggest that the DCB domain does not bind immunophilin in vivo nor is its interaction with another DCB domain essential for ARF-GEF function.
Heterotypic Interaction of the DCB Domain with Other Parts of the Protein Is Essential for GNOM Function
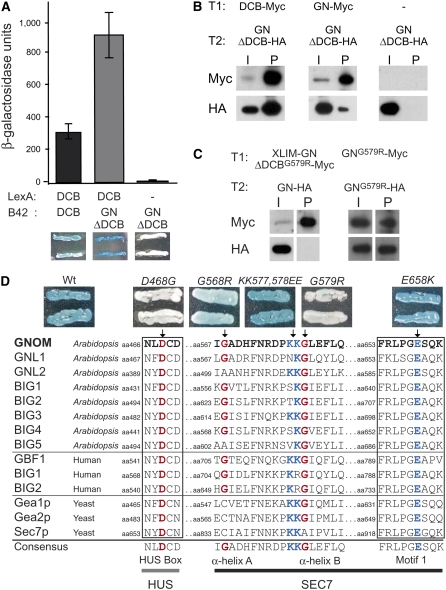
To assess its essential, unidentified function, the DCB domain was tested for interaction with other domains of the GNOM protein by yeast two-hybrid assay. Surprisingly, the DCB domain interacted not only with itself (homotypic interaction) but also remarkably strongly with GNOM lacking the DCB domain (GNOMΔDCB; heterotypic interaction) (Figure 3A). To determine whether the heterotypic interaction also occurs in vivo, we coexpressed fragments of GNOM protein in transgenic plants. Coimmunoprecipitation of both DCB-Myc and full-length GNOM-Myc with GNOMΔDCB-HA revealed the heterotypic interaction (Figure 3B), even though the interaction of GNOMΔDCB with full-length GNOM was reduced, which might suggest that an intramolecular interaction of full-length GNOM is preferred to an intermolecular interaction with GNOMΔDCB. Attempts by yeast two-hybrid analysis to map the DCB-interacting region to any specific domain within GNOMΔDCB were unsuccessful (see Supplemental Figure 1B online), which might be attributed to improper folding of single domains when taken out of their normal context or to more than one domain being involved in heterotypic interaction. To delineate the interacting region and to assess the functional significance of the heterotypic interaction, we analyzed the impact of amino acid exchange mutations that are known to compromise large ARF-GEF function (Figure 3D): E658K and G579R mutations characterized in GNOM and the HUS box mutation (D468G) characterized in the yeast ARF-GEF Gea2p. The HUS box mutation disrupts a conserved motif N(Y/F)DC(D/N) of the HUS domain and severely affects viability (Mouratou et al., 2005; Park et al., 2005). Yeast two-hybrid interaction analysis of the DCB domain with mutated GNOMΔDCB revealed that the E658K mutation had no deleterious effect. By contrast, the G579R mutation and the HUS box mutation eliminated the heterotypic interaction (Figure 3D). Coimmunoprecipitation experiments demonstrated the biological significance of the yeast two-hybrid results for the G579R mutant GNOM protein in planta (Figure 3C). Specifically, XLIM-GNOMG579RΔDCB did not coprecipitate with full-length GNOM. By contrast, full-length GNOM proteins that carried the G579R mutation were still able to dimerize via their DCB domains. Thus, the heterotypic interaction critically depends on specific amino acid residues in the C-terminal part of the HUS domain and the adjacent N-terminal part of the SEC7 domain. The mutations of these residues cause severe mutant phenotypes (Busch et al., 1996; Park et al., 2005), strongly suggesting that the heterotypic interaction is essential for large ARF-GEF function.
Figure 3.
The DCB Domain Mediates Heterotypic Interaction with GNOMΔDCB.
(A) Interaction of the DCB domain with itself (left panel) or GNOMΔDCB (center panel); negative control, GNOMΔDCB alone (right panel). Yeast two-hybrid interaction assay: quantitative assays each of at least six independent transformants. Error bars indicate sd (top panel). Color assay: blue staining indicates interaction (bottom panel).
(B) Coimmunoprecipitation of Myc- and HA-tagged GNOM variants from protein extracts of doubly transgenic (T1, transgene 1; T2, transgene 2) plants with anti-Myc beads. I, input; P, precipitate. Interaction of GNOMΔDCB-HA with DCB-Myc (left panel) and GNOM-Myc (center panel); negative control, GNOMΔDCB-HA alone (right panel).
(C) Coimmunoprecipitation of Myc- and HA-tagged GNOM variants from protein extracts of doubly transgenic (T1, transgene 1; T2, transgene 2) plants with anti-Myc beads. I, input; P, precipitate. No interaction of XLIM-GNOMG579R-Myc with GNOM-HA (left panel) and interaction of GNOMG579R-Myc with GNOMG579R-HA (right panel).
(D) Interaction of the DCB domain with GNOMΔDCB harboring amino acid exchange mutations: D468G, G568R, G579R, KK577,578EE, and E658K; positive control, nonmutant GNOMΔDCB (Wt). Yeast two-hybrid interaction color assay: blue staining indicates interaction (top panel). Conserved amino acid residues of eukaryotic large ARF-GEFs (bottom panel). Amino acid sequence alignment of the HUS box and relevant regions of the SEC7 domain of large ARF-GEFs from Arabidopsis, Homo sapiens, and Saccharomyces cerevisiae (yeast) (ClustalW). Mutations in GNOM that affect or do not affect heterotypic interaction are indicated, and the corresponding amino acid residues are highlighted in red or blue, respectively.
The N-Terminal Part of the SEC7 Domain Is Required for Heterotypic Interaction but Not for the Catalytic Exchange Reaction
The E658K mutation destroys the Glu finger in motif 1 of the SEC7 domain required for GDP release from the ARF substrate, resulting in catalytic inactivity (Beraud-Dufour et al., 1998; Cherfils et al., 1998). The G579R mutation and the mutation of a newly isolated gnom allele G568R reside in the N-terminal part of the SEC7 domain comprising the first two α-helices (αA and αB) clearly not in contact with the ARF GTPase (see Supplemental Figure 4 online; Renault et al., 2003). To assess whether the integrity of the two α-helices in the N-terminal part of the SEC7 domain is required for the heterotypic interaction, we analyzed the G568R mutation along with double point mutations of two conserved Lys residues directly adjacent to the G579R mutation (KK577,578EE), residing in an intervening loop between the two α-helices (Figure 3D; see Supplemental Figure 4 online). Compared with G579R, the G568R mutation affected the heterotypic interaction only moderately (Figure 3D). Interestingly, this correlates well with its weaker mutant phenotype compared with the complete loss-of-function phenotype of G579R (Figure 4A). By contrast, the KK577,578EE mutation did not affect the heterotypic interaction (Figure 3D).
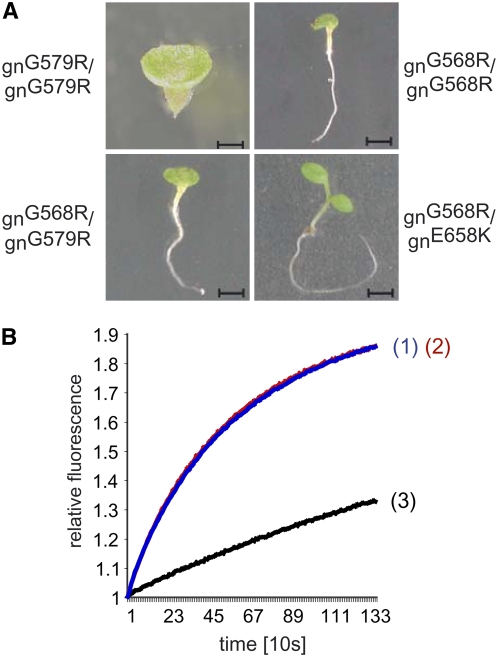
Figure 4.
The N-Terminal Part of SEC7 Is Required for the Heterotypic Interaction but Not for the Catalytic Exchange Reaction.
(A) Interallelic complementation assay. Seedling phenotypes of gnom homozygous mutants (top panels): gnomG579R (left; bar = 0.05 mm), gnomG568R (right; bar = 2.5 mm); and gnom trans-heterozygotes (lower panel): gnomG568R/gnomG579R (left; bar = 2.5 mm) and gnomG568R/gnomE658K (right; bar = 3 mm).
(B) In vitro catalytic exchange activity of the SEC7 domain of BIG3 harboring a single amino acid exchange mutation. The GDP/GTP exchange activity of His-SEC7BIG3 (1; blue) and His-SEC7BIG3G626R (2; red) were measured on ARF1. Negative control: ARF1 alone (3; black).
The alleles gnomG579R and gnomE658K were shown to complement each other, which already suggested that the N-terminal part of the SEC7 domain is required for a function unrelated to catalytic activity (Busch et al., 1996). Consistent with this, the newly isolated weak allele gnomG568R also complemented the catalytically inactive gnomE658K, whereas gnomG568R did not complement gnomG579R, suggesting that the latter two mutations affect the same noncatalytic function (Figure 4A). To obtain biochemical confirmation for this conclusion, we attempted to assess the catalytic activity of the SEC7 domain of GNOM harboring the G579R mutation. As this recombinant protein was insoluble, we instead used the SEC7 domain of the GNOM homolog BIG3 (previously named BIG2), which had been shown to be catalytically active on ARF1 (Nielsen et al., 2006). We introduced the G579R-homologous mutation into the highly conserved SEC7 domain of BIG3 (SEC7BIG3G626R; Figure 3D). SEC7BIG3G626R catalyzed the GDP/GTP exchange on ARF1 as efficiently as wild-type SEC7BIG3 (Figure 4B). Thus, the N-terminal part of the SEC7 domain mediates a noncatalytic, fundamental aspect of large ARF-GEF function.
The N-Terminal Part of the SEC7 Domain Is Also Required for Membrane Association of GNOM
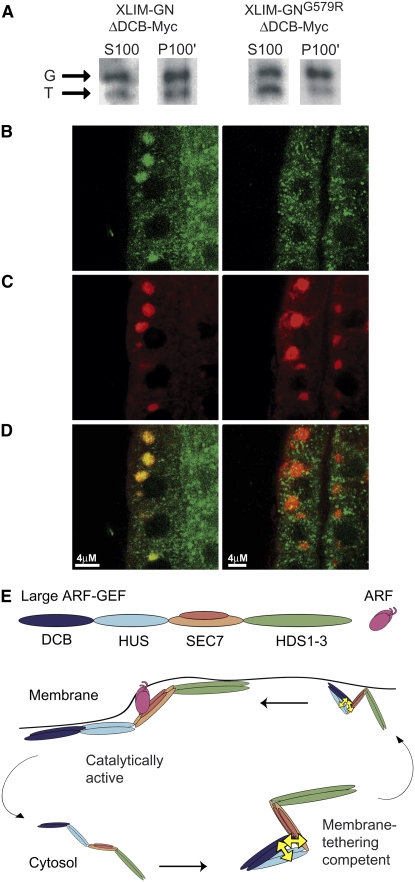
The catalytic GDP/GTP exchange reactions of ARF-GEFs depend on the translocation of the ARF-GEF and its ARF substrate from the cytosol to the membrane. Therefore, we analyzed whether impaired heterotypic interaction compromises membrane association of GNOM. As shown above, XLIM-GNOMG579RΔDCB was unable to complement the catalytically inactive GNOME658K protein (Figure 2D), in contrast with the nonmutant variant XLIM-GNOMΔDCB (Figure 2B). These two truncated GNOM proteins were analyzed for their ability to associate with membranes by subcellular fractionation of protein extracts. Similar to endogenous GNOM, XLIM-GNOMΔDCB partitioned between soluble (S100) and membrane (P100') fractions, whereas the ratio of partitioning was altered in the mutant variant XLIM-GNOMG579RΔDCB, with the majority of the truncated protein localizing in the soluble fraction (Figure 5A). There was still XLIM-GNOMG579RΔDCB detectable in the membrane fraction, which might be due to contamination of the highly concentrated P100' fraction with soluble proteins (see Supplemental Figure 5 online). Alternatively, there might be residual mutant protein associated with the membrane. To assess membrane association of GNOMG579RΔDCB independently, we additionally localized the transgenic proteins in seedling roots treated with the fungal toxin Brefeldin A (BFA). BFA traps ARF exchange intermediate ARF•GDP-SEC7 on membranes (Peyroche et al., 1999; Steinmann et al., 1999; Niu et al., 2005), leading to the accumulation of GNOM in large ARF1-positive BFA compartments (Geldner et al., 2003; Paciorek et al., 2005), which makes it very easy to detect membrane association of GNOM in cells. Like full-length GNOM protein, XLIM-GNOMΔDCB colocalized with ARF1 in BFA compartments. By contrast, the same truncated protein carrying the G579R mutation, XLIM-GNOMG579RΔDCB, was not detectable in ARF1-positive BFA compartments (Figures 5B to 5D). We ensured that the expression levels of both transgenic proteins were comparable, and the BFA sensitivity of the SEC7 domain was not impaired by the mutation (see Supplemental Figure 6 online). We therefore conclude that the heterotypic interaction of the DCB domain with GNOMΔDCB promotes the association of GNOM with membranes. This conclusion is moreover consistent with the observation that HUS box mutations, which we showed also to compromise the heterotypic interaction, affect membrane association of the yeast homolog Gea2p (Park et al., 2005).
Figure 5.
The N-Terminal Part of the SEC7 Domain Is Required for Membrane Association of GNOM.
(A) to (D) Membrane association of XLIM-GNOMΔDCB-Myc and XLIM-GNOMG579RΔDCB-Myc expressed as transgenes in wild-type seedlings.
(A) Subcellular fractionation of soluble (S100) and membrane (P100') fractions detected with anti-SEC7-antiserum; P100' was loaded in 10-fold excess of S100. Arrows: endogenous full-length GNOM (G); internal control, transgenically produced truncated GNOM (T). Note difference in S100/P100' ratio of GNOMΔDCB and GNOMG579RΔDCB.
(B) to (D) Subcellular localization of XLIM-GNOMΔDCB-Myc (left panels) and XLIM-GNOMG579RΔDCB-Myc (right panels) in BFA-treated seedling roots.
(B) Myc staining of transgenic proteins (green).
(C) ARF1 staining of BFA compartments (red).
(D) Double labeling of Myc and ARF1 (overlay).
(E) Regulation of large ARF-GEF function by heterotypic interaction. Cyclic changes of ARF-GEF conformation from a form competent for membrane tethering (right) to an open catalytically active form that is effectively membrane associated (left), which is followed by dissociation from the membrane and reestablishment of the heterotypically interacting form. Double-headed arrows (yellow) indicate the heterotypic interaction of the DCB domain with the HUS and SEC7 domains. The model depicts the heterotypic interaction as intramolecular and the large ARF-GEF as a constitutive dimer but does not pretend to reflect the actual as yet unknown structural changes of GNOM.
DISCUSSION
A central regulatory step for large ARF-GEFs is their reversible recruitment to membranes, the mechanism of which has remained elusive. Among several conserved noncatalytic domains whose functions are ill defined, the N-terminal DCB domain of GNOM had been implicated in dimerization and binding of CYP19-4 (CYCLOPHILIN 5; Grebe et al., 2000). Our results now reveal an essential role for the DCB domain in vivo. Surprisingly, it is not the DCB-mediated dimerization that is required for GNOM function but rather a novel heterotypic interaction of the DCB domain with the C-terminal part of GNOM, which is required for membrane association. These findings provide an intriguing molecular explanation for the puzzling interallelic complementation of GNOM alleles and the strong phenotype of a noncatalytic mutation: Dimerization via DCB domains restores GNOM activity by physically clasping the complementary residual functions of two proteins defective in a single domain: heterotypic interaction and catalytic activity. Our results demonstrate that GNOM exists predominantly as dimer formed by DCB–DCB interaction in vivo. Interestingly, the putative substrate ARF appears to form dimers by structural and biochemical analyses (Amor et al., 1994; Greasley et al., 1995; Zhao et al., 1999). Hence, rather than being required for ARF-GEF function, dimerization of the ARF and its exchange factor seems to play a structural role in the nucleotide exchange process and might facilitate their local accumulation and scaffolding of effector proteins at specific sites on the membrane.
A recent study of mammalian large ARF-GEFs also provided evidence for dimerization via DCB–DCB interaction and for heterotypic interaction (Ramaen et al., 2007). Interestingly, the reported heterotypic interaction was confined to the DCB and HUS domains, in contrast with the heterotypic interaction of GNOM domains. Thus, the involvement of the SEC7 domain might represent a plant-specific feature of large ARF-GEF function. Moreover, the available gnom mutations in our study enabled us to demonstrate the biological relevance of domain interactions. No such studies have been performed for mammalian large ARF-GEFs.
Our results furthermore demonstrate that the heterotypic interaction of the DCB domain with other conserved domains is essential for GNOM function. A single amino acid exchange mutation in either the SEC7 or the HUS domain abolishes the heterotypic interaction and, in addition, impairs membrane association (Park et al., 2005; this study). Although large ARF-GEFs have no distinct membrane association domain, the GDP/GTP exchange on ARF substrates occurs at the membrane. The heterotypic interaction shown here might provide a protein conformation required for membrane association. This is consistent with the previous observation that GNOM is released from the membrane by urea in a concentration-dependent manner but neither by high salt nor alkaline treatment (Steinmann et al., 1999). We propose regulated changes in the heterotypic interaction as a conserved mechanism to control membrane association and activity of large ARF-GEFs (Figure 5E). In our model, the conformation of the heterotypically interacting ARF-GEF acts as a prerequisite for membrane tethering, whereas effective membrane association might be achieved by a conformational change that exposes hydrophobic surfaces for interaction. These proposed conformational changes cannot occur in the heterotypic interaction–deficient mutant protein GNOMG579R, which accounts for its nonfunctionality, although N-terminal dimerization and catalytic exchange activity are not compromised. However, N-terminal dimerization with the noncatalytic variant GNOME658K restores its membrane association and, thus, function. ARF-GEF is depicted as a constitutive dimer, which accounts for the complementing behavior of the full-length mutant proteins, GNOME658K and GNOMG579R, as well as the biochemical results. Constitutive dimerization of the DCB domains in turn implies that the heterotypic interaction may occur within each of the two monomers.
A similar mechanism of conformation-dependent protein function has been discussed for the distantly related ARF-GEF RalF from Legionella (Amor et al., 2005). Such conformational changes might be regulated by phosphorylation and dephosphorylation, as this has been correlated with membrane association of the mammalian large ARF-GEFs BIG1 and BIG2 (Kuroda et al., 2007). Interestingly, viruses appear to take advantage of the heterotypic interaction of large ARF-GEFs at the membrane to interfere with membrane trafficking of the host. Specifically, the enteroviral membrane protein 3A influences membrane association dynamics of the mammalian large ARF-GEF GBF1 by interacting with its N terminus (Wessels et al., 2006a, 2006b; Belov et al., 2007), which appears to involve the integral DCB-HUS structure of GBF1 rather than the individual domains (Ramaen et al., 2007). In summary, we identified a novel heterotypic interaction of large ARF-GEFs that provides a molecular mechanism for membrane association. This regulatory step might play a central role in membrane trafficking by determining when and where ARF GTPases are activated.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana growth conditions and gnom alleles G579R (B4049) and E658K (emb30-1) have been described (Busch et al., 1996). Sequencing of the ethyl methanesulfonate–induced gnomG568R allele revealed a change of codon 568 from GGA (G) to AGA (R).
Binary Vector Constructs, Generation of Transgenic Plants, and Complementation Analysis
GNOM constructs are based on the genomic fragment GNXbaIwt-myc (Geldner et al., 2003). The 3xMyc-tag was substituted by 1xHA-tag, where indicated. Primer extension PCR deleted the DCB domain (1 to 232 amino acids) and introduced restriction sites at the start codon. XLIM (1 to 58 amino acids) and XLDB (290 to 350 amino acids) (Ung et al., 2001) were PCR amplified and introduced into the restriction sites, in the case of XLDB, a 2xHA-tag was added C-terminally. G579R and E658K mutations were introduced by site-directed mutagenesis (Sawano and Miyawaki, 2000). To generate the DCB-Myc construct (1 to 246 amino acids), restriction sites used for insertion of the C-terminal 3xMyc-tag were inserted by primer extension PCR on the cDNA vector c96 (Busch et al., 1996). GNOM constructs were cloned into pGreenII conferring BASTA or hygromycin resistance.
PCR-amplified CDS of At CYP19-4 was cloned into the ribosomal protein S5A promoter expression cassette (Weijers et al., 2005) and C-terminally 3xMyc tagged. gnom heterozygous transgenic seedlings were selected on 50 μg/mL of kanamycin (Roth), 15 μg/mL of hygromycin, or 30 μg/mL of phosphinothricin (Duchefa). Complementation was analyzed by transgenic expression, GNOM genomic background, and gnom segregation rates in T2-T5 generations.
Yeast Two-Hybrid Interaction Assays
Assay and constructs of DCB (1 to 246 amino acids), SEC7 (551 to 818 amino acids), HDS1-3 (818 to 1451 amino acids) domains, and CYP19-4 are as described (Grebe et al., 2000). GNOMΔDCB (232 to 1451 amino acids) and HUS domain (232 to 548 amino acids) were PCR amplified and cloned into pJG4-5 and pEG202. D468G, G568R, KK577,578EE, G579R, and E658K mutations were introduced by site-directed mutagenesis.
Preparation of Native Protein Extracts
Seedlings (0.5 g) were homogenized (potter) in 1.5 mL of extraction buffer (50 mM Tris/HCl, pH 8.0, 150 mM NaCl, protease inhibitor cocktail [Sigma-Aldrich], and 1 mM PMSF) and centrifuged twice (10,000g for 30 min at 4°C).
Coimmunoprecipitation
Protein extracts containing 1% Triton X-100 were incubated with 20 μL of bed volume of equilibrated rabbit anti-Myc agarose beads (Sigma-Aldrich) at 4°C overnight. Beads were washed four times with extraction buffer and bound proteins eluted by boiling in 50 μL of 2× Laemmli buffer.
Subcellular Fractionation
Protein extracts were centrifuged at 100,000g for 60 min at 4°C, yielding supernatant S100. The pellet was incubated with 1.3 mL of extraction buffer for 30 min and centrifuged, and the pellet (P100') was resolved in 50 μL of 2× Laemmli buffer.
Size-Exclusion Chromatography
Protein extracts were separated as described (Schwechheimer et al., 2002). Protein size distribution was analyzed with a gel filtration calibration kit (Amersham Biosciences). The fractions were precipitated using Stratagene Clean resin and analyzed by SDS-PAGE and protein gel blotting and the signals quantified (Bio-Rad QuantityOne software).
SDS-PAGE and Protein Gel Blotting
SDS-PAGE and protein gel blotting were performed as described (Lauber et al., 1997). Antibodies and dilutions were as follows: anti-Myc 9E10 (Santa Cruz) 1:600, rabbit anti-SEC7 (Steinmann et al., 1999) 1:6000, rabbit anti-POR (Steinborn et al., 2002) 1:5000, anti-mouse or anti-rabbit alkaline phosphatase–conjugated antibodies (Novagen and Jackson Immunoresearch) 1:5000, and peroxidase-conjugated mouse anti-HA (Roche) 1:1000. Detection was performed with the CDP-Star (Tropix) or BM-chemiluminescence blotting substrate (Roche).
Expression Vector Constructs, Purification of Proteins, and Analysis of Catalytic Exchange Activity
Construction of expression vectors, production of recombinant proteins, and fluorescence measurements were performed as described (Nielsen et al., 2006). G626R and L741M mutations were introduced into the SEC7 domain of BIG3 by site-directed mutagenesis.
Whole-Mount Immunofluorescence and Confocal Microscopy
Immunofluorescence analysis and confocal microscopy were performed as described (Lauber et al., 1997). Rabbit anti-ARF1 antibody (Ritzenthaler et al., 2002) was diluted 1:1000. Seedlings were treated with 200 μM BFA for 120 min, where indicated.
Immunogold Labeling and Electron Microscopy
Immunogold labeling and electron microscopic analysis of ultrathin thawed cryosections were performed as described (Völker et al., 2001). The 9E10 mouse anti-Myc antibody was diluted 1:200. Goat anti-mouse IgG coupled to Nanogold was silver enhanced with HQSilver for 8 min (Nanoprobes).
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL data libraries under the following accession numbers: Arabidopsis genes GNOM (At1g13980), GNL1 (At5g39500), GNL2 (At5g19610), BIG1 (At4g38200), BIG2 (At3g60860), BIG3 (At1g01960), BIG4 (At4g35380), BIG5 (At3g43300), ARF1 (At2g47170), CYP19-4 (At2g29960); Xenopus leavis XLIM (AAB70190), Xl XLDB (NP_001080549); Homo sapiens BIG1 (NP_006412), Hs BIG2 (NP_006411), Hs GBF1 (NP_004184), Hs ARNO (X99753.1); Saccharomyces cerevisiae Gea1p (NP_012565), Sc Gea2p (NP_010892), Sc Sec7p (P11075).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Domain Interactions of the GNOM Protein.
Supplemental Figure 2. Functional Analysis of GNOM Mutant Proteins.
Supplemental Figure 3. CYP19-4 Localizes to the Secretory Pathway.
Supplemental Figure 4. The N-terminal α-Helices A and B of the SEC7 Domain Are Not in Contact with the ARF Substrate.
Supplemental Figure 5. Antibody Detection of Proteins in Subcellular Fractions.
Supplemental Figure 6. G579R and the Homologous Mutation in BIG3 Do Not Interfere with ARF-GEF Expression Level and BFA Sensitivity, Respectively.
Supplemental Table 1. Segregation Rates of Complementing Allele Combinations.
Supplementary Material
Acknowledgments
We thank David G. Robinson for anti-ARF1 antiserum, Andreas Maier and Claus Schwechheimer for support with chromatographic analysis, and Niko Geldner for critical discussions. This work was funded by the Human Frontier Science Program and Deutsche Forschungsgemeinshaft (SFB 446).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Gerd Jürgens (gerd.juergens@zmbp.uni-tuebingen.de).
Online version contains Web-only data.
References
- Amor, J.C., Harrison, D.H., Kahn, R.A., and Ringe, D. (1994). Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature 372 704–708. [DOI] [PubMed] [Google Scholar]
- Amor, J.C., Swails, J., Zhu, X., Roy, C.R., Nagai, H., Ingmundson, A., Cheng, X., and Kahn, R.A. (2005). The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J. Biol. Chem. 280 1392–1400. [DOI] [PubMed] [Google Scholar]
- Belov, G.A., Altan-Bonnet, N., Kovtunovych, G., Jackson, C.L., Lippincott-Schwartz, J., and Ehrenfeld, E. (2007). Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 81 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraud-Dufour, S., Robineau, S., Chardin, P., Paris, S., Chabre, M., Cherfils, J., and Antonny, B. (1998). A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J. 17 3651–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg, N., Baraldi, E., Nilges, M., and Saraste, M. (1999). The PH superfold: A structural scaffold for multiple functions. Trends Biochem. Sci. 24 441–445. [DOI] [PubMed] [Google Scholar]
- Busch, M., Mayer, U., and Jürgens, G. (1996). Molecular analysis of the Arabidopsis pattern formation of gene GNOM: gene structure and intragenic complementation. Mol. Gen. Genet. 250 681–691. [DOI] [PubMed] [Google Scholar]
- Chantalat, S., Courbeyrette, R., Senic-Matuglia, F., Jackson, C.L., Goud, B., and Peyroche, A. (2003). A novel Golgi membrane protein is a partner of the ARF exchange factors Gea1p and Gea2p. Mol. Biol. Cell 14 2357–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils, J., Menetrey, J., Mathieu, M., Le Bras, G., Robineau, S., Beraud-Dufour, S., Antonny, B., and Chardin, P. (1998). Structure of the Sec7 domain of the Arf exchange factor ARNO. Nature 392 101–105. [DOI] [PubMed] [Google Scholar]
- Cox, R., Mason-Gamer, R.J., Jackson, C.L., and Segev, N. (2004). Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell 15 1487–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N. (2003). The Role of the Arabidopsis GNOM Gene in Auxin Transport and Embryonic Axis Formation. PhD dissertation (Tübingen, Germany: Eberhard Karls University).
- Geldner, N., Anders, N., Wolters, H., Keicher, J., Kornberger, W., Muller, P., Delbarre, A., Ueda, T., Nakano, A., and Jürgens, G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230. [DOI] [PubMed] [Google Scholar]
- Gillingham, A.K., and Munro, S. (2007). The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23 579–611. [DOI] [PubMed] [Google Scholar]
- Greasley, S.E., Jhoti, H., Teahan, C., Solari, R., Fensome, A., Thomas, G.M., Cockcroft, S., and Bax, B. (1995). The structure of rat ADP-ribosylation factor-1 (ARF-1) complexed to GDP determined from two different crystal forms. Nat. Struct. Biol. 2 797–806. [DOI] [PubMed] [Google Scholar]
- Grebe, M., Gadea, J., Steinmann, T., Kientz, M., Rahfeld, J.U., Salchert, K., Koncz, C., and Jürgens, G. (2000). A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Y.J., Albers, M.W., Lane, W.S., Bierer, B.E., Schreiber, S.L., and Burakoff, S.J. (1991). Molecular cloning of a membrane-associated human FK506- and rapamycin-binding protein, FKBP-13. Proc. Natl. Acad. Sci. USA 88 6677–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, F., Moss, J., and Vaughan, M. (2007). Regulation of brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1) and BIG2 activity via PKA and protein phosphatase 1gamma. Proc. Natl. Acad. Sci. USA 104 3201–3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau, S. (2005). Funktionelle Charakterisierung der Dimerisierungs- und Cyclophilin bindenden Domäne der ARF GDP/GTP-Austauschfaktoren GNOM, GNL1 und GNL2 von Arabidopsis thaliana. Diploma thesis (Tübingen, Germany: Eberhard Karls University).
- Lauber, M.H., Waizenegger, I., Steinmann, T., Schwarz, H., Mayer, U., Hwang, I., Lukowitz, W., and Jürgens, G. (1997). The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J. Cell Biol. 139 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour, S.J., Skaug, J., Zhao, X.H., Giordano, J., Scherer, S.W., and Melancon, P. (1999). p200 ARF-GEP1: A Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc. Natl. Acad. Sci. USA 96 7968–7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, U., Büttner, G., and Jürgens, G. (1993). Apical-basal pattern formation in the Arabidopsis embryo: Studies on the role of the gnom gene. Development 117 149–162. [Google Scholar]
- Mouratou, B., Biou, V., Joubert, A., Cohen, J., Shields, D.J., Geldner, N., Jürgens, G., Melancon, P., and Cherfils, J. (2005). The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics 6 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, M., Albrethsen, J., Larsen, F.H., and Skriver, K. (2006). The Arabidopsis ADP-ribosylation factor (ARF) and ARF-like (ARL) system and its regulation by BIG2, a large ARF-GEF. Plant Sci. 171 707–717. [Google Scholar]
- Nigam, S.K., Jin, Y.J., Jin, M.J., Bush, K.T., Bierer, B.E., and Burakoff, S.J. (1993). Localization of the FK506-binding protein, FKBP 13, to the lumen of the endoplasmic reticulum. Biochem. J. 294 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, T.K., Pfeifer, A.C., Lippincott-Schwartz, J., and Jackson, C.L. (2005). Dynamics of GBF1, a Brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell 16 1213–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciorek, T., Zazimalova, E., Ruthardt, N., Petrasek, J., Stierhof, Y.D., Kleine-Vehn, J., Morris, D.A., Emans, N., Jürgens, G., Geldner, N., and Friml, J. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 435 1251–1256. [DOI] [PubMed] [Google Scholar]
- Padilla, P.I., Chang, M.J., Pacheco-Rodriguez, G., Adamik, R., Moss, J., and Vaughan, M. (2003). Interaction of FK506-binding protein 13 with brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1): Effects of FK506. Proc. Natl. Acad. Sci. USA 100 2322–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.K., Hartnell, L.M., and Jackson, C.L. (2005). Mutations in a highly conserved region of the Arf1p activator GEA2 block anterograde Golgi transport but not COPI recruitment to membranes. Mol. Biol. Cell 16 3786–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche, A., Antonny, B., Robineau, S., Acker, J., Cherfils, J., and Jackson, C.L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3 275–285. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Paris, S., and Jackson, C.L. (1996). Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 384 479–481. [DOI] [PubMed] [Google Scholar]
- Ramaen, O., Joubert, A., Simister, P., Belgareh-Touze, N., Olivares-Sanchez, M.C., Zeeh, J.C., Chantalat, S., Golinelli-Cohen, M.P., Jackson, C.L., Biou, V., and Cherfils, J. (2007). Interactions between conserved domains within homodimers in the BIG1, BIG2, and GBF1 Arf guanine nucleotide exchange factors. J. Biol. Chem. 282 28834–28842. [DOI] [PubMed] [Google Scholar]
- Renault, L., Guibert, B., and Cherfils, J. (2003). Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426 525–530. [DOI] [PubMed] [Google Scholar]
- Ritzenthaler, C., Nebenfuhr, A., Movafeghi, A., Stussi-Garaud, C., Behnia, L., Pimpl, P., Staehelin, L.A., and Robinson, D.G. (2002). Reevaluation of the effects of brefeldin A on plant cells using tobacco Bright Yellow 2 cells expressing Golgi-targeted green fluorescent protein and COPI antisera. Plant Cell 14 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, T., Niwa, Y., Ashida, H., Tanaka, K., Kawamukai, M., Matsuda, H., and Nakagawa, T. (1999). Expression of a gene for cyclophilin which contains an amino-terminal endoplasmic reticulum-targeting signal. Plant Cell Physiol. 40 77–87. [DOI] [PubMed] [Google Scholar]
- Sawano, A., and Miyawaki, A. (2000). Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28 E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., and Deng, X.W. (2002). Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14 2553–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell, D.E., Leu, W.M., Gillmor, C.S., Xia, G., Feldmann, K.A., and Chua, N.H. (1994). EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77 1051–1062. [DOI] [PubMed] [Google Scholar]
- Shin, H.W., and Nakayama, K. (2004). Guanine nucleotide-exchange factors for arf GTPases: Their diverse functions in membrane traffic. J. Biochem. (Tokyo) 136 761–767. [DOI] [PubMed] [Google Scholar]
- Steinborn, K., Maulbetsch, C., Priester, B., Trautmann, S., Pacher, T., Geiges, B., Küttner, F., Lepiniec, L., Stierhof, Y.-D., Schwarz, H., Jürgens, G., and Mayer, U. (2002). The Arabidopsis PILZ group genes encode tubulin-folding cofactor orthologs required for cell division but not cell growth. Genes Dev. 16 959–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmann, T., Geldner, N., Grebe, M., Mangold, S., Jackson, C.L., Paris, S., Galweiler, L., Palme, K., and Jürgens, G. (1999). Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science 286 316–318. [DOI] [PubMed] [Google Scholar]
- Ung, T.L., Cao, C., Lu, J., Ozato, K., and Dever, T.E. (2001). Heterologous dimerization domains functionally substitute for the double-stranded RNA binding domains of the kinase PKR. EMBO J. 20 3728–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völker, A., Stierhof, Y.D., and Jürgens, G. (2001). Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J. Cell Sci. 114 3001–3012. [DOI] [PubMed] [Google Scholar]
- Weijers, D., Benkova, E., Jager, K.E., Schlereth, A., Hamann, T., Kientz, M., Wilmoth, J.C., Reed, J.W., and Jürgens, G. (2005). Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 24 1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, E., Duijsings, D., Lanke, K.H., van Dooren, S.H., Jackson, C.L., Melchers, W.J., and van Kuppeveld, F.J. (2006. a). Effects of picornavirus 3A proteins on protein transport and GBF1-dependent COP-I recruitment. J. Virol. 80 11852–11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels, E., Duijsings, D., Niu, T.K., Neumann, S., Oorschot, V.M., de Lange, F., Lanke, K.H., Klumperman, J., Henke, A., Jackson, C.L., Melchers, W.J., and van Kuppeveld, F.J. (2006. b). A viral protein that blocks Arf1-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev. Cell 11 191–201. [DOI] [PubMed] [Google Scholar]
- Yamaji, R., Adamik, R., Takeda, K., Togawa, A., Pacheco-Rodriguez, G., Ferrans, V.J., Moss, J., and Vaughan, M. (2000). Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc. Natl. Acad. Sci. USA 97 2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L., Helms, J.B., Brunner, J., and Wieland, F.T. (1999). GTP-dependent binding of ADP-ribosylation factor to coatomer in close proximity to the binding site for dilysine retrieval motifs and p23. J. Biol. Chem. 274 14198–14203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.