Abstract
The glandular trichomes (lupulin glands) of hop (Humulus lupulus) synthesize essential oils and terpenophenolic resins, including the bioactive prenylflavonoid xanthohumol. To dissect the biosynthetic processes occurring in lupulin glands, we sequenced 10,581 ESTs from four trichome-derived cDNA libraries. ESTs representing enzymes of terpenoid biosynthesis, including all of the steps of the methyl 4-erythritol phosphate pathway, were abundant in the EST data set, as were ESTs for the known type III polyketide synthases of bitter acid and xanthohumol biosynthesis. The xanthohumol biosynthetic pathway involves a key O-methylation step. Four S-adenosyl-l-methionine–dependent O-methyltransferases (OMTs) with similarity to known flavonoid-methylating enzymes were present in the EST data set. OMT1, which was the most highly expressed OMT based on EST abundance and RT-PCR analysis, performs the final reaction in xanthohumol biosynthesis by methylating desmethylxanthohumol to form xanthohumol. OMT2 accepted a broad range of substrates, including desmethylxanthohumol, but did not form xanthohumol. Mass spectrometry and proton nuclear magnetic resonance analysis showed it methylated xanthohumol to 4-O-methylxanthohumol, which is not known from hop. OMT3 was inactive with all substrates tested. The lupulin gland-specific EST data set expands the genomic resources for H. lupulus and provides further insight into the metabolic specialization of glandular trichomes.
INTRODUCTION
Humans have long used wild and cultivated hops (Humulus lupulus; Cannabaceae) for their medicinal properties and as a key ingredient in brewing beer (Hornsey, 2003). The female inflorescences (cones) of hop are rich in terpenoid essential oils and terpenophenolic resins. In addition to the terpenophenolic acylphloroglucinols (e.g., humulone) that give beer its characteristic bitter flavor, hop cones also contain ∼1% of xanthohumol, a prenylchalcone with potent cancer preventive properties (Stevens and Page, 2004). Xanthohumol has been shown to exert cytoprotective effects through the induction of phase 2 proteins that function to detoxify carcinogens and metabolize oxidative radicals and also shows antioxidant and free radical scavenging properties (Miranda et al., 2000a, 2000b). Recent studies demonstrate that xanthohumol inhibits proliferation of fibrosarcoma cells (Goto et al., 2005) and induces apoptosis in prostate cancer cells (Colgate et al., 2006). Therefore, this phytochemical may be useful for both the prevention and treatment of certain types of cancer.
Xanthohumol and other hop terpenophenolics accumulate primarily in peltate glandular trichomes, termed lupulin glands, which are visible as yellow structures at the base of bracteoles in hop cones (Figures 1A and 1B). Lupulin glands are composed of a disk of biosynthetic secretory cells and a subcuticular cavity in which terpenophenolics and essential oils (i.e., monoterpenes and sesquiterpenes) are stored (Oliveira and Pais, 1990). Initially concave and cup-like, the trichomes develop a peaked appearance as the subcuticular cavity fills during ripening (Figures 1C and 1D).
Figure 1.
Morphology of Hop Cones and Lupulin Glands.
(A) Cones of hop cultivar Taurus. Cones are ∼5 cm in length.
(B) Longitudinal section of a hop cone showing lupulin glands at the base of bracteoles.
(C) A light microscopy image of ripe lupulin glands. Bar = 500 μm.
(D) Scanning electron micrograph of a ripe lupulin gland showing the peaked appearance of the filled subcuticular sac. Bar = 100 μm.
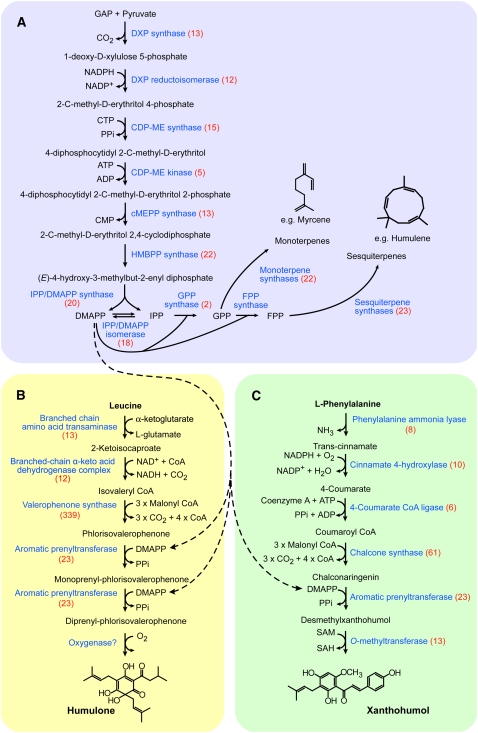
The biosynthesis of xanthohumol and other plant terpenophenolics is not completely understood. Such compounds have a mixed biosynthetic origin (Figure 2) and are composed of a polyketide-derived core modified with isoprenoid-derived prenyl side chains (Page and Nagel, 2006). Xanthohumol originates in phenylpropanoid metabolism, with chalcone synthase (CHS) catalyzing the condensation of coumaroyl CoA with malonyl CoA to form chalconaringenin (Figure 2C). A trichome-specific CHS gene, chs_h1, has been cloned from hop (Novak et al., 2003). Prenylation of chalconaringenin yields desmethylxanthohumol, which is methylated to form xanthohumol. A reverse order for these two reactions is also possible, but the presence of desmethylxanthohumol in hop cones (Stevens et al., 1997) and the absence of 6′-O-methylchalconaringenin suggest that prenylation occurs first. The last step in the pathway, methylation at the 6′-OH of desmethylxanthohumol yielding xanthohumol, is likely performed by an S-adenosyl-l-methionine (SAM)–dependent O-methyltransferase (OMT). Neither the aromatic prenyltransferase nor the OMT of the xanthohumol pathway has been previously identified.
Figure 2.
The Three Major Biosynthetic Pathways Active in Lupulin Glands.
Enzyme names are shown in blue. Each enzyme is annotated with the number of corresponding ESTs shown in parentheses. Dashed arrows indicate the role of DMAPP for prenylation in the bitter acid and xanthohumol pathways.
(A) The terpenoid pathway in hop lupulin glands. This pathway includes the MEP pathway for synthesis of IPP and DMAPP and also provides myrcene and humulene, the main terpenoids in hop lupulin glands. The mevalonate pathway is not shown because it appears to contribute little to the formation of lupulin gland terpenoids.
(B) The bitter acid pathway in hop lupulin glands. The biosynthesis of humulone, the main α-acid found in hop trichomes, is shown. The nature of the aromatic prenylation and final oxidation steps are not known.
(C) The xanthohumol pathway in hop lupulin glands. The aromatic prenyltransferase is not identified.
We used EST analysis of lupulin glands from high-xanthohumol hop cultivars to investigate the biosynthesis of xanthohumol and other trichome metabolites. The lupulin gland-specific EST data set was analyzed for cDNAs that encode known biosynthetic enzymes as well as enzymes from pathways that supply metabolic precursors. With the goal of identifying the desmethylxanthohumol 6′-O-methyltransferase from H. lupulus, three candidate OMTs were expressed as recombinant proteins and their enzymatic function tested. One of these, OMT1, was found to methylate desmethylxanthohumol to form xanthohumol. The identification of a specific desmethylxanthohumol OMT clarifies the xanthohumol pathway and provides the basis for metabolic engineering of prenylchalcone biosynthesis in hop.
RESULTS
Xanthohumol Accumulates in Lupulin Glands
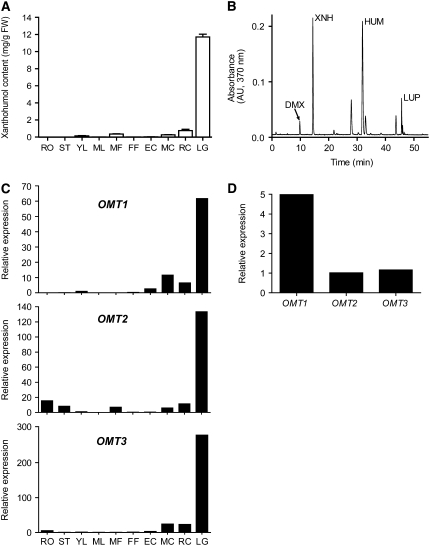
We used HPLC to measure the distribution of xanthohumol in different tissues of the H. lupulus cultivar Taurus, which has been reported to contain 0.95% by dry weight (bdw) xanthohumol in cones (Biendl, 2002/2003) (Figure 3A). In addition to xanthohumol, the compounds desmethylxanthohumol, humulone, and lupulone were also detected but not quantified (Figure 3B). Xanthohumol accumulated to its highest levels in lupulin glands, where it was found at 11.7 mg/g fresh weight, and ripe cones contained more xanthohumol than early-stage or midstage cones. Xanthohumol was found in significant amounts in male flowers, presumably due to the presence of lupulin glands on anthers (Nickerson et al., 1988). Leaves contained low but detectable levels of xanthohumol. Removing leaf glands by abrasion led to an 88% (n = 3) reduction in the amount of xanthohumol present in leaves (see Supplemental Figure 1 online), indicating that the lupulin gland is the primary site of xanthohumol accumulation in leaves.
Figure 3.
Quantification of Xanthohumol and Hl OMT Gene Expression Analysis of Different Hop Tissues.
(A) Amounts of xanthohumol occurring in different tissues of the cultivar Taurus measured using HPLC. Values represent mean ± sd (n = 3). RO, roots; ST, stem; YL, young leaf; ML, mature leaf; MF, male flower; FF, female flower; EC, early-stage cone; MC, mid-stage cone; RC, ripe cone; LG, lupulin gland; FW, fresh weight.
(B) A representative chromatogram obtained by HPLC analysis of a Taurus lupulin gland sample. DMX, desmethylxanthohumol; XNH, xanthohumol; HUM, humulone; LUP, lupulone.
(C) Real-time PCR analysis of transcript levels of OMT1, OMT2, and OMT3 in Taurus tissues. Each sample contained pooled material from several collections of each tissue from multiple plants. Expression values were normalized with GAPDH amplification and are displayed relative to the expression level in young leaves.
(D) Comparison of the relative expression of OMT1, OMT2, and OMT3 in lupulin glands using RT-PCR.
Genes Encoding Enzymes of Terpenoid and Terpenophenolic Biosynthesis Are Expressed in Lupulin Glands
We constructed two standard and two normalized cDNA libraries from glands isolated from the H. lupulus cultivars Taurus and Nugget; the latter contains 0.69% bdw xanthohumol (Biendl, 2002/2003). The normalized libraries were used to deepen the EST coverage beyond highly expressed transcripts. The cDNA libraries were used for random 5′ single-pass sequencing to produce an EST data set of 4953 putative unique transcripts (see Supplemental Table 1 online). As a means of integrating the lupulin gland transcriptome with the biosynthetic activities in these cells, we extracted information on the number of ESTs corresponding to identifiable enzymes for the three major pathways (terpenoid, bitter acid, and xanthohumol) (Figure 2). This analysis was performed manually by keyword searches of the top five protein matches as determined by BLASTx.
In lupulin glands, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) function both as precursors for terpenoid biosynthesis and as the source of the prenyl side chains of terpenophenolics (Figure 2A). The complete plastidic methylerythritol 4-phosphate (MEP) pathway for synthesis of IPP and DMAPP was represented, with a total of 100 ESTs corresponding to the seven MEP pathway enzymes. By contrast, only 10 ESTs matched any of the six enzymes of the cytosolic mevalonate (MVA) pathway. Because of its small contribution to terpenoid metabolism in hop trichomes, the MVA pathway is not shown in Figure 2. A large contig consisting of 18 ESTs that correspond to IPP isomerase (IDI) was also present. The translated amino acid sequence of the coding region of the IDI contig contained a predicted chloroplast transit peptide, which suggests that it functions in the MEP pathway. Despite the requirement of geranyl diphosphate and farnesyl diphosphate for the synthesis of monoterpenes and sesquiterpenes, respectively, only two geranyl diphosphate synthase (GPPS) ESTs and one farnesyl diphosphate synthase were present. One GPPS EST matched GPPS small subunit proteins from Antirrhinum majus, Mentha × piperita, and Clarkia breweri, while the other was more similar to the A. majus GPPS large subunit and geranylgeranyl diphosphate synthases. The presence of ESTs corresponding to both subunits suggests that H. lupulus may possess a heterodimeric GPPS (Burke et al., 1999; Tholl et al., 2004). Six terpene synthases were identified: a 22-EST contig and a singleton (one EST) were predicted to encode sesquiterpene synthases, and three contigs (16, three, and two ESTs) and one singleton corresponded to monoterpene synthases.
Humulone and lupulone are derived from primary metabolism by the two-step degradation of Leu to form isovaleryl CoA (Goese et al., 1999) (Figure 2B). We identified ESTs for both a branched-chain amino acid transaminase and a branched-chain α-keto acid dehydrogenase E1 β-subunit of the Leu degradation pathway. Isovaleryl CoA is used by valerophenone synthase (VPS) to form phlorisovalerophenone (Paniego et al., 1999). VPS was the second most abundant transcript in the EST data set, with 339 ESTs. The humulone pathway contains two aromatic prenylation steps, and we identified 23 ESTs that were annotated as small-molecule prenyltransferases. This number excludes sequences predicted to encode protein prenyltransferases and short-chain prenyltransferases, such as geranylgeranyl diphosphate synthase and farnesyl diphosphate synthase. The small molecule prenyltransferases ESTs constituted four contigs (14, two, two, and two ESTs) and three singletons. The final step in the humulone pathway is the oxidative conversion of deoxyhumulone to humulone. The class of enzyme catalyzing this oxidation is not clear; therefore, we could not assign ESTs to this step. Cytochrome P450s perform many oxidations in secondary metabolic pathways and may also function in the oxidation of deoxyhumulone. Forty-one ESTs, comprising nine contigs and 10 singletons, were predicted to represent cytochrome P450s.
Xanthohumol and other prenylchalcones arise from Phe via the action of several enzymes, including Phe ammonia lyase, cinnamate 4-hydroxlase, and 4-coumarate CoA ligase (Figure 2C). A large contig (61 ESTs) matched the lupulin gland-specific CHS, CHS_H1 (Matousek et al., 2002). The prenyltransferase enzyme that transfers DMAPP to chalconaringenin to form desmethylxanthohumol is unknown, although the seven prenyltransferases described above are also candidates for this enzyme. The postulated final step in the xanthohumol pathway is the methylation of desmethylxanthohumol, and we describe the functional characterization of candidate OMTs below.
Miscellaneous Biosynthetic Enzymes
Transcripts showing sequence similarity to other known biosynthetic enzymes were present in the data set, but because of the incomplete knowledge about metabolic activities in lupulin glands, it was not possible to assign their functions to certain pathways. In addition to VPS and chs_h1, several other type III polyketide synthases were abundant in the ESTs. Forty-nine ESTs corresponded to CHS2 and 11 to CHS4 (Okada et al., 2004). CHS3, which appears to be nonfunctional (Okada et al., 2004), was not detected. An intriguing result was the high number of ESTs for chalcone isomerase (CHI)–like proteins, which accounted for three large contigs (46, 32, and 24 ESTs) and two singletons. CHI catalyzes the intramolecular cyclization of chalcones to flavanones in flavonoid biosynthesis and is therefore a ubiquitous enzyme in plants. Given that CHI would convert xanthohumol to isoxanthohumol, the occurrence of only trace amounts of isoxanthohumol and other flavanones in lupulin glands indicates that these proteins may not function as true CHIs. CHI-like proteins have been suggested to have enzymatic activity beyond chalcone isomerization or to possess noncatalytic functions, such as serving as flavonoid carriers or stabilizers (Gensheimer and Mushegian, 2004; Ralston et al., 2005).
Few other ESTs showed matches to enzymes of flavonoid and/or anthocyanin biosynthetic pathways. Five ESTs were found that correspond to flavonol synthase, two singletons for flavonoid 3-O-glucosyltransferase, one for leucoanthocyanidin dioxygenase, and one for a putative leucoanthocyanidin reductase, which is involved in the biosynthesis of catechin, a precursor for condensed tannins. Since condensed tannins, composed of both catechin and epicatechin units, are abundant in cone tissue (Taylor et al., 2003) but have not been reported in lupulin glands, the latter EST may have arisen from the contamination of the trichome preparation with cone-derived cells. SAM is the universal methyl donor in plant metabolism and is required for methyl transfer reactions in primary and secondary metabolism (Roje, 2006). An important role for SAM in lupulin glands is in the methylation of desmethylxanthohumol (Figure 2C). A key enzyme in this pathway, SAM synthetase, was among the most highly expressed transcripts (35 ESTs).
Identification of OMT Enzymes
We were especially interested in ESTs that could encode the H. lupulus enzyme that methylates desmethylxanthohumol to form xanthohumol (or, as noted above, the methylation of chalconaringenin to yield 6′-O-methylchalconaringenin). Seven related cDNAs (five contigs and two singletons) representing 40 ESTs were predicted to encode enzymes that methylate low molecular weight metabolites and are designated Hl OMT1 through OMT7 (Table 1). Other H. lupulus methyltransferases that catalyze reactions in primary metabolism (including the methylation of RNA, amino acids, and those functioning in the biosynthesis of SAM) were excluded from this analysis. A 13-member contig, CMT4, matched salicylic acid carboxyl methyltransferase and other enzymes of the SABATH family of plant small molecule methyltransferases (D'Auria et al., 2003). OMT5 and OMT6 showed high similarity to enzymes that methylate caffeoyl CoA and other phenylpropanoids and are therefore likely involved in lignin biosynthesis.
Table 1.
Methyltransferase Enzymes Represented by ESTs from H. lupulus Lupulin Glands
| Name | Top BLASTx Matcha | Accession No. | Identity (%)b | ESTs |
|---|---|---|---|---|
| OMT1 | (R,S)-reticuline 7-OMT (Papaver somniferum) | AAQ01668 | 50 | 13 |
| OMT2 | Caffeic acid OMT (Rosa chinensis) | BAC78828 | 55 | 1 |
| OMT3 | Orcinol OMT (Rosa hybrid) | CAH05081 | 74 | 4 |
| CMT4 | Salicylic acid carboxyl MT(Hoya carnosa) | CAI05934 | 44 | 13 |
| OMT5 | Caffeoyl CoA OMT (Broussonetia papyrifera) | AAT37172 | 95 | 6 |
| OMT6 | Caffeoyl CoA OMT (Arabidopsis thaliana) | AAG52015 | 56 | 2 |
| OMT7 | Caffeic acid OMT (Medicago sativa) | AAB46623 | 87 | 1 |
BLASTx comparison to the National Center for Biotechnology Information (NCBI) nonredundant database on August 25, 2006.
As determined by amino acid comparison of translated nucleotide consensus sequence.
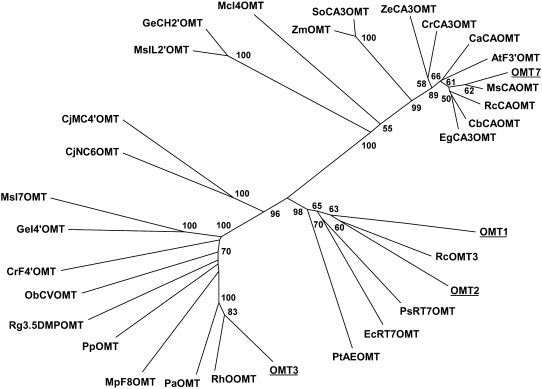
We focused on four enzymes, OMT1, OMT2, OMT3, and OMT7, that grouped together with type 1 OMTs (Noel et al., 2003) involved in secondary metabolic pathways, including flavonoid and chalcone-methylating enzymes. An unrooted tree clustering of these four enzymes with 28 other plant OMTs of known and putative enzyme activities is shown in Figure 4. The most abundant OMT was OMT1 (13 members), which showed high similarity to an (R,S)-reticuline 7-OMT from Papaver somniferum involved in alkaloid biosynthesis. OMT1 and OMT2 are 49% identical on the protein level and show 35 to 55% identity to OMT enzymes of benzylisoquinoline alkaloid biosynthesis (Ounaroon et al., 2003; Fujii et al., 2007) or lignin biosynthesis (Li et al., 1997). OMT3 clusters with enzymes that methylate small phenolic compounds of biosynthetic pathways leading to volatiles (Lavid et al., 2002; Burga et al., 2005) or are involved in flavonoid metabolism (Willits et al., 2004). It shares 50 to 70% protein sequence identity with the members of this cluster. The singleton OMT7, which was similar to caffeic acid methylating enzymes, was not studied further because it was uncovered relatively late in EST sequencing.
Figure 4.
Unrooted Similarity Tree of Plant OMT Proteins Constructed Using the Neighbor-Joining Method.
Bootstrap values from a minimum of 1000 trials are shown. Protein names and accession numbers are listed in Methods.
We used RT-PCR analysis to measure the relative gene expression of Hl OMT1, OMT2, and OMT3 in different hop tissues (Figure 3C). All three showed a similar tissue-specific expression pattern, and this pattern resembled the relative abundance of xanthohumol in these same tissues (Figure 3A). The OMT1 transcript was most abundant in lupulin glands and was also present in early-, mid-, and late-stage cones. OMT2 mRNA was found at high levels in lupulin glands and at lower levels in diverse tissues, such as cones, male flowers, and roots. Comparing the relative expression levels in lupulin glands, OMT1 was the most abundant OMT transcript (Figure 3D). The correlation of the high expression levels of OMT1, as evidenced by both RT-PCR and EST counts, and its specific expression in the lupulin glands where xanthohumol is synthesized provided evidence that is was the mostly likely candidate to synthesize xanthohumol.
Expression and Assay of Recombinant OMT1, OMT2, and OMT3
The Hl OMT1 cDNA was 1240 nucleotides with a 1059-nucleotide open reading frame (ORF) encoding a 39.2-kD (352–amino acid) protein. The full-length sequence of OMT2 was obtained using rapid amplification of cDNA ends (RACE) PCR. The 1302-bp OMT2 cDNA contained a 1083-bp ORF encoding a 39.8-kD (360–amino acid) protein. The 1284-bp OMT3 cDNA contained an ORF of 1134 bp encoding a 42.1-kD (377–amino acid) protein. The three enzymes were expressed as his-tag fusion proteins in insect cells. To determine enzyme activity, insect cell lysate was used directly in assays containing 14C-SAM and one of 34 different substrates (Table 2). Control assays with no substrate (ethanol only) or with lysate of noninfected insect cells were performed and showed no activity. OMT1 methylated desmethylxanthohumol and xanthogalenol (3′ prenyl-4′ O-methylchalconaringenin); the latter is an unusual prenylchalcone present in wild North American hops and their descendants (Stevens et al., 2000). No activity was detected with caffeic acid, prenylflavanones from hops (e.g., 8-prenylnaringenin), simple phenols, or phenylpropanoids. OMT2 showed less selectivity in its substrate preferences and methylated a diverse group of phenylpropanoid-derived compounds, including the hop chalcones chalconaringenin, desmethylxanthohumol, and xanthohumol. OMT3 was inactive with all substrates tested (Table 2).
Table 2.
Enzymatic Methylation of Substrates by Hl OMT1, OMT2, and OMT3
| Substrate | Compound Class | OMT1a | OMT2b | OMT3 |
|---|---|---|---|---|
| Chalconaringenin | Chalcone | 5 | 40 | 0 |
| Desmethylxanthohumol | Chalcone | 100 | 25 | 0 |
| Xanthohumol | Chalcone | 0 | 36 | 0 |
| Xanthogalenol | Chalcone | 445 | 6 | 0 |
| Isoliquiritigenin | Chalcone | 6 | 100 | 0 |
| Butein | Chalcone | 0 | 50 | 0 |
| 2′,4-Dihydroxychalcone | Chalcone | 5 | 45 | 0 |
| 6-Prenylnaringenin | Flavanone | 0 | 0 | 0 |
| 8-Prenylnaringenin | Flavanone | 0 | <2 | 0 |
| Isoxanthohumol | Flavanone | 0 | 0 | 0 |
| Naringenin | Flavanone | 0 | 3 | 0 |
| Quercetin | Flavonol | 0 | 0 | 0 |
| Taxifolin | Dihydroflavonol | 0 | 0 | 0 |
| Kaempferol | Flavanol | 0 | 5 | 0 |
| Apigenin | Flavone | 0 | 3 | 0 |
| Pelargonidin | Anthocyanidin | 0 | 2 | 0 |
| Resveratrol | Stilbene | 0 | 87 | 0 |
| Genistein | Isoflavone | 0 | 25 | 0 |
| (+)-Catechin | Catechin | 0 | 0 | 0 |
| (-)-Epicatechin | Catechin | 0 | 0 | 0 |
| Aesculetin | Coumarin | 0 | 0 | 0 |
| Ellagic acid | Polyhenol | 0 | 0 | 0 |
| Guaiacol | Simple phenol | 0 | 42 | 0 |
| Eugenol | Simple phenol | 0 | 15 | 0 |
| Orcinol | Simple phenol | 0 | 15 | 0 |
| Catechol | Simple phenol | 8 | 12 | 0 |
| Resorcinol | Simple phenol | 5 | 11 | 0 |
| Phloroglucinol | Simple phenol | 0 | <2 | 0 |
| p-Coumaric acid | Phenylpropanoid | 0 | 2 | 0 |
| Sinapic acid | Phenylpropanoid | 0 | <2 | 0 |
| Ferulic acid | Phenylpropanoid | 0 | <2 | 0 |
| Vanillic acid | Phenylpropanoid | 0 | <2 | 0 |
| Caffeic acid | Phenylpropanoid | 7 | <2 | 0 |
| Salicylic acid | Phenylpropanoid | 0 | 0 | 0 |
100% activity set for desmethylxanthohumol.
100% activity set for isoliquiritigenin.
OMT1 and OMT2 were purified as his-tag fusion proteins using a cobalt affinity resin. Yields from insect cell cultures were typically 12 mg (OMT1) and 33 mg (OMT2) of protein per liter of culture. The enzymes were pure as determined by SDS-PAGE with Coomassie blue staining. Gel filtration chromatography showed that OMT1 and OMT2 had native molecular masses of 76 and 64 kD, respectively, indicating that both proteins are homodimers. Using desmethylxanthohumol as the acceptor substrate, OMT1 had maximal activity at pH 9.0 over the temperature range of 30 to 37°C. OMT2 had pH optimum of 8.5 and a temperature optimum of 39°C with xanthohumol. Neither enzyme required bivalent cations (Mg2+, Fe2+, Zn2+, Cu2+, Ca2+, and Co2+) for activity.
We determined the kinetic properties of OMT1 and OMT2 with substrates that are known to be present in hops (Table 3). OMT1 was tested only with desmethylxanthohumol since the amount of xanthogalenol available was very limited. These assays were complicated by the spontaneous isomerization of the chalconaringenin and desmethylxanthohumol at the basic pH of the methylation reactions, thereby decreasing the amount of substrate available. OMT1 showed a moderate affinity for desmethylxanthohumol (Km of 18 μM) and a low affinity for SAM (Km of 286 μM). S-adenosyl homocysteine (SAH) inhibited the reaction with a Ki of 98 μM with respect to SAM; xanthohumol was a competitive inhibitor with respect to desmethylxanthohumol with a Ki of 2 μM. We did not have enough 4-O-methylxanthohumol to perform inhibition experiments with OMT2 but found that this enzyme was inhibited by SAH with a Ki of 5 μM with respect to SAM.
Table 3.
Kinetic Properties of Hl OMT1 and OMT2
| Km Substrate (μM)a | Vmax Substrate (pkat/mg)a | Vmax/Km | |
|---|---|---|---|
| OMT1 | |||
| Desmethylxanthohumol | 18 ± 2 | 55 ± 1 | 3.1 |
| SAM | 286 ± 23b | 56 ± 1 | 0.2 |
| OMT2 | |||
| Chalconaringenin | 237 ± 19 | 914 ± 21 | 3.9 |
| Desmethylxanthohumol | 23 ± 6 | 468 ± 34 | 20.3 |
| Xanthohumol | 31 ± 3 | 451 ± 14 | 14.4 |
| Resveratrol | 19 ± 1 | 908 ± 10 | 48.5 |
| SAM | 34 ± 3c | 402 ± 9 | 11.8 |
Values represent mean ± se (n = 3).
Determined with desmethylxanthohumol as cosubstrate.
Determined with xanthohumol as cosubstrate.
OMT2 methylated resveratrol with the highest efficiency, as determined by Vmax/Km ratios. Resveratrol is present in Taurus and Nugget cones (Jerkovic and Collin, 2007), but it is not clear if this compound occurs in lupulin glands or in cone tissues. Methylated derivatives of resveratrol have not been reported from hop. Furthermore, methylated resveratrol was not identified in transgenic hops that accumulate large amounts of resveratrol in leaves and cones (Schwekendiek et al., 2007).
Identification of Enzyme Reaction Products of OMT1 and OMT2
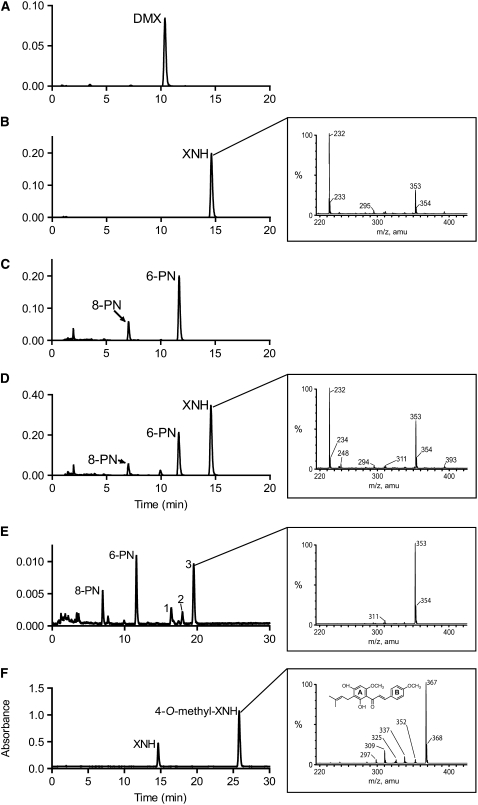
Desmethylxanthohumol has four hydroxyl groups that can be methylated, and it was important to establish the regioselectivity of OMT1. Figures 5A and 5B show the elution of desmethylxanthohumol and xanthohumol standards, respectively. As shown in Figure 5C, HPLC analysis of a reaction of OMT1 and desmethylxanthohumol without SAM found that desmethylxanthohumol spontaneously isomerized to 6-prenylnaringenin (6-PN; 11.7 min) and 8-prenylnaringenin (8-PN; 7.0 min), but no methylation products were formed. This was determined by comparison of retention time with authentic standards and liquid chromatography–mass spectrometry (LC-MS) of the product peaks. Inclusion of SAM in the assay resulted in an additional peak (14.6 min) that had the same retention time and UV spectrum as xanthohumol (Figure 5D). A definitive identification was made by negative-ion electrospray ionization (ESI)-LC-MS analysis of the reaction mixture (Figure 5D, inset), which showed that the product had the same mass as xanthohumol ([M-H]− mass-to-charge ratio [m/z] 353). Therefore, OMT1 is an SAM:desmethylxanthohumol 6′-O-methyltransferase that catalyzes xanthohumol formation.
Figure 5.
HPLC and LC-MS Analysis of Product Formation by Recombinant Hl OMT1 and OMT2.
(A) Chromatogram of authentic desmethylxanthohumol (DMX) standard analyzed at an absorbance of 330 nm.
(B) Chromatogram of authentic xanthohumol (XNH) standard at 330 nm. Inset: negative-ion ESI-LC-MS analysis of the xanthohumol standard.
(C) HPLC analysis of the methylation of desmethylxanthohumol by OMT1 in the absence of the methyl donor SAM. The chromatogram was obtained at 330 nm.
(D) HPLC analysis of the methylation of desmethylxanthohumol by OMT1 in the presence of SAM. OMT1 methylates desmethylxanthohumol to form xanthohumol (14.6 min). 6-PN and 8-PN are formed by isomerization of the substrate. The chromatogram was obtained at 330 nm. Inset: negative-ion ESI-LC-MS analysis of the 14.6 min peak produced by methylation of desmethylxanthohumol by OMT1.
(E) HPLC analysis of the methylation of desmethylxanthohumol by OMT2. Desmethylxanthohumol spontaneously isomerizes to form 6-PN (11.7 min) and 8-PN (7.0 min) in the OMT assay. The identities of peaks 1 to 3 are described in Results. The chromatogram was extracted at 330 nm. Inset: negative-ion ESI-LC-MS analysis of the 19.5 min peak corresponding to a monomethylated prenylnaringenin-type flavanone (m/z 353).
(F) HPLC analysis of the methylation of xanthohumol by OMT2. Inset: negative-ion ESI-LC-MS analysis of the product peak corresponding to 4-O-methylxanthohumol (m/z 367).
Recombinant OMT2 did not form xanthohumol from desmethylxanthohumol (Figure 5E) but did methylate chalconaringenin, desmethylxanthomol, xanthohumol, and resveratrol to give products that had increased retention times compared with their substrates. These products showed increased masses of 14 atomic mass units, indicating that each was methylated once (data shown for xanthohumol, Figure 5F). In the reaction of OMT2 with desmethylxanthohumol, 6- and 8-prenylnaringenin and several reaction products with increased retention times relative to 6- and 8-prenylnaringenin were detected. The UV spectra of the peaks at 16.4 min (peak 1, Figure 5E) and 19.5 min (peak 3, Figure 5E) were identical to 6- and 8-prenylnaringenin (maxima at 228 to 230, 294, and 338 nm), indicating that they were also flavanones. LC-MS analysis showed that they had masses of [M-H]− m/z 353, suggesting they are the monomethyl derivatives of 6- and 8-prenylnaringenin. The minor product at 17.9 min (peak 2, Figure 5E) had a mass of [M-H]− m/z 353 but a chalcone-like UV spectrum (UVmax at 371 nm). Since OMT2 did not methylate 6-PN and 8-PN (Table 3), we reason that the flavanones at 16.4 and 19.5 min form by isomerization of a methylated chalcone precursor, such as the compound eluting at 17.9 min. Methylation at the 4′-OH of desmethylxanthohumol would yield xanthogalenol. However, the retention time of xanthogalenol did not match that of the minor chalcone product (17.9 min) formed by the OMT2-catalyzed methylation of desmethylxanthohumol. The retention times of the flavanones at 16.4 and 19.5 min were also different from the two flavanones that are produced by isomerization of xanthogalenol (16.7 and 21.6 min). Methylation at the 2′-OH of desmethylxanthohumol by OMT2 can be excluded as 2′-O-methyldesmethylxanthohumol would only undergo cyclization at C-6′ and result in only one flavanone.
To demonstrate conclusively the methyltransferase activity of OMT2, we performed a large-scale reaction with xanthohumol as substrate and isolated the product by semipreparative HPLC. The compound had a maximum absorption of 368 nm and a mass of [M-H]− m/z 367, which corresponds to the addition of a single methyl group. Comparing the proton nuclear magnetic resonance (1H-NMR) spectra of the isolated product with the parent compound, xanthohumol, showed the disappearance of a singlet at δ 10.1 ppm (4-OH position) in the xanthohumol spectrum and a new methyl signal at δ 3.80 ppm (see Supplemental Table 2 online). An additional difference was a shift in the resonances of the B-ring protons (Δ0.1 ppm for H-2, 6; Δ0.18 ppm and H-3, 5), while the A-ring proton (H-5′) was unchanged. A nuclear Overhauser effect (NOE) difference experiment was performed to conclusively demonstrate that OMT2 methylates xanthohumol at the 4-OH of the B-ring. Irradiation of the 4-OCH3 protons, resonating at 3.80 ppm, produced an enhancement of the resonance corresponding to H-3/H-5 (7.01 ppm). Likewise, irradiation of the H-3/H-5 protons gave an enhancement for 4-OCH3. Irradiation of the 6′-OCH3 (3.85 ppm) signal enhanced the signal of H-5′ (6.07 ppm). OMT2 methylates xanthohumol at the 4-OH group to yield 4-O-methylxanthohumol (2′,4′-dihydroxy-4,6′-dimethoxy-3′-prenylchalcone). These results show that OMT2 is a putative SAM:xanthohumol 4-O-methyltransferase that also accepts other phenolic substrates. We did not determine the structures of the methylation products from chalconaringenin, desmethylxanthomol, and resveratrol, but it is likely that OMT2 methylates these substrates at the 4-OH position.
DISCUSSION
The physiological and metabolic specialization of glandular trichomes and their high expression of biosynthetic enzymes have made them valuable targets for genomic investigations of natural product biosynthesis. EST data sets derived from trichome-specific cDNA libraries have been used to identify enzymes leading to terpenoids in mint (Mentha × piperita; Lange et al., 2000), phenylpropenes in basil (Ocimum basilicum; Gang et al., 2001), methylketones in tomato (Solanum lycopersicum; Fridman et al., 2005), and sesquiterpenes in Artemisia annua (Teoh et al., 2006). Based on the successful application of EST genomics to other trichome systems and the accumulation of bitter acids and xanthohumol in lupulin glands, we chose this approach to study terpenophenolic biosynthesis in hop. We selected high-xanthohumol H. lupulus cultivars for trichome isolation as well as cDNA library normalization to increase the likelihood of identifying enzymes of xanthohumol biosynthesis. The lupulin gland EST data set proved to be a rich source of ESTs corresponding to known and predicted enzymes in terpenophenolic and terpenoid biosynthesis. More than 5% of these ESTs could be assigned to pathways leading to such metabolites and VPS was represented a remarkable 339 times (3.2% of the ESTs). The 4953 unique transcripts identified in this study, representing >3 Mb of sequence information, is a significant new genomic resource for H. lupulus.
Given the interest in the bioactivity of xanthohumol and the lack of information about its biosynthesis we focused on the identification of the desmethylxanthohumol 6′-OMT. We had previously suggested that prenylation preceded methylation (Stevens and Page, 2004), but this assertion lacked experimental evidence. The identification of OMT1 as a desmethylxanthohumol 6′-OMT clarifies the order of reactions in the xanthohumol pathway. In support of this finding, 6′-O-methylchalconaringenin is not known from hop and none of the three OMT enzymes we assayed methylated chalconaringenin to form this compound. OMT2 accepted chalconaringenin as a substrate, but based on its methylation of xanthohumol at the 4-OH position, does not methylate at the equivalent 6′-OH of any substrate. The correct order of enzymatic steps of the xanthohumol pathway is shown in Figure 2C.
The kinetic properties of OMT1 were similar to those observed with isoflavone OMTs from alfalfa (Medicago sativa; Km of 20 and 150 μM for daidzein and SAM, respectively; Ki of SAH of 35 μM) and other legumes (Edwards and Dixon, 1991; Maxwell et al., 1992), although the affinity of OMT1 for SAM was almost twofold lower. Edwards and Dixon (1991) suggested that the activity of the isoflavone OMTs is regulated by the availability of their isoflavone substrates, and OMT1 may also be rate limited by the pool of desmethylxanthohumol in the cellular compartment where it is active. The activity of OMT2, which had similar Km values for desmethylxanthohumol, xanthohumol, resveratrol, and SAM, is likely not regulated in this manner. The nearly sevenfold higher catalytic efficiency of OMT2 with desmethylxanthohumol compared with OMT1 would seem to indicate that OMT2 is a better candidate for the xanthohumol-forming OMT than OMT1. However, product characterization studies clearly show that OMT2 methylates desmethylxanthohumol at the 4-OH position and does not form xanthohumol. The large difference in Vmax/Km ratios between the two enzymes may be due to the relative instability of recombinant OMT1 compared with OMT2: OMT2 was stable for weeks at 4°C and longer when stored with 50% glycerol at −20°C, while OMT1 quickly lost activity at both temperatures or when stored in dilute solutions.
Our results did not allow us to determine the endogenous substrate of the protein encoded by OMT2. Recombinant OMT2 showed methylation activity with 13 of the 33 substrates tested, all of which had a free 4-OH group. We conclusively identified the product formed by methylation of xanthohumol as 4-O-methylxanthohumol. 4-O-methylxanthohumol is not a novel natural product, as rats fed xanthohumol excrete this compound (Nookandeh et al., 2004), but 4-O-methylxanthohumol or chalcones bearing 4-O-methyl groups have not been reported from hop. Only one EST for OMT2 was identified in the EST data set, which may suggest that the methylation products are not abundant metabolites. OMT2's preference for chalcones (Table 3), its catalytic efficiency with desmethylxanthohumol and xanthohumol, and the diversity of this class of metabolites in hops could indicate that it forms a minor methylated chalcone that has not yet been isolated. Given its high affinity and Vmax/Km ratio for resveratrol, it is also possible that OMT2's in vivo function is to methylate resveratrol but that this compound is not present in lupulin glands because the stilbene pathway is blocked at one or more steps. With no hint of enzyme activity, it is impossible to assign function to OMT3.
The differences in the substrate specificity of Hl OMT1 and OMT2 are noteworthy. Like the only other known chalcone OMT, the isoliquiritigenin 2′-O-methyltransferase from M. sativa (Maxwell et al., 1993), OMT1 showed a remarkable specificity for desmethylxanthohumol and for one other chalcone, xanthogalenol. OMT2 methylated diverse substrates. Several type 1 OMTs with high substrate and/or regiospecificity have been described before, for example, a 4′-O-methyltransferase isolated from Catharanthus roseus only methylated flavonoids with the B-ring configuration of homoeriodyctiol and a specific A-ring substitution pattern (Schroder et al., 2004). By contrast, many OMTs exhibit a broad substrate spectrum and may accept substrates from different compound classes (Gauthier et al., 1998; Chiron et al., 2000). The structural basis for the difference between OMT1 and OMT2 is not clear. The two enzymes are of similar size (OMT1, 39.2 kD; OMT2, 39.8 kD), and both are homodimers, but their amino acid sequences showed only 49% identity. The crystal structures of several OMTs, including the chalcone OMT from M. sativa, have been solved (Zubieta et al., 2001, 2002, 2003; Ferrer et al., 2005). It would be of interest to compare the structure of Hl OMT1 with the alfalfa chalcone OMT and to determine how the enzyme accommodates the hydrophobic prenyl side chain in the active site.
The biosynthesis of xanthohumol and other prenylchalcones represents a deviation from the standard flavonoid pathway in plants. Plants form chalconaringenin as the first intermediate in the flavonoid pathway, but in most cases this compound is rapidly cyclized to naringenin by CHI. This standard route is functional in hop tissues, such as leaves and cones, as shown by their content of flavonols (Sägesser and Deinzer, 1996), but it appears to be almost entirely absent in lupulin glands. There were a few enzymes of flavonoid biosynthesis in the EST data set (e.g., five ESTs corresponded to flavonol synthase), but given the lack of flavanones in the trichomes (Figure 3B), we consider these to be from a trace amount of nontrichome tissue. Chalconaringenin and desmethylxanthohumol are highly unstable, as demonstrated by the latter's facile isomerization to 6- and 8-prenylnaringenin in the OMT assays (Figure 5C). The 6′-O-methylation of desmethylxanthohumol by OMT1 serves to inhibit isomerization by favoring the interaction of the remaining free hydroxyl group with the nearby keto functionality; xanthohumol is much more stable than its precursor. A similar role has been described for the isoliquiritigen 2′-O-methyltransferase from M. sativa, which stabilizes a chalcone through A-ring methylation.
Zubieta et al. (2001) have previously noted that methylation patterns serve to determine product outcomes in plant secondary metabolism; therefore, OMTs serve as key switches at branch points of metabolic pathways. Another function of O-methylation is to modify the biological activity of end-product metabolites. Methylation of the 6′-OH group of chalcones, such as desmethylxanthohumol, prevents intramolecular cyclization via their α,β-unsaturated carbonyl groups, preserving their reactivity toward nucleophilic residues in proteins. Liu et al. (2005) have shown that the α,β-unsaturated carbonyl of xanthohumol reacts with Cys residues on the mammalian sensor protein Keap1, and this reactivity may be important for other types of bioactivity. The ecological role of prenylchalcones is not known, but it is tempting to speculate that their localization in surface structures that occur on female cones and their reactive chalcone structures indicate that they serve to prevent herbivores from consuming developing seeds. Methylation serves to increase lipophilicity, thereby potentially reducing the loss of prenylchalcones during heavy rains.
While methylation of desmethylxanthohumol increases the stability of this prenylchalcone, the mechanism by which chalconaringenin and desmethylxanthohumol are stabilized in planta is not known. One possibility is that metabolic channeling of intermediates via a metabolon prevents chalcone cyclization. The advantages of the formation of a metabolon would be severalfold. It would enable the quick conversion of the labile chalcone intermediates into more stable compounds and thereby prevent their diffusion into the surrounding cellular environment and subsequent isomerization. It would also allow for control of metabolic crosstalk and substrate availability between different pathways that compete for substrates, for example, the availability of DMAPP for both the humulone and xanthohumol pathways. Although OMT1 is a soluble enzyme, it may transiently associate with other enzymes in the xanthohumol pathway as has been suggested for the enzymes of the phenylpropanoid and the flavonoid pathways (Winkel-Shirley, 1999; Winkel, 2004).
Our EST analysis also sheds light on the isoprenoid origin of the terpenophenolics. Recognizing that measuring gene expression using EST frequency has limitations (Audic and Claverie, 1997; Park et al., 2006), it is informative that 100 ESTs corresponded to MEP pathway enzymes and only 10 for the MVA pathway. Further evidence for the predominance of the former is provided by the presence of a chloroplast transit peptide in the predicted amino acid sequence of the contig encoding IPP isomerase, suggesting that it functions in the plastid. Monoterpenes are synthesized via the MEP pathway, and recent evidence shows that sesquiterpenes, at least in some cases, are as well (Dudareva et al., 2005). The prevalence of MEP pathway indicates that the majority of terpenoid and prenylated metabolites found in hop trichomes are formed via this pathway. This is supported by a 13C-labeling study of humulone formation that found that its prenyl side chains have their origin in deoxyxylulose (Goese et al., 1999). The lower abundance of MVA transcripts does not rule out that this pathway plays a minor role in lupulin gland metabolism. Dudareva et al. (2005) concluded that the contributions of the MEP and MVA pathways may differ according to developmental stage or cell type. The high expression of the MEP pathway in the mint and basil trichome ESTs (Lange et al., 2000; Gang et al., 2001) and our own similar findings for hop suggests that this may be a general condition in secretory tissues synthesizing large amounts of terpenoids.
Selective breeding of cultivated hops has led to substantial increases in the content of terpenophenolics, with super-high-alpha varieties containing up to 24% bdw α-acids, 8% β acids, and >1% bdw xanthohumol in cones. The discovery of enzymes responsible for terpenophenolic synthesis in hop could create new opportunities for improvement of this valuable crop. The hop lupulin gland EST resource described in this study will accelerate both gene discovery and molecular breeding efforts for hops.
METHODS
Collection of Hop Samples
Hop (Humulus lupulus) samples from the varieties Taurus and Nugget were collected near Mainburg and Baasdorf, Germany, in 2002, 2003, and 2005. Lupulin glands were isolated from dissected cones by agitation in liquid nitrogen. The material was filtered through a 1-mm metal sieve and a 500-μm nylon screen to remove cone material. The glands were recovered from the liquid nitrogen and stored at −80°C. Microscopic examination showed gland preparations contained small amounts of nonglandular trichomes (cystolith hairs) but no cone tissue.
HPLC Analysis of Xanthohumol Levels
Samples of different hop tissues were frozen in liquid nitrogen and ground to a powder. A 150-mg sample was extracted with 400 μL of 75% acetonitrile and 25% water (containing 0.1% trifluoroacetic acid [TFA] [v/v]) and filtered through a 0.45-μm SpinX column (Costar). Each tissue was analyzed in triplicate. HPLC analysis was performed on a Waters 2695 separations module with a Sunfire C18 reversed phase column (150 × 4.6-mm i.d.; 3.5 μm) at a temperature of 30°C. The gradient elution system consisted of 50% water (0.1% TFA [v/v]):50% acetonitrile changing to 20% water (0.1% TFA [v/v]):80% acetonitrile over 40 min at 1 mL/min. Injection volume was 20 μL. Xanthohumol was detected at 370 nm with photodiode array detection. Quantification was performed using peak area by comparison to a standard curve (r2 0.999).
cDNA Library Construction and EST Sequencing
Total RNA was isolated from frozen lupulin glands either using buffered phenol-chloroform-isoamyl alcohol containing aurintricarboxylic acid followed by a cleanup with an RNeasy plant mini kit (Qiagen) or using an RNeasy plant mini kit directly. Aurintricarboxylic acid was removed by Sephadex G-50 gel filtration (Skidmore and Beebee, 1989). mRNA was purified using an Oligotex kit (Qiagen) or a Dynabeads mRNA purification kit (Invitrogen). A cDNA library (HLUPJN1) was constructed from 1 μg of mRNA from Taurus lupulin glands collected in 2002 and 2003 using a SMART cDNA library kit (Clontech). cDNAs were ligated into λTriplEx2 and packaged with MaxPlax Lambda packaging extract (Epicentre Technologies) using Escherichia coli strain XL1 Blue MRF'. Libraries from lupulin glands isolated from cones of Taurus (HLUPLC1; cones collected in 2003) and Nugget (HLUTR2CH and HLUTR3CH; cones collected in 2005) were constructed from 2 to 3 μg of mRNA using a cDNA library synthesis kit (Stratagene). cDNAs were ligated into the plasmid vector pBluescript SK+ and electroporated into E. coli DH10B T1 phage-resistant cells (Invitrogen). Library normalization was performed to reduce the number of highly abundant transcripts present in the standard cDNA libraries. HLUPLC1 was normalized before sequencing using a hydroxyapatite-based method (C0t 2.5 and 5) (Bonaldo et al., 1996). HLUTR2CH was normalized to produce library HLUTR3CH. cDNAs from the library HLUPJN1 were amplified from phage plaques using the vector primers 5′-CAAGCTCCGAGATCTGGACGAGC-3′ and 5′-ATACGACTCACTATAGGGCGAATTGGCC-3′. PCR products were purified prior to 5′ sequencing with the primer 5′-CTCGGGAAGCGCGCCATTG-3′ and BigDye terminators (Applied Biosystems). BigDye reactions from library HLUPJN1 were resolved on an ABI 3100 sequencer (Applied Biosystems). Bacterial colonies from libraries HLUPLC1, HLUTR2CH, and HLUTR3CH were grown overnight in 96-well plates and aliquots used for TempliPhi amplifications (GE Healthcare). TempliPhi products were sequenced with the T3 primer. BigDye reactions from libraries HLUPLC1, HLUTR2CH, and HLUTR3CH were resolved on an ABI 3730xl sequencer (Applied Biosystems).
Bioinformatics
ESTs were processed and annotated using a high-performance cluster computer. Phred was used to read DNA sequencer trace data, call bases, and assign quality values (Ewing et al., 1998). Quality assessment, confidence reassurance, and calculation of trimming points were performed using Lucy (Chou and Holmes, 2001). Crossmatch (www.phrap.org) was used to identify and mask contaminating cloning, vector, and bacterial sequences and Vmatch (www.vmatch.de) to identify repetitive elements by comparison of the sequences to The Institute for Genomic Research (TIGR) plant repeat databases (www.tigr.org/tdb/e2k1/plant.repeats). Poly(A/T) tails were identified using TIGR Gene Indices cleaning protocols (Quackenbush et al., 2001) and trimmed using SeqClean (www.tigr.org/tdb/tgi/software). Only sequences exceeding 100 bp in length after trimming were used for clustering. TGICL software was used for clustering and assembly of the final data set (Pertea et al., 2003), and the resulting clusters were assembled using CAP3 assembly program (Huang and Madan, 1999). Sequences were annotated by BLASTx comparison to the NCBI nonredundant database.
RACE PCR Cloning of OMT2
RACE was used to obtain the 5′ end of the OMT2 cDNA. Lupulin gland mRNA was reverse transcribed using a GeneRacer kit (Invitrogen). RACE PCR reactions were performed with a GeneRacer 5′ primer (5′-CGACTGGAGCACGAGGACACTGA-3′) and 5′ OMT2r primer (5′-CGGCAGTGGGGATCCATTCAAAC-3′). A nested reaction with the GeneRacer 5′ nested primer (5′-GGACACTGACATGGACTGAAGGAGTA-3′) and 5′ OMT2r-nested primer (5′-CATGGGACACTAGTCCGGTGTACGTT-3′) was also performed. The blunt-end PCR product was incubated with dATP and Taq polymerase, cloned into the vector pGEM-T (Promega), and sequenced.
Real-Time PCR Analysis of Gene Expression
RNA for RT-PCR was isolated from pooled samples of hop tissues collected in the field (EZNA plant RNA kit; Omega Bio-Tek) and genomic DNA removed by DNase I (Qiagen). RNA quality was assessed using the RNA 600 LabChip of the 2100 Agilent Bioanalyzer. First-strand cDNA was synthesized from 1 μg of total RNA using the QuantiTect Reverse Transcription kit (Qiagen) using a mixture of oligo(dT) and random primers. Gene-specific primers were as follows: OMT1f (5′-TAAAGGAACAGTGGTGGACGTTG-3′), OMT1r (5′-ACCGCATCAGCACTAGGAATTGA-3′), OMT2f (5′-TCCAGGGAATCCGGAGTTCAACAA-3′), OMT2r (5′-CGACAACATGGGGTAGATCATAG-3′), OMT3f (5′-TTTGGACCCTGCTTCATCGCA-3′), OMT3r (5′-AGGAGGGCTGGTCTTCGAAATGTA-3′), GAPDHf (5′-ACCGGAGCCGACTTTGTTGTTGAA-3′), and GAPDHr (5′-TCGTACTCTGGCTTGTATTCCTTC-3′). The expected size of qPCR products ranged from 165 to 215 bp. RT-PCR analyses were performed using SYBR Green I (Platinum SYBR Green qPCR Super Mix-UDG kit; Invitrogen) on an Mx3000P instrument (Stratagene). ROX was used as a reference dye in a final concentration of 50 nM. All reactions were run in duplicate using PCR conditions: 50°C for 2 min; 40 cycles of 95°C for 2 min, 55°C for 1 min, and 72°C for 20 s). A melting curve analysis was performed to check for specific product amplification (95°C for 1 min, 55°C for 30 s, and 95°C for 30 s). Controls lacking template for all primer pairs and controls lacking reverse transcription controls for all cDNAs were included in each run. Ct values were calculated using MxPro QPCR software v. 3.00 (Stratagene). The 2−ΔΔCT method was used for relative gene expression analysis (Livak and Schmittgen, 2001).
Expression of Recombinant OMTs in Insect Cells
The ORFs of OMT1-3 were amplified from lupulin gland first-strand cDNA using the primers orfOMT1f (5′-CGTTGAATTCATGGAATCTCTAAGAGGCCAAGA-3′), orfOMT1r (5′-GTGCAAGCTTTCACACTAGAAAGGCCTCAATAA-3′), orfOMT2f (5′-GAGAATTCATGGAGTTGGCACGGAATGATCA-3′), orfOMT2r (5′-GTCAAGCTTTCATTGTGGATAGGCTTCAATGAC-3′), orfOMT3f (5′-TAGGATCCATGGAAGTAGAGCAAGT-3′), and orfOMT3r (5′-GCGCGTCGACCTAATTCTTTGTCAAAGA-3′). The primers were designed to contain sites for EcoRI in the forward primer and HindIII in the reverse primer. PCR products were digested, cloned into pFastBacHTa (Invitrogen), and transformed into E. coli DH10Bac (Invitrogen). All cloning procedures were verified by sequencing. Bacmid DNA was isolated and transfected into Sf9 insect cells to generate recombinant baculovirus. The primary viral stock was amplified fivefold to produce a high titer viral stock (P5) that was used to infect Sf9 insect cell cultures for protein expression. Expression cultures were grown in SF-900 II SFM media (Invitrogen) either as adherent cultures (5 mL) in T25 flasks or as suspension cultures (200 mL; 76 rpm) in 500-mL spinner flasks. Expression cultures (∼2 × 106 cells/mL) were infected with P5 viral stock and grown for 72 h at 28°C before harvesting.
Assay of OMT Activity Using 14C-SAM
Substrates were dissolved in ethanol to a concentration of 50 mM. The standard assay (50 μL) contained 1 mM substrate, 7.6 μM 14C-SAM (52.8 mCi/mmol; PerkinElmer), and 10 μL of crude insect cell lysate in 50 mM Tris-HCl, pH 7.5, containing 2 mM DTT. The reaction was incubated for 30 min at 30°C and terminated by addition of 2.5 μL of 6 n HCl. The 14C-labeled methylated products were extracted with 100 μL of ethyl acetate and 20-μL aliquots measured by liquid scintillation counting. Assays were repeated at least once. Control assays containing ethanol instead of substrate, lysate from insect cells not expressing recombinant protein, or insect cells infected with empty baculovirus were also performed.
Purification and Biochemical Characterization of Recombinant OMT1 and OMT2
Insect cells were harvested after 72 h of infection and lysed in binding buffer (20 mM Tris-HCl, pH 8.0, 7.5 mM imidazole, and 0.5 M NaCl, 0.5% [v/v] protease inhibitor cocktail [Set III; Calbiochem]) by sonication. Cell debris was removed by centrifugation and the supernatant bound to cobalt affinity resin (Talon; Clontech), washed twice in wash buffer (20 mM Tris-HCl, pH 8.0, 10% [v/v] glycerol, 7.5 mM imidazole, 150 mM NaCl), and eluted in elution buffer (20 mM Tris-HCl, pH 8.0, 20% [v/v] glycerol, 100 mM imidazole, and 150 mM NaCl). The elution buffer was exchanged using Sephadex-25 gel filtration with storage buffer (100 mM sodium glycinate, pH 9.0, for OMT1 and pH 8.5 for OMT2, 50% [v/v] glycerol, 2 mM DTT, and 50 mM NaCl) and the protein concentrated using a 10k Amicon Ultra centrifugal filter (Millipore). Purified proteins were stored at −20°C. Protein purity was determined by SDS-PAGE gel electrophoresis with Coomassie Brilliant Blue staining.
OMT assays were performed under linear product formation conditions with respect to time and protein concentration. The enzyme assays (50 μL) were performed with 100 μM SAM, 1.9 μM 14C-SAM (52.8 mCi/mmol), and 200 to 400 μM phenolic substrate in 100 mM Tris-HCl, pH 7.5, and 2 mM DTT at 30°C. Temperature optima were determined across a temperature range from 0 to 60°C. Enzymatic activity was tested in various buffers spanning a pH range from 4.0 to 10.0 using the following buffers: sodium citrate (pH 4.0, 4.5, 5.0, 5.5, and 6.0), potassium phosphate (pH 5.7, 6.0, 6.5, 7.0, and 7.5), Bis-Tris-HCl (6.0, 6.5, and 7.0), Tris-HCl (7.0, 7.5, 8.0, and 8.5), and sodium glycinate (pH 8.5, 9.0, 9.5, and 10.0). Kinetic parameters were determined by varying substrate concentrations at a fixed concentration of the second substrate under optimum temperature and buffering conditions. Kinetic constants were determined by fitting initial velocity versus substrate concentration to the Michaelis-Menten equation using nonlinear regression (GraphPad Prism). All assays were performed in triplicate except Ki determinations.
Identification of Reaction Products by LC-MS and 1H-NMR
Purified OMT1 (38 μg) was incubated with 2 mM SAM and 150 μM desmethylxanthohumol in 100 μL of 0.1 M sodium glycinate, pH 9.0, for 80 min at 34°C with 120 μmol desmethylxanthohumol added after 40 min. Purified OMT2 (31.5 μg) was incubated with 400 μM SAM and 1.6 mM chalconaringenin, 80 μM desmethylxanthohumol, 200 μM xanthohumol, or 500 μM resveratrol in 0.1 M sodium glycinate, pH 8.5, at 39°C. Control reactions lacking SAM were also performed. Incubation times varied from 20 min to 2 h with additional substrate (1.6 mM chalconaringenin, 80 μM desmethylxanthohumol, 120 μM xanthohumol, or 400 μM resveratrol) added at the halfway point. The reactions were stopped with 4 μL of 6 n HCl and extracted with ethyl acetate. The ethyl acetate phase was dried, resuspended in 30 μL acetonitrile, and analyzed by HPLC.
HPLC separation of reaction products was performed as described for xanthohumol quantification. A Waters Alliance 2695 chromatography system coupled to a ZQ 2000 mass detector and a 2996 photodiode array detector controlled by MassLynx software was used for LC-MS analyses. Separations were performed on a Waters Sunfire RP C18 (150 × 2.1 mm i.d.; 3.5 μm) at 0.2 mL/min at a temperature of 35°C. The solvent system consisted of solvent A, 0.15% acetic acid in 10% acetonitrile (aqueous, [v/v]), and solvent B, 0.14% acetic acid in 100% acetonitrile. The elution gradient was: (1) 0 to 28 min, 60% A to 40% B to 40% A to 60% B; 28 to 31 min, 40% A to 60% B to 100% B; 31 to 39 min, 100% B. Injection volumes were 0.5 to 10 μL. The mass detector parameters (ESI) were set to capillary (kV) 2.70, cone (V) −15 to −30 over a mass range of 215 to 425, extractor (V) −3.50 and radio frequency lens (V) −0.7. Photodiode array detection was performed over the range 200 to 400 nm, with specific detection at 290 and 370 nm.
For product identification by 1H and NOE NMR, a 4-mL reaction of OMT2 with xanthohumol was prepared as above. The dried residue was dissolved in acetonitrile and separated on a Waters SunFire Prep C18 (250 × 10 mm i.d.; 5 μm) column using a mobile phase of 30% water (0.1% [v/v] trifluoracetic acid) and 70% acetonitrile at a flow rate of 3 mL/min. The product peak was collected, dried in vacuo, and lyophilized. The 1H NMR spectra of xanthohumol and 4-O-methylxanthohumol, as well as the NOE experiments on the latter, were obtained on a Bruker Avance DRX 500 MHz spectrometer at ambient temperature in DMSO-d6.
Similarity Tree of OMTs
A similarity tree for comparison of the four OMTs from the lupulin gland EST data set and 28 related, functionally defined, or putative plant OMTs was constructed using amino acid sequences by the neighbor-joining method with TreeCon for Windows, v.1.3b (Van de Peer et al., 1994). Distance calculation was done by Poisson correction, and insertions-deletions were not taken into account. The full alignment for the tree is included in Supplemental Data Set 1 online.
Accession Numbers
Sequence data for the H. lupulus genes described in this article can be found in the GenBank/DDBJ/EMBL data libraries under the following accession numbers: OMT1 (EU309725), OMT2 (EU309726), OMT3 (EU309727), CMT4 (EU309728), OMT5 (EU309729), OMT6 (EU309730), and OMT7 (EU309731). Protein names from Figure 4, with accession numbers in parentheses, are as follows: Hl OMT1, Humulus lupulus OMT1; Rc OMT3, Rosa chinensis var spontanea caffeic acid OMT (BAC78828)*; Hl OMT2, Humulus lupulus OMT2; Ps RT7OMT, Papaver somniferum (R,S)-reticuline 7-OMT (AAQ01668)*; Ec RT7OMT, Eschscholzia californica reticuline 7-OMT (BAE79723)*; PtAEOMT, Pinus taeda hydroxycinnamic acid/hydroxycinnamoyl-CoA ester OMT (AAC49708)*; Hl OMT3, Humulus lupulus OMT3; Rh OOMT, Rosa hybrid cultivar orcinol OMT (AAM23004)*; Pa OMT, Prunus armeniaca OMT (AAB71213); Mp F8OMT, Mentha piperita flavonoid 8-OMT (AAR09600)*; Pp OMT, Pyrus pyrifolia OMT (BAA86059); Rg 3.5DMPOMT, Ruta graveolens 3,5-dimethoxyphenol OMT (AAX82431)*; Ob CVOMT, Ocimum basilicum chavicol OMT (AAL30423)*; Cr F4′OMT, Catharanthus roseus flavonoid 4′-OMT (AAR02420)*; Ge I4′OMT, Glycyrrhiza echinata 2,7,4′-trihydroxyisoflavanone 4′-OMT (BAC58011); Ms I7OMT, Medicago sativa isoflavone OMT (AAC49928)*; Cj NC6OMT, Coptis japonica (R,S)-norcoclaurine 6-OMT (Q9LEL6)* ; Cj MC4′OMT, Coptis japonica 3′-hydroxy-N-methylcoclaurine 4′-OMT (Q9LEL5)* ; Ge CH2′OMT, Glycyrrhiza echinata chalcone 2′-OMT (BAA13683)* ; Ms IL2′OMT, Medicago sativa isoliquiritigenin 2′-OMT (AAB48059)*; Mc I4OMT, Mesembryanthemum crystallinum inositol 4-OMT (P45986)*; Zm OMT, Zea mays OMT (P47917); So CA3OMT, Saccharum officinarum caffeic acid 3-OMT (O82054)*; Ze CA3OMT, Zinnia elegans caffeic acid 3-OMT (Q43239)*; Cr CA3OMT, Catharanthus roseus caffeic acid 3-OMT (Q8W013)*; Ca CAOMT, Chrysosplenium americanum caffeic acid OMT (AAA86982)*; At F3′OMT, Arabidopsis thaliana flavonol 3′-OMT (AAB96879)*; Hl OMT7, Humulus lupulus OMT7; Ms CAOMT, Medicago sativa caffeic acid OMT (P28002)*; Rc CAOMT, Rosa chinensis caffeic acid OMT (CAD29457)*; Cb CAOMT, Clarkia breweri caffeic acid OMT (AAB71141)*; Eg CA3OMT, Eucalyptus gunni caffeic acid 3-OMT (P46484)*. Asterisks indicate OMTs with known catalytic activity.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Xanthohumol Levels in Leaves with Intact Glandular Trichomes and with Trichomes Removed.
Supplemental Table 1. Summary of Hop Lupulin Gland ESTs.
Supplemental Table 2. 1H-NMR Spectra of Xanthohumol and 4-O-Methylxanthohumol (δ in ppm, J in Hz).
Supplemental Data Set 1. Amino Acid Alignment of Plant O-Methyltransferases Used to Generate Figure 4.
Supplementary Material
Acknowledgments
We thank Feng Wang for cDNA library construction, the National Research Council–Plant Biology Institute DNA Service Unit for DNA sequencing, Jacek Nowak and Kannan Vijayan for bioinformatics, and Richard Hughes and Greg Bishop for LC-MS analysis. Jitao Zou and Joan Krochko commented on an earlier draft of the manuscript. We thank Toni Kutchan and the staff of the Leibniz Institute for Plant Biochemistry for their support during the initial stage of this project. We also thank Martin Biendl and Timo Lambertsen (Mainburg) and Hopfenhof Regner (Edderitz) for facilitating collection of hop samples. Funding was provided by S.S. Steiner and the National Research Council of Canada. This is manuscript NRCC# 48443.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jonathan E. Page (jon.page@nrc-cnrc.gc.ca).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Audic, S., and Claverie, J.M. (1997). The significance of digital gene expression profiles. Genome Res. 7 986–995. [DOI] [PubMed] [Google Scholar]
- Biendl, M. (2003 2002). Research on the xanthohumol content of hops. Hopfenrundschau International 2002/2003 72–75. [Google Scholar]
- Bonaldo, M.F., Lennon, G., and Soares, M.B. (1996). Normalization and subtraction: Two approaches to facilitate gene discovery. Genome Res. 6 791–806. [DOI] [PubMed] [Google Scholar]
- Burga, L., Wellmann, F., Lukacin, R., Witte, S., Schwab, W., Schroder, J., and Matern, U. (2005). Unusual pseudosubstrate specificity of a novel 3,5-dimethoxyphenol O-methyltransferase cloned from Ruta graveolens L. Arch. Biochem. Biophys. 440 54–64. [DOI] [PubMed] [Google Scholar]
- Burke, C.C., Wildung, M.R., and Croteau, R. (1999). Geranyl diphosphate synthase: Cloning, expression, and characterization of this prenyltransferase as a heterodimer. Proc. Natl. Acad. Sci. USA 96 13062–13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiron, H., Drouet, A., Claudot, A.C., Eckerskorn, C., Trost, M., Heller, W., Ernst, D., and Sandermann, H., Jr. (2000). Molecular cloning and functional expression of a stress-induced multifunctional O-methyltransferase with pinosylvin methyltransferase activity from Scots pine (Pinus sylvestris L.). Plant Mol. Biol. 44 733–745. [DOI] [PubMed] [Google Scholar]
- Chou, H.H., and Holmes, M.H. (2001). DNA sequence quality trimming and vector removal. Bioinformatics 17 1093–1104. [DOI] [PubMed] [Google Scholar]
- Colgate, E.C., Miranda, C.L., Stevens, J.F., Bray, T.M., and Ho, E. (2006). Xanthohumol, a prenylflavonoid derived from hops induces apoptosis and inhibits NF-kappaB activation in prostate epithelial cells. Cancer Lett. 246 201–209. [DOI] [PubMed] [Google Scholar]
- D'Auria, J.C., Chen, F., and Pichersky, E. (2003). The SABATH family of methyltransferases in Arabidopsis thaliana and other plant species. Rec. Adv. Phytochem. 37 253–283. [Google Scholar]
- Dudareva, N., Andersson, S., Orlova, I., Gatto, N., Reichelt, M., Rhodes, D., Boland, W., and Gershenzon, J. (2005). The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 102 933–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, R., and Dixon, R.A. (1991). Isoflavone O-methyltransferase activities in elicitor-treated cell suspension cultures of Medicago sativa. Phytochemistry 30 2597–2606. [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8 175–185. [DOI] [PubMed] [Google Scholar]
- Ferrer, J.L., Zubieta, C., Dixon, R.A., and Noel, J.P. (2005). Crystal structures of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol. 137 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, E., Wang, J., Iijima, Y., Froehlich, J.E., Gang, D.R., Ohlrogge, J., and Pichersky, E. (2005). Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17 1252–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, N., Inui, T., Iwasa, K., Morishige, T., and Sato, F. (2007). Knockdown of berberine bridge enzyme by RNAi accumulates (S)-reticuline and activates a silent pathway in cultured California poppy cells. Transgenic Res. 16 363–375. [DOI] [PubMed] [Google Scholar]
- Gang, D.R., Wang, J., Dudareva, N., Nam, K.H., Simon, J.E., Lewinsohn, E., and Pichersky, E. (2001). An investigation of the storage and biosynthesis of phenylpropenes in sweet basil. Plant Physiol. 125 539–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier, A., Gulick, P.J., and Ibrahim, R.K. (1998). Characterization of two cDNA clones which encode O-methyltransferases for the methylation of both flavonoid and phenylpropanoid compounds. Arch. Biochem. Biophys. 351 243–249. [DOI] [PubMed] [Google Scholar]
- Gensheimer, M., and Mushegian, A. (2004). Chalcone isomerase family and fold: No longer unique to plants. Protein Sci. 13 540–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goese, M., Kammhuber, K., Bacher, A., Zenk, M.H., and Eisenreich, W. (1999). Biosynthesis of bitter acids in hops. A (13)C-NMR and (2)H-NMR study on the building blocks of humulone. Eur. J. Biochem. 263 447–454. [DOI] [PubMed] [Google Scholar]
- Goto, K., Asai, T., Hara, S., Namatame, I., Tomoda, H., Ikemoto, M., and Oku, N. (2005). Enhanced antitumor activity of xanthohumol, a diacylglycerol acyltransferase inhibitor, under hypoxia. Cancer Lett. 219 215–222. [DOI] [PubMed] [Google Scholar]
- Hornsey, I.S. (2003). A History of Beer and Brewing. (Cambridge, UK: Royal Society of Chemistry).
- Huang, X., and Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerkovic, V., and Collin, S. (2007). Occurrence of resveratrol and piceid in American and European hop cones. J. Agric. Food Chem. 55 8754–8758. [DOI] [PubMed] [Google Scholar]
- Lange, B.M., Wildung, M.R., Stauber, E.J., Sanchez, C., Pouchnik, D., and Croteau, R. (2000). Probing essential oil biosynthesis and secretion by functional evaluation of expressed sequence tags from mint glandular trichomes. Proc. Natl. Acad. Sci. USA 97 2934–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavid, N., et al. (2002). O-methyltransferases involved in the biosynthesis of volatile phenolic derivatives in rose petals. Plant Physiol. 129 1899–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Popko, J.L., Zhang, X.H., Osakabe, K., Tsai, C.J., Joshi, C.P., and Chiang, V.L. (1997). A novel multifunctional O-methyltransferase implicated in a dual methylation pathway associated with lignin biosynthesis in loblolly pine. Proc. Natl. Acad. Sci. USA 94 5461–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G., Eggler, A.L., Dietz, B.M., Mesecar, A.D., Bolton, J.L., Pezzuto, J.M., and van Breemen, R.B. (2005). Screening method for the discovery of potential cancer chemoprevention agents based on mass spectrometric detection of alkylated Keap1. Anal. Chem. 77 6407–6414. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Matousek, J., Novak, P., Briza, J., Patzak, J., and Niedermeierova, H. (2002). Cloning and characterisation of chs-specific DNA and cDNA sequences from hop (Humulus lupulus L.). Plant Sci. 162 1007–1018. [Google Scholar]
- Maxwell, C.A., Edwards, R., and Dixon, R.A. (1992). Identification, purification, and characterization of S-adenosyl-L-methionine:isoliquiritigenin 2′-O-methyltransferase from alfalfa (Medicago sativa L.). Arch. Biochem. Biophys. 293 158–166. [DOI] [PubMed] [Google Scholar]
- Maxwell, C.A., Harrison, M.J., and Dixon, R.A. (1993). Molecular characterization and expression of alfalfa isoliquiritigenin 2′-O-methyltransferase, an enzyme specifically involved in the biosynthesis of an inducer of Rhizobium meliloti nodulation genes. Plant J. 4 971–981. [DOI] [PubMed] [Google Scholar]
- Miranda, C.L., Aponso, G.L., Stevens, J.F., Deinzer, M.L., and Buhler, D.R. (2000. a). Prenylated chalcones and flavanones as inducers of quinone reductase in mouse Hepa 1c1c7 cells. Cancer Lett. 149 21–29. [DOI] [PubMed] [Google Scholar]
- Miranda, C.L., Stevens, J.F., Ivanov, V., McCall, M., Frei, B., Deinzer, M.L., and Buhler, D.R. (2000. b). Antioxidant and prooxidant actions of prenylated and nonprenylated chalcones and flavanones in vitro. J. Agric. Food Chem. 48 3876–3884. [DOI] [PubMed] [Google Scholar]
- Nickerson, G.B., Williams, P.A., and Haunold, A. (1988). Composition of male hop oil. J. Am. Soc. Brew. Chem. 46 14–17. [Google Scholar]
- Noel, J.P., Dixon, R.A., Pichersky, E., Zubieta, C., and Ferrer, J.-L. (2003). Structural, functional, and evolutionary basis for methylation of plant small molecules. Rec. Adv. Phytochem. 37 37–58. [Google Scholar]
- Nookandeh, A., Frank, N., Steiner, F., Ellinger, R., Schneider, B., Gerhauser, C., and Becker, H. (2004). Xanthohumol metabolites in faeces of rats. Phytochemistry 65 561–570. [DOI] [PubMed] [Google Scholar]
- Novak, P., Matousek, J., and Briza, J. (2003). Valerophenone synthase-like chalcone synthase homologues in Humulus lupulus. Biol. Plant. 46 375–381. [Google Scholar]
- Okada, Y., Sano, Y., Kaneko, T., Abe, I., Noguchi, H., and Ito, K. (2004). Enzymatic reactions by five chalcone synthase homologs from hop (Humulus lupulus L.). Biosci. Biotechnol. Biochem. 68 1142–1145. [DOI] [PubMed] [Google Scholar]
- Oliveira, M.M., and Pais, M.S. (1990). Glandular trichomes of Humulus lupulus var. Brewer's gold (hops): Ultrastructural aspects of peltate trichomes. J. Submicrosc. Cytol. Pathol. 22 241–248. [Google Scholar]
- Ounaroon, A., Decker, G., Schmidt, J., Lottspeich, F., and Kutchan, T.M. (2003). (R,S)-reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum - cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J. 36 808–819. [DOI] [PubMed] [Google Scholar]
- Page, J.E., and Nagel, J. (2006). Biosynthesis of terpenophenolics in hop and cannabis. In Integrative Plant Biochemistry, J.T. Romeo, ed (Oxford, UK: Elsevier), pp. 179–210.
- Paniego, N.B., Zuurbier, K.W., Fung, S.Y., van der Heijden, R., Scheffer, J.J., and Verpoorte, R. (1999). Phlorisovalerophenone synthase, a novel polyketide synthase from hop (Humulus lupulus L.) cones. Eur. J. Biochem. 262 612–616. [DOI] [PubMed] [Google Scholar]
- Park, S., Sugimoto, N., Larson, M.D., Beaudry, R., and van Nocker, S. (2006). Identification of genes with potential roles in apple fruit development and biochemistry through large-scale statistical analysis of expressed sequence tags. Plant Physiol. 141 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, G., Huang, X., Liang, F., Antonescu, V., Sultana, R., Karamycheva, S., Lee, Y., White, J., Cheung, F., Parvizi, B., Tsai, J., and Quackenbush, J. (2003). TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 19 651–652. [DOI] [PubMed] [Google Scholar]
- Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., Parvizi, B., Pertea, G., Sultana, R., and White, J. (2001). The TIGR Gene Indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralston, L., Subramanian, S., Matsuno, M., and Yu, O. (2005). Partial reconstruction of flavonoid and isoflavonoid biosynthesis in yeast using soybean type I and type II chalcone isomerases. Plant Physiol. 137 1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roje, S. (2006). S-adenosyl-L-methionine: Beyond the universal methyl group donor. Phytochemistry 67 1686–1698. [DOI] [PubMed] [Google Scholar]
- Sägesser, M., and Deinzer, M. (1996). HPLC-ion spray-tandem mass spectrometry of flavonol glycosides in hops. J. Am. Soc. Brew. Chem. 54 129–134. [Google Scholar]
- Schroder, G., Wehinger, E., Lukacin, R., Wellmann, F., Seefelder, W., Schwab, W., and Schroder, J. (2004). Flavonoid methylation: A novel 4′-O-methyltransferase from Catharanthus roseus, and evidence that partially methylated flavanones are substrates of four different flavonoid dioxygenases. Phytochemistry 65 1085–1094. [DOI] [PubMed] [Google Scholar]
- Schwekendiek, A., Spring, O., Heyerick, A., Pickel, B., Pitsch, N.T., Peschke, F., de Keukeleire, D., and Weber, G. (2007). Constitutive expression of a grapevine stilbene synthase gene in transgenic hop (Humulus lupulus L.) yields resveratrol and its derivatives in substantial quantities. J. Agric. Food Chem. 55 7002–7009. [DOI] [PubMed] [Google Scholar]
- Skidmore, A.F., and Beebee, T.J. (1989). Characterization and use of the potent ribonuclease inhibitor aurintricarboxylic acid for the isolation of RNA from animal tissues. Biochem. J. 263 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, J.F., Ivancic, M., Hsu, V., and Deinzer, M.L. (1997). Prenylflavonoids from Humulus lupulus. Phytochemistry 44 1575–1585. [DOI] [PubMed] [Google Scholar]
- Stevens, J.F., and Page, J.E. (2004). Xanthohumol and related prenylflavonoids from hops and beer: To your good health! Phytochemistry 65 1317–1330. [DOI] [PubMed] [Google Scholar]
- Stevens, J.F., Taylor, A.W., Nickerson, G.B., Ivancic, M., Henning, J., Haunold, A., and Deinzer, M.L. (2000). Prenylflavonoid variation in Humulus lupulus: Distribution and taxonomic significance of xanthogalenol and 4′-O-methylxanthohumol. Phytochemistry 53 759–775. [DOI] [PubMed] [Google Scholar]
- Taylor, A.W., Barofsky, E., Kennedy, J.A., and Deinzer, M.L. (2003). Hop (Humulus lupulus L.) proanthocyanidins characterized by mass spectrometry, acid catalysis, and gel permeation chromatography. J. Agric. Food Chem. 51 4101–4110. [DOI] [PubMed] [Google Scholar]
- Teoh, K.H., Polichuk, D.R., Reed, D.W., Nowak, G., and Covello, P.S. (2006). Artemisia annua L. (Asteraceae) trichome-specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett. 580 1411–1416. [DOI] [PubMed] [Google Scholar]
- Tholl, D., Kish, C.M., Orlova, I., Sherman, D., Gershenzon, J., Pichersky, E., and Dudareva, N. (2004). Formation of monoterpenes in Antirrhinum majus and Clarkia breweri flowers involves heterodimeric geranyl diphosphate synthases. Plant Cell 16 977–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer, Y., Van den Broeck, I., De Rijk, P., and De Wachter, R. (1994). Database on the structure of small ribosomal subunit RNA. Nucleic Acids Res. 22 3488–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willits, M.G., Giovanni, M., Prata, R.T., Kramer, C.M., De Luca, V., Steffens, J.C., and Graser, G. (2004). Bio-fermentation of modified flavonoids: An example of in vivo diversification of secondary metabolites. Phytochemistry 65 31–41. [DOI] [PubMed] [Google Scholar]
- Winkel, B.S. (2004). Metabolic channeling in plants. Annu. Rev. Plant Biol. 55 85–107. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (1999). Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 107 142–149. [Google Scholar]
- Zubieta, C., He, X.Z., Dixon, R.A., and Noel, J.P. (2001). Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 8 271–279. [DOI] [PubMed] [Google Scholar]
- Zubieta, C., Kota, P., Ferrer, J.L., Dixon, R.A., and Noel, J.P. (2002). Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 14 1265–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta, C., Ross, J.R., Koscheski, P., Yang, Y., Pichersky, E., and Noel, J.P. (2003). Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15 1704–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.