SUMMARY
Keap1/Nrf2 signaling defends organisms against the detrimental effects of oxidative stress, and has been suggested to abate its consequences, including aging-associated diseases like neurodegeneration, chronic inflammation, and cancer. Nrf2 is a prominent target for drug discovery, and Nrf2-activating agents are in clinical trials for cancer chemoprevention. However, aberrant activation of Nrf2 by keap1 somatic mutations may contribute to carcinogenesis and promote resistance to chemotherapy. To evaluate potential functions of Keap1 and Nrf2 for organismal homeostasis, we characterized the pathway in Drosophila. We demonstrate that Keap1/Nrf2 signaling in the fruitfly is activated by oxidants, induces antioxidant and detoxification responses, and confers increased tolerance to oxidative stress. Importantly, keap1 loss-of-function mutations extend the lifespan of Drosophila males, supporting a role for Nrf2 signaling in the regulation of longevity. Interestingly, cancer chemopreventive drugs potently stimulate Drosophila Nrf2 activity, suggesting the fruitfly as an experimental system to identify and characterize such agents.
INTRODUCTION
Nrf2 (NF-E2-Related Factor 2) is a transcription factor of the leucine zipper family, and Keap1 (Kelch-like ECH-Associated Protein 1) is its specific repressor. Keap1/Nrf2 signaling mediates cellular responses to oxidative stresses and electrophilic xenobiotics (Motohashi and Yamamoto, 2004). Studies in keap1 and nrf2 knockout mice have demonstrated the crucial role of the Keap1/Nrf2 module as a multi-organ protector in vivo. Nrf2 signaling defends the organism against the sequelae of oxidative stress, including aging-related diseases like neurodegeneration, chronic inflammation, and cancer (Lee et al., 2005; Motohashi and Yamamoto, 2004). Thus, Nrf2 is a prominent target in the discovery of preventive and therapeutic modalities for such diseases. Importantly, compounds that activate Nrf2 have been shown to prevent cancer in animal models, and they are currently evaluated in human clinical trials for cancer chemoprevention (the prevention of cancer in high risk individuals through the use of non-toxic natural or synthetic agents) (Sporn and Liby, 2005; Yu and Kensler, 2005; Zhang et al., 1997). On the other hand, the recent discovery of keap1 somatic mutations in cancer cell lines and human cancer samples suggests that the aberrant activation of Nrf2 signaling may also contribute to carcinogenesis and promote resistance to chemotherapy (Padmanabhan et al., 2006; Singh et al., 2006). Thus, a better understanding of the Keap1/Nrf2 pathway and of its roles in health and disease is urgently needed.
In unstressed conditions Nrf2 is tethered to its cytoplasmic inhibitor Keap1, an actin-binding protein (Figure 1A). Keap1 suppresses the activity of Nrf2 by sequestering it in the cytoplasm, and also by targeting it for proteasomal degradation. In addition to serving as an inhibitor, Keap1 can function as a sensor of oxidants and electrophiles, which react with its redox-sensitive cysteine residues (Zhang, 2006). Oxidative stresses or electrophilic xenobiotics abolish the inhibition of Nrf2 by Keap1 (Itoh et al., 2004). Nrf2 is then stabilized and accumulates in the nucleus, where it binds to the Antioxidant Response Element (ARE) in the enhancers of its target genes (Jaiswal, 2004). Experiments in vertebrate systems indicate that Nrf2 activates transcription as a dimer with a small Maf (Musculo-Aponeurotic Fibrosarcoma) protein (Itoh et al., 1997). Binding of the small Maf/Nrf2 dimer to ARE sequences results in the coordinated transcriptional up-regulation of a battery of antioxidant enzymes and detoxifying proteins. This regulated adaptive response has been elegantly termed “the electrophile counter-attack” (Prestera et al., 1993); it includes thioredoxins and glutathione-synthesizing enzymes (which maintain the redox balance), glutathione S-transferases (which detoxify xenobiotics), molecular chaperones, and proteasome subunits (which remove damaged macromolecules). In addition, the basal Nrf2 activity that is present under non-stressed conditions maintains the housekeeping expression of the same antioxidant and detoxification genes (Lee et al., 2005; Motohashi and Yamamoto, 2004). Thus, Keap1/Nrf2 signaling regulates the basal and inducible activity of a cell defense network.
Figure 1. Nrf2 and Keap1 homologues are conserved in Drosophila.
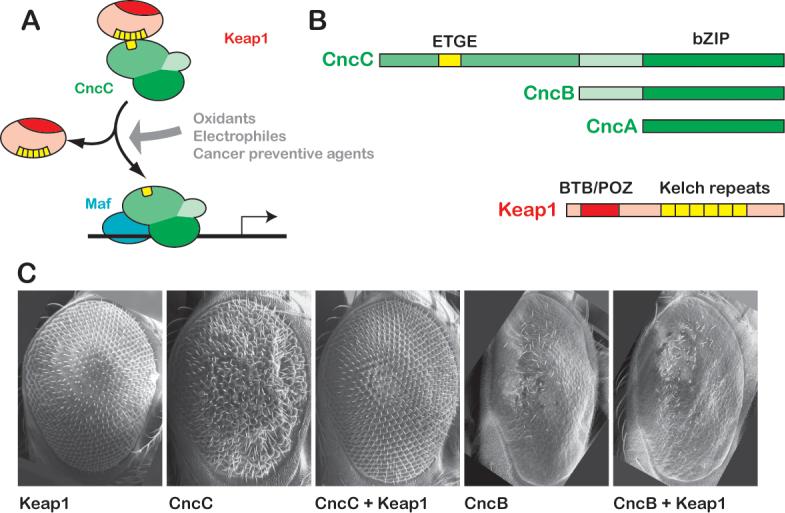
A. The Keap1/Nrf2 signaling pathway. In basal conditions, Keap1 binds to Nrf2 and inhibits its activity. Oxidative stressors, electrophilic xenobiotics, and cancer chemopreventive agents releave this inhibition. Stabilized Nfr2 then accumulates in the nucleus and dimerizes with a small Maf protein to transcriptionally activate a battery of cell protective genes. B. Nrf2 and Keap1 homologues are present in Drosophila. The cnc locus of Drosophila encodes three protein products. All contain the bZIP region that mediates dimerization and DNA binding. Only the longest isoform, CncC, contains domains predicted to bind Keap1, including the ETGE motif, and is thus potentially a Nrf2 homologue. The Drosophila Keap1 protein shows a high degree of sequence similarity to its vertebrate Keap1 counterparts (sequence alignments for CncC and Keap1 proteins are shown in Supplemental Fig 1). Conserved domains include the BTB/POZ domain required for dimerization and 6 Kelch repeats for binding to Nrf2 and anchoring to actin. C. Over-expression of Drosophila Keap1 can inhibit CncC activity in vivo. Expression of CncC in the developing Drosophila eye from a UAS transgene under the control of sepGal4 causes a reproducible aberrant phenotype. Over-expression of Keap1 under the same conditions (or with GMRGal4, data not shown) has no phenotypic effect, but it can completely suppress the effects of CncC over-expression. The activity of the shorter CncB isoform, which lacks the putative Keap1-interacting domain, is not inhibited by Keap1 co-expression. The GMRGal4 driver was used to express CncB (and Keap1) in the rightmost two panels of C., because expression by sepGal4 did not produce a phenotype at 25°C.
The oxidative stress theory of aging predicts that bolstering the organism's antioxidant defenses may retard the aging process and extend lifespan (Harman, 1956). This hypothesis has received considerable support in recent years, mainly by studies in model organisms showing that the over-expression of antioxidant genes can augment oxidative stress tolerance and promote longevity (Dugan and Quick, 2005; Orr et al., 2005). By extension, signaling pathways that regulate antioxidant responses are plausible regulators of the aging process (Wang et al., 2003). On the other hand, some of the model organism studies showing lifespan extension by “longevity genes” have been performed using short-lived control strains; this suggests that the observed lifespan extensions may represent a rescue of pathology rather than a genuine effect on lifespan (Sohal et al., 2002). Thus, the oxidative stress theory of aging warrants further experimental tests in model organisms, by using healthy, long-lived control strains, and by exploring the roles of additional antioxidant response pathways.
Model organisms could be employed to test the hypothesis that Nrf2 signaling may regulate lifespan. Keap1, Nrf2, and small Maf homologues are well conserved in Danio rerio, and studies in zebrafish have provided important insights into Keap1/Nrf2 signaling (Kobayashi et al., 2002; Takagi et al., 2004); however, zebrafish are not yet broadly developed as a platform for aging studies. C. elegans, a well-established model for lifespan experiments, possesses an Nrf2-related protein (SKN-1) which confers resistance against certain pro-oxidant xenobiotics. However, the biochemical mechanisms underlying its regulation and function are somewhat different from those of Nrf2 in vertebrates: most notably, the worm does not possess Keap1 and small Maf homologues. SKN-1 is apparently regulated mainly by phosphorylation, and binds DNA as a monomer (An and Blackwell, 2003; Blackwell et al., 1994; Inoue et al., 2005). Recently, this distant relative of mammalian Nrf2 was found to be required for lifespan extension by caloric restriction in the worm (Bishop and Guarente, 2007). Whether increasing SKN-1 activity extends lifespan in non-restricted conditions was not addressed; also, C. elegans cannot be used to test the role of Keap1 in longevity, because it lacks a keap1 homologue.
Although Drosophila melanogaster is a well-established model for aging research, Keap1/Nrf2 signaling has to our knowledge not yet been characterized in this organism. This is surprising, given the fact that Nrf2 belongs to the cap'n'collar (cnc) subfamily of leucine zippers, named after the cnc gene of Drosophila, and that Keap1 is a member of the Kelch family of actin-binding proteins, also named after the fruitfly's Kelch protein (a component of the egg chambers). We decided to employ Drosophila as a model system to investigate the role of Keap1/Nrf2 signaling in organismal responses to oxidative stress and the regulation of lifespan. We demonstrate that the fruitfly possesses functional homologues of Keap1 and Nrf2. Our data show that the Drosophila Keap1 and Nrf2 proteins comprise a cell protective module that responds to oxidants and cancer chemopreventive agents, induces antioxidant and detoxification responses, confers increased tolerance to oxidative stress, and regulates longevity.
RESULTS
Nrf2 and Keap1 homologues are conserved in Drosophila
Three RNA isoforms are transcribed off the Drosophila cnc locus, designated cncA, cncB, and cncC (McGinnis et al., 1998). These isoforms encode three different proteins, which share their C-terminal regions and thus comprise the same DNA-binding domain. However, the three Cnc translation products differ at their N-termini: CncC encompasses CncB, which in turn encompasses CncA (Figure 1B). The CncB isoform is required together with a small Maf subunit for the development of embryonic head structures (Veraksa et al., 2000). The unique N-terminus of CncC predicts a different role for this isoform: it shows similarity to the Neh2 domain of Nrf2, which contains the Keap1-binding ETGE motif and an upstream hydrophobic region (Supplemental Figure 1A). Based on this distinctive homology, it has been suggested that CncC might be the Drosophila counterpart of Nrf2 (Kobayashi et al., 2002). As a first step to addressing this idea and the potential role of CncC in antioxidant responses, we examined its expression in Drosophila. In larvae, cncC mRNA is most abundantly expressed in the alimentary canal, and shows a strong, distinctive staining pattern (Supplemental Figure 2A-D). This is reminiscent of the broad expression of mammalian Nrf2 in the digestive tract (Chan et al., 1996; Wakabayashi et al., 2003), which, like the skin and airways, is a major frontier where the organism comes into direct contact with its environment. cncC mRNA expression was also detected in the Malpighian tubules (which are detoxification organs) and in the salivary glands (Supplemental Figure 2C, E); but not in imaginal discs or in the fat body. cncC mRNA is also present in adult female and male flies as determined by RT-PCR (not shown), which is consistent with a role in the homeostasis of the mature organism.
The Drosophila genome also harbors a likely homologue of vertebrate keap1 genes. This gene, CG3962, and its product have not yet been functionally characterized. Multiple sequence alignments show that the predicted Drosophila Keap1 (dKeap1) protein has striking similarity with its mammalian and zebrafish homologues (Supplemental Figure 1B). Similar to cncC, dkeap1 mRNA is expressed in the alimentary canal and in the Malpighian tubules of third instar larvae and adult flies, and it shows a similar distribution as cncC but with a weaker, more diffuse staining pattern. keap1 mRNA is also detected in the salivary glands, the brain, and the ring gland, but not in imaginal discs or in the fat body (Supplemental Figure 2F-I). These findings demonstrate that the candidate Drosophila homologues of Nrf2 and Keap1 are expressed in the fly's digestive tract, which represents the first line of defense to ingested environmental stressors, and in the Malpigghian tubules, which are major sites of detoxification. Expression of keap1 and cncC in the adult gut and Malpigghian tubules was verified by RT-PCR on dissected adult tissues (not shown).
Consistent with the conservation of the Keap1-binding motif in CncC (Supplemental Figure 1A), the physical association between Drosophila Keap1 and CncC has previously been suggested by a genome-wide yeast two-hybrid experiment (Giot et al., 2003). This interaction predicts that Keap1 should act as a negative regulator of CncC in vivo. To test this hypothesis, we examined whether these proteins interact genetically when over-expressed in the Drosophila eye (Figure 1C). The compound eye of Drosophila comprises about 800 ommatidia or facets arranged in a regular hexagonal pattern. Genetic manipulation of cell signaling during eye development often causes distinctive, but generally non-lethal phenotypes, making the eye a convenient system for the analysis of regulatory interaction between different genes. Over-expression of Keap1 in the eye using the eye-specific sepGal4 driver does not distort the wild-type pattern (Figure 1C). In contrast, over-expression of CncC with the same driver disrupts the ommatidial arrangement, resulting in a rough eye appearance. Co-expression of Keap1 completely suppresses this CncC gain-of-function (g-o-f) phenotype, restoring the eye to a wild-type morphology. Although the basis of the CncC over-expression phenotype is not yet well understood, its rescue by Keap1 co-expression suggests that Drosophila Keap1 may inhibit the activity of CncC. This effect requires the CncC-specific N-terminal domain, which includes the predicted Keap1-binding region. The shorter CncB isoform lacks this N-terminal domain, and does not interact with Keap1 in this assay: over-expression of CncB in the eye with the GMRGal4 driver results in ommatidial loss and a smooth eye appearance, a phenotype that is not modified by the co-expression of Keap1. These data suggest an inhibitory role of Keap1 on CncC, which is reminiscent of the regulatory function of vertebrate Keap1 on Nrf2.
CncC regulates the stress response gene gstD1 and the keap1 gene
Nrf2 regulates the basal and inducible expression of antioxidant and detoxifying genes (Lee et al., 2005; Motohashi and Yamamoto, 2004). To test whether CncC has functional homology to Nrf2, we first examined whether CncC can also regulate the transcription of such genes. gstD1 is a prototypical oxidative stress response gene; it encodes a well known detoxification enzyme (Sawicki et al., 2003) and its transcription unit is preceded by a consensus ARE sequence (Figure 3A). mRNA in situ hybridization was used to examine whether the over-expression of CncC in embryos is sufficient to induce expression of gstD1 mRNA. CncC was over-expressed using the engrailedGal4 (enGal4) driver, which is active in a distinctive, easily recognizable striped pattern in the epidermis, and also in the dorsal part of the hindgut. Induction of gstD1 mRNA expression in response to ectopic expression of CncC was readily apparent (Figure 2A). Interestingly, we found that over-expression of CncC can also induce the expression of dkeap1 mRNA (Figure 2A).
Figure 3. An oxidative stress-responsive enhancer is regulated by Keap1/CncC.
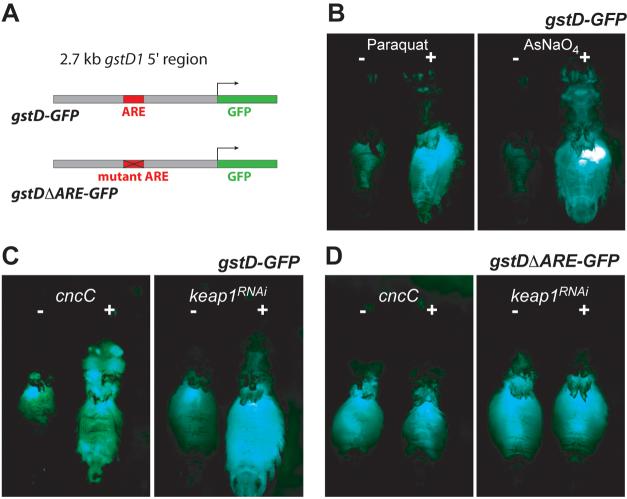
A. Structure of the gstD-GFP reporter transgene. The genomic sequence upstream of the gstD1 gene harbors an ARE sequence, and was used to control the expression of GFP in transgenic reporter flies. The gstDΔARE-GFP reporter is identical, except that the ARE consensus sequence has been disrupted by base substitutions (see methods). B. The transcriptional activity of the gstD enhancer was potently induced in the gut and in other tissues by oxidants. Animals were exposed to 20 mM Paraquat or 1 mM sodium meta-arsenite in sucrose solution, and GFP fluorescence was monitored after 16 hours. The flies' wings and legs were dissected away to facilitate handling and to expose the abdomen. C. The gstD enhancer responds to Keap1/CncC signaling. The activity of the gstD reporter can be induced by over-expression of CncC or by RNAi-mediated knock-down of Keap1. Keap1 RNAi and CncC expression was driven by the ubiquitously expressed RU486-inducible tubGSGal4 driver; thus the two animals shown in any of the panels are genetically identical and differ only by being fed RU486 or mock treated for 48 hours before analysis. RU486 feeding by itself has no effect on reporter activity (not shown). D. The transcriptional activation of the gstD enhancer by Keap1/CncC signaling is mediated by an ARE. Mutation of the ARE abolishes enhancer activation by both CncC over-expression and Keap1 knock-down.
Figure 2. CncC regulates stress response genes and the keap1 gene.
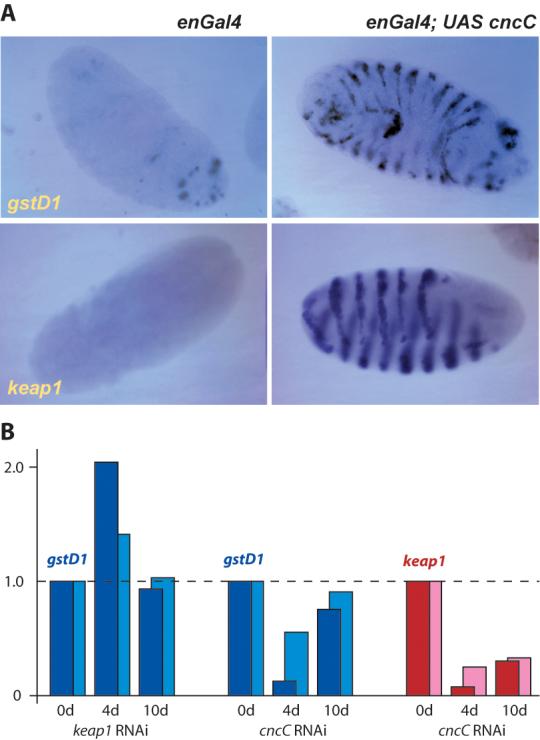
A. The over-expression of CncC in the epidermis of the embryo using the enGal4 driver induces the glutathione S-transferase D1 gene, a well known antioxidant and detoxification gene (detected by mRNA in situ hybridization). As negative controls, other members of the gstD gene family were not induced in parallel experiments (not shown). The Drosophila keap1 gene can also be up-regulated by CncC over-expression.
B. Validation of gstD1 and keap1 as CncC target genes by real-time RT-PCR. These experiments employ transgenic expression of RNAi targeting either keap1 or cncC mRNA under the control of tubGSGal4, which permits the conditional knockdown of CncC or Keap1 in adult flies. Two separate driver lines with independent transgene insertions were used; tubGS5 (dark blue and dark red bars) consistently yielded slightly stronger effects than tubGS10 (light blue and light red). The expression of UAS keap1RNAi or UAS cncCRNAi (as indicated at the bottom of the histogram) was activated by feeding adults with RU486 as detailed in the methods section. Sibling control flies from the same culture where treated with food containing an equivalent amount of ethanol solvent. The resultant changes in the mRNA levels of gstD1 and keap1 were examined by real time RT-PCR after 4 and 10 days of conditional knockdown.
To confirm the identification of gstD1 and keap1 as CncC target genes, and to derive quantitative and temporally resolved information about their regulation, we performed real-time RT-PCR experiments in CncC and Keap1 loss-of-function (l-o-f) conditions (Figure 2B). CncC and Keap1 were conditionally knocked-down in adult flies by RNAi (by expressing transgenes with inverted repeats corresponding to their respective coding segments, UAS cncCRNAi or UAS keap1RNAi). These genetic manipulations employed the conditional driver tubulin-GeneSwitch-Gal4 (tubGSGal4), which is ubiquitously expressed, but is active only upon the dietary administration of RU486 (Scott Pletcher, personal communication). The efficiency of knockdown was validated by real-time RT-PCR (Supplemental Figure 3). The mRNA levels of gstD1 and keap1 were examined by real-time RT-PCR after 4 and 10 days of RU486 administration (Figure 2B). gstD1 behaved as a bona fide Keap1/CncC target gene: its levels were reduced by CncC knockdown and increased by Keap1 knockdown after 4 days of treatment. gstD1 levels tended to return to baseline after 10 days. Consistent with these data, gstD1 levels were also increased in flies heterozygous for either of two keap1 mutations (Figure 6B). Importantly, keap1 levels were dramatically reduced by CncC knockdown; taken together with the inhibitory function of Keap1 on CncC (Figure 1C), and the induction of keap1 by CncC (Figure 2A) these findings confirm the presence of an auto-regulatory loop in the pathway. These observations are consistent with the recent finding that keap1 is a target gene of Nrf2 in mammalian cell culture systems: stress releases Nrf2 from Keap1 inhibition, Nrf2 trans-activates keap1, and Keap1 promotes the degradation of Nrf2 (Lee et al., 2007). Our findings show that this auto-regulatory loop is an ancient mechanism conserved in evolution, further highlighting Keap1 as a crucial node in the pathway.
Figure 6. keap1 heterozygosity confers oxidative stress resistance and lifespan extension.
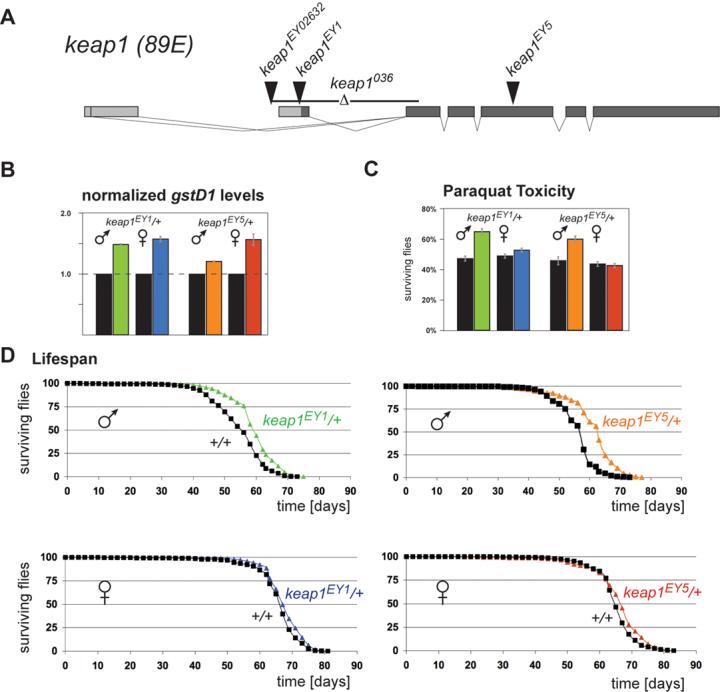
A. Structure of the Drosophila keap1 gene and nature of loss-of-function alleles. Alternative keap1 transcripts differ at their 5′ ends. Exons are depicted as boxes, with coding segments indicated by dark shading. EY02632 is a homozygous viable allele; keap1036, keap1EY1, and keap1EY5 are larval lethal alleles (see Supplemental Method). B. Elevated gstD1 expression levels in keap1 heterozygotes. mRNA levels of gstD1 in 1 day old flies were quantified separately for males and females by real-time RT-PCR. Control flies were siblings of the heterozygotes and were homozygous wild-type for keap1. Data shown are mean ± SEM of three experiments performed in duplicate. C. Partial loss of Keap1 increases Paraquat resistance. Male heterozygous keap1EY1/+ and keap1EY5/+ flies show a significantly higher survival rate 16 hours after a Paraquat challenge than their otherwise genetically identical wild-type siblings. Data are represented as mean ± SEM of four experiments performed in triplicate. Females do not display this effect. D. keap1 heterozygosity extends lifespan. Under standard culture conditions, male keap1EY1/+ and keap1EY5/+ flies live significantly longer than their sibling controls. Data are represented as the percentage of flies that are alive at each age. 500-700 flies of each genotype and gender were assayed (see methods for experimental details).
An oxidative stress-responsive enhancer is regulated by Keap1/CncC via an ARE
To address the role of Keap1/CncC in antioxidant and detoxification responses of adult flies, we constructed reporter transgenes that express GFP or β-galactosidase (LacZ) under the control of a 2.7 kb genomic sequence upstream of the gstD1 gene. In flies carrying the gstD-GFP reporter, GFP activity can be monitored in adults at basal conditions and after exposure to oxidative stressors (Figure 3). In unstressed conditions, GFP activity was detected mostly in the gut, which is reminiscent of the cncC expression pattern. Reporter activity is markedly induced when these flies are exposed to various oxidants, including Paraquat (a free radical generator), arsenic (a heavy metal), diethyl-maleate (DEM, a glutathione-depleting agent), and hydrogen peroxide, a bona fide oxidant (Figure 3B, and data not shown). These findings demonstrate that the cloned genomic sequence represents an enhancer responsive to diverse oxidative stressors, and that the gstD-GFP reporter fly lines are valuable tools for the live monitoring of antioxidant responses.
To investigate whether the genetic activation of Keap1/CncC signaling elicits a transcriptional effect that mimics gene induction after exposure to oxidants, we analyzed the transcriptional response of the gstD enhancer to the over-expression of CncC, or to the knock-down of Keap1 by RNA interference (RNAi) in adult flies. Reporter activity was induced in CncC g-o-f conditions and in Keap1 l-o-f conditions, supporting a role of these proteins as regulators of antioxidant gene expression (Figure 3C). Consistent results were obtained in larvae: the gstD-GFP reporter could be induced in the epidermis and dorsal hindgut by knocking down keap1, and the gstD-lacZ reporter could be induced in brains with keap1 mutant clones (Supplemental Figure 4). The activation of reporter activity upon suppression of keap1 expression in several tissues of larvae and adults suggests that Nrf2 function is latently present in a broad range of Drosophila cell types and organs.
The finding that Keap1/CncC signaling activates the gstD-GFP transgene indicates that reporter activity is regulated by one or more functional ARE sequences in the cloned enhancer. Indeed, the gstD enhancer harbors a 9 bp sequence (TGACcggGC) that perfectly matches the consensus core ARE sequence (TGAYnnnGC). Moreover, this 9 bp core element lies at the center of a 20 bp sequence (TCAgcATGACcggGCAaaaa), which also conforms perfectly to the extended ARE consensus sequence (TMAnnRTGAYnnnGCRwwww; (Nioi et al., 2003). Since this cloned genomic fragment does not contain any other ARE-like sequences, we hypothesized that this element mediates trans-activation by the Keap1/CncC pathway. To test this idea, we generated transgenic reporter flies bearing a version of the enhancer with mutated ARE (gstDΔARE-GFP). In contrast to the wild-type gstD-GFP reporter, the ARE-mutated version could not be induced by either CncC over-expression or Keap1 knock-down (Figure 3D). Finally, we showed that a lacZ reporter transgene driven by a synthetic multimer ARE is also activated in CncC g-o-f or Keap1 l-o-f conditions, proving that the ARE is sufficient for transcriptional activation by Keap1/CncC (Supplemental Figure 5). These findings demonstrate that an ARE sequence mediates the trans-activation of the oxidative stress-responsive gstD enhancer by Keap1/CncC signaling, supporting the functional conservation of AREs in the Drosophila genome.
The cancer chemopreventive Nrf2 activator oltipraz regulates the gstD enhancer via CncC and the ARE
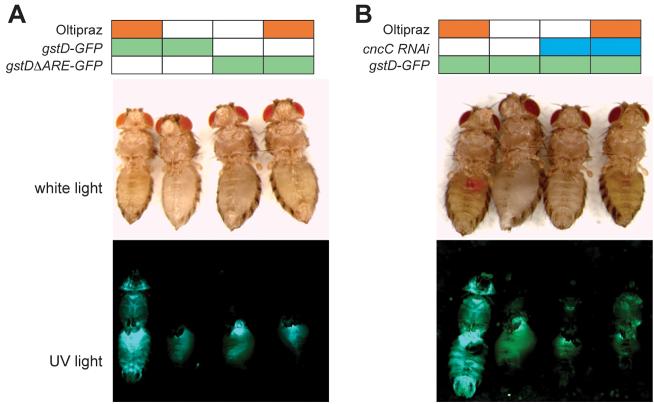
Oltipraz, a synthetic dithiolthione compound, is effective as a cancer chemopreventive agent in rodent models, and it is evaluated in human clinical trials for cancer chemoprevention (Glintborg et al., 2006; Kensler et al., 1999; Zhang et al., 1997). We tested whether dietary administration of oltipraz would activate the gstD-GFP reporter in flies, and if such induction would require CncC and the ARE. Adult flies were allowed to feed on food supplemented with oltipraz or on control food, and reporter activity was examined after 48 hours. Oltipraz-treated flies showed marked activation of the gstD-GFP reporter in the gut as well as other regions of the body (Figure 4). This induction was reduced when CncC was knocked-down, and it was abolished when the ARE was mutated. Similar results were obtained after exposing reporter flies to tBHQ (another Nrf2 inducer), and to the potent oxidant DEM (not shown). These findings demonstrate that CncC is essential for ARE-mediated transcriptional responses, and thus validate the functional homology of CncC and Nrf2. Moreover, the results prove the feasibility of in vivo pharmacological manipulation of the Nrf2 pathway in Drosophila using chemical probes like oltipraz. Thus, Drosophila could be used as a platform for the discovery or preclinical testing of novel Nrf2 activators.
Figure 4. The cancer chemopreventive agent oltipraz regulates the gstD enhancer via CncC and the ARE.
A. Activation of the gstD enhancer by oltipraz requires the ARE. Feeding oltipraz to flies activates the gstD-GFP reporter. However, mutation of the ARE completely abolishes reporter induction by oltipraz. The activity of the gstD-GFP reporter was monitored by inspection of the respective animal under UV light (bottom panel). The same animals are also shown in white light illumination. B. Activation of the gstD enhancer by oltipraz requires CncC. CncC activity was reduced by RU486-inducible knock-down of CncC for 4 days before oltipraz administration. Targeting CncC by RNAi reduced both the basal activity of the gstD enhancer, and its activation by oltipraz.
CncC regulates the organism's resistance to oxidative stress
Having established that Keap1/Nrf2 signaling elicits transcriptional antioxidant and detoxification responses in the fruitfly, we examined its importance for organismal homeostasis. If CncC initiates an “electrophile counter-attack” in Drosophila, then CncC g-o-f conditions might be expected to confer increased tolerance to oxidative stress; conversely, loss of CncC function should decrease oxidative stress tolerance. To test these hypotheses, we conditionally over-expressed or knocked-down CncC. We then compared the flies' oxidative stress tolerance to that of their sibling controls (Figure 5). Over-expression of CncC significantly increased the survival of both female and male flies after exposure to a semi-lethal dose of Paraquat (94% vs 31% for females, p<0.001; 62% vs 45% for males, p=0.003). Conversely, knocking-down CncC significantly decreased the survival of both sexes after Paraquat exposure (35% vs 65% for females, p=0.008; 53% vs 72% for males, p=0.02). These findings demonstrate that CncC is critical for the defense of Drosophila against oxidative stress, and that survival after an oxidative stress challenge can be augmented by the pre-emptive activation of CncC signaling.
Figure 5. CncC mediates the resistance of Drosophila to oxidative stress.
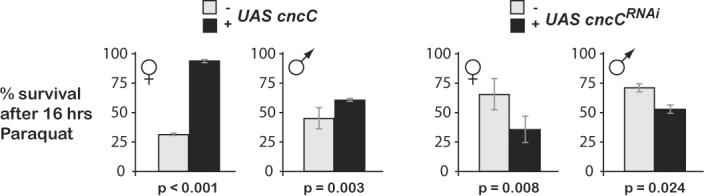
A. Over-expression of CncC before a Paraquat challenge increases the flies' survival rate. Flies bearing tubGSGal4 and UAS cncC transgenes were maintained on RU486-containing or control food for 4 days before being challenged with a semi-lethal dose of Paraquat. The survival rate of RU486-fed female and male flies 16 hours after the start of Paraquat exposure was significantly higher than that of their sibling controls, suggesting that CncC over-expression increases oxidative stress tolerance. B. RNAi-mediated knock-down of CncC before a Paraquat challenge reduces the flies' survival rate. Flies bearing tubGSGal4 and UAS cncCRNAi transgenes were maintained on RU486-containing or control food for 4 days before being challenged with Paraquat. The survival rate of RU486-fed female and male flies after 16 hours was significantly lower than that of their sibling controls, suggesting that CncC is required for normal oxidative stress tolerance. All data are represented as mean ± SEM of three experiments performed in triplicate. p-values are shown below each histogram. The flies' gender and specific genetic background can profoundly influence their tolerance to oxidative stress. Therefore, the dose of Paraquat for each experiment was adjusted to achieve (approximately) a 50% death rate in each control group after 16 hours. Moreover, only flies within a panel are directly comparable, because they are of the same gender and genetically identical, and differ only in having being fed RU486 for 4 days before the stress. RU486 feeding by itself has no effect on the Paraquat resistance of wild-type w1118 flies, or on any of the tubGSGal4, UAS cncC, and UAS cncCRNAi stocks when out-crossed to the w1118 background (not shown).
keap1 heterozygosity confers oxidative stress resistance and lifespan extension
The regulation of keap1 by CncC (Figure 2) and the inhibitory effect of Keap1 on CncC (Figure 1C) suggested a negative feedback loop in Keap1/CncC signaling, highlighting Keap1 as a crucial node of the pathway. To further investigate the role of Keap1, we used two independent mutant alleles that disrupt coding exons of keap1: keap1EY1 and keap1EY5 (shown in Figure 6A and described in detail in the Supplemental Data). Significantly, steady state levels of gstD1 mRNA are elevated in flies heterozygous for either keap1 allele (Figure 6B), suggesting that loss of one copy of the keap1 gene can result in a gain of CncC function in vivo. The oxidative stress tolerance and lifespan of keap1 heterozygotes were compared to those of their otherwise genetically identical wild-type siblings. Male keap1EY1/+ and keap1EY5/+ flies showed significantly increased survival rates after exposure to Paraquat (Figure 6C). Importantly, male keap1 heterozygotes also lived significantly longer (8-10% extension of median lifespan, p<0.0001) than their otherwise genetically identical siblings (Figure 6D and Supplemental Table 1). Female keap1 heterozygotes did not show significant differences in either Paraquat resistance or longevity. These findings demonstrate that partial l-o-f of the Nrf2 pathway's negative regulator has significant beneficial effects on the oxidative stress tolerance and longevity of male Drosophila. To our knowledge, this is the first evidence for a role of Keap1 in lifespan regulation.
DISCUSSION
Here we show that the Drosophila CncC and Keap1 proteins are genuine Nrf2 pathway components, regulate ARE-mediated transcription and detoxification gene expression, and are crucial for the organism's defense against oxidative stress. These findings should accelerate genetic approaches to the functional characterization of the Nrf2 pathway aimed at the comprehensive elucidation of its roles in health and disease. In this respect, the lifespan extension of keap1 heterozygous male flies is an example of new insight derived from Drosophila. Very recently, the Nrf2-related SKN-1 protein was found to be required for longevity induced by dietary restriction in C.elegans (Bishop and Guarente, 2007). This finding supports the concept that Cnc family members have an evolutionarily conserved function in lifespan regulation. Our results suggest that this role extends beyond calorie restriction, and establish a function for Keap1 (which is not conserved in worms) in this context. It was previously reported that the transcriptional activity of Nrf2 decreases in aging mice (Suh et al., 2004). Based on these findings, it will be very interesting to investigate the role of Keap1/Nrf2 in the aging of vertebrates.
The effect of keap1 heterozygosity on stress resistance and longevity shows a marked sexual dimorphism, with significant, albeit modest, effects discernible only in males. Although the basis for this sex difference is presently unknown, sexually dimorphic genetic effects on Drosophila lifespan are not uncommonly reported, for example in the case of chico (Clancy et al., 2001; Tu et al., 2002). Also other genes with effects on aging, such as p53 or puc, affect males and females differently (Bauer et al., 2005; Wang et al., 2005). It is important to note that the sexual dimorphism in the longevity of keap1 heterozygotes is reflected in their Paraquat sensitivity. In males, removal of one gene copy of keap1 increases Paraquat resistance, whereas females do not show this effect (Fig. 6C). While these findings imply that females react differently to keap1 heterozygosity, such a difference is not manifested at the level of gstD1 expression, which is significantly elevated in keap1 heterozygotes of both sexes (Fig. 6B). These data suggest that although keap1 heterozygosity increases stress response gene levels in both sexes, this translates into a benefit for Paraquat tolerance and longevity only in male flies. Importantly, when CncC is over-expressed Paraquat resistance is enhanced in both males and females (Fig. 5). Taken together, the data suggest that females can also benefit from increased CncC activity, but keap1 heterozygosity is not sufficient to yield stress protection and extend lifespan in females. Wild-type female flies generally live longer and are more stress resistant than males, and this effect is also seen in our study. Figure 6D and Supplemental Table 1 show that the extended lifespan of male keap1 heterozygotes is very similar to that of female flies. Given the modest, albeit significant, benefits observed in males, we propose that keap1 heterozygosity is not sufficient to increase stress tolerance and extend lifespan beyond certain biological limits.
The involvement of Nrf2 in both cancer prevention and lifespan regulation adds to the intricate links between cancer and aging. In addition to its role in cancer prevention, the antioxidant and cell protective properties of Nrf2 make it an important target for drug discovery in the prevention and treatment of oxidative stress- and aging-related diseases, including neurodegenerative disorders (van Muiswinkel and Kuiperij, 2005). Many Drosophila models of neurodegeneration are available, rendering the fruitfly an excellent system for the preclinical testing of this strategy (Bilen and Bonini, 2005). Thus, our characterization of Keap1/Nrf2 signaling in Drosophila facilitates studies that will examine the preventive and/or therapeutic effects of Nrf2-activating cancer chemopreventive agents in neurodegenerative diseases. Studies utilizing oltipraz, in particular, are especially appealing, since it is a relatively safe agent that has been used for the treatment of schistosomiasis (a parasitic disease) and is tested in human clinical trials for cancer chemoprevention (Kensler et al., 1999; Zhang et al., 1997).
EXPERIMENTAL PROCEDURES
Drosophila lines
Flies were reared at 25°C. sepGal4 and GMRGal4 flies were from Marek Mlodzik. tubGSGal4 flies express RU486-regulated Gal4 under the control of the tubulin enhancer (Scott Pletcher, personal communication); we used insertions on the second (tubGS10) and third chromosome (tubGS5). engrailedGal4 and EY02632 flies were from the Bloomington Stock Center. 4xARE-lacZ (originally named 4C (Veraksa et al., 2000)) and UAS cncB flies were from William McGinnis. To create lines expressing Keap1 and CncC, the coding segments of each gene were amplified by PCR from adult cDNA and cloned into pUAST (Brand and Perrimon, 1993). For RNAi-mediated knock-down of Keap1 and CncC, lines expressing inverted repeats corresponding to the parts of each gene's coding segment were created by PCR amplification from genomic DNA and cloning into pWIZ (Lee and Carthew, 2003). Primers and cloning details are available upon request. Transgenic flies were generated by standard procedures in a w1118 background. Independent insertions of UAS cncCRNAi or UAS keap1RNAi transgenes on the major autosomes were combined to increase knock-down efficiency. The gstD-GFP and gstD-lacZ reporter lines were constructed by amplifying a 2708 bp genomic fragment that lies between the gstD1 and gstD2 genes by PCR and cloning it into pGreen H-Pelican and pH-Pelican, respectively (Barolo et al., 2000). To create the ARE-mutant version of the reporter (gstDΔARE-GFP), the ARE sequence in the cloned gstD enhancer was mutagenized to an XhoI restriction site (TGACCG>CTCGAG). Somatic mutant clones were generated by the MARCM system (Wu and Luo, 2006) using the chromosomes eyFlp ; actGal4>>UASGFP ; FRT82B tubGal80, and FRT82B or FRT82B keap1036.
X-gal staining, mRNA in situ hybridization, and real-time RT-PCR
X-Gal staining of embryos was performed as previously described (Su et al., 1998). Since endogenous CncC is zygotically expressed from late embryonic stages onwards (McGinnis et al., 1998), the Keap1 RNAi experiments were performed in third instar larvae. Dissected larval tissues were fixed in 1% glutaraldehyde in PBS, and stained at 37°C in 10 mM sodium phosphate buffer pH 7.2, 150 mM NaCl, 1 mM MgCl2, 10 mM K4[Fe(CN)6], 10 mM K3[Fe(CN)6], and 0.1% Triton-X-100.
mRNA in situ hybridization was performed as previously described (Jasper et al., 2001). Briefly, genomic DNA fragments corresponding to parts of the coding segments of cncC and keap1 were amplified by PCR and cloned into PCRII-TOPO (Invitrogen, Carlsbad, CA). Digoxigenin-labeled cncC and keap1 -specific RNA probes were prepared by in vitro transcription with the DIG RNA labeling kit (Roche, Palo Alto, CA). The hybridization and detection procedure was performed as previously described (Jasper et al., 2001). RNA extraction from whole flies or dissected adult guts and the real-time RT-PCR procedure were performed as previously described (Wang et al., 2003).
Chemicals and fluorescent reporter assays
Paraquat, DEM, hydrogen peroxide, tBHQ, and RU486 were from Sigma-Aldrich (St. Louis, MO), sodium meta-arsenite from J.T.Baker (Phillipsburg, NJ), and oltipraz from LKT Labs (St. Paul, MN). To examine the effects of oxidative stressors on gstD enhancer activity, 1-day old homozygous gstD-GFP reporter flies were mated for one day, starved for 3 hours in vials, and then fed a solution of 5% sucrose ± Paraquat (20 mM), sodium meta-arsenite (1 mM), hydrogen peroxide (3%) or DEM (20 mM; the test and control solutions for DEM also contained 10% ethanol). To assess whether Keap1 and CncC regulate the gstD enhancer via the ARE, flies bearing both a tubGSGal4 driver and a wild-type or mutant reporter transgene were generated by genetic recombination and crossed to the appropriate UAS lines. The genetically identical progeny of such crosses were separated by gender after one day of mating, and each gender was further split into two groups that were maintained for 48 hours on either RU486-containing (320 μM) or control food (corresponding amount of ethanol) (McGuire et al., 2004). To assess the effects of oltipraz on reporter activity, 2-day old flies of the appropriate genotypes were maintained for 48 hours on food supplemented with oltipraz. To examine the role of CncC in mediating the effects of oltipraz, oltipraz administration was preceded by a 4-day period of feeding on RU486-containing food to knock-down CncC by RNAi.
Paraquat resistance assays
Resistance of adult flies to an oxidative challenge was assessed as previously described (Wang et al., 2003). To evaluate the role of CncC in Paraquat tolerance, 1 day old flies of the indicated genotypes were mated for one day, split into females and males, and then kept on RU486-containing (320 μM) or control food for 4 days (flipped on fresh food after the first 2 days). Groups of 60 flies were then starved for 3 hours in vials, and then fed a solution of 5% sucrose ± a semi-lethal dose of Paraquat (10-20 mM). Survivors were scored after 16 hours. To evaluate the oxidative stress resistance of keap1 heterozygotes, flies of the appropriate genotypes were treated in the same manner except for the RU486 feeding period. Percentages of surviving flies were compared by t-test using SPSS 9.0 (SPSS Inc., Chicago, IL). From each genotype and gender, more than 1000 flies were scored.
Lifespan measurements
Flies carrying the mini-white-marked keap1EY1 or keap1EY5 mutation were backcrossed more than 12 times into the w1118 background, and then heterozygous mutant males were crossed to y1,w1118 virgins. The lifespan (and oxidative stress tolerance) of mutant and wild-type progeny of this cross was compared as previously described (Wang et al., 2003). Briefly, female and male progeny of the cross were collected 1 day after hatching and allowed to mate in bottles for 1 day. Next, females and males were separated and transferred to vials (30 flies per vial). Flies were transferred to fresh vials every other day (daily at old ages), at which time the number of dead flies was recorded. From each genotype and gender, 500-700 flies were scored for longevity. Survival curves were analyzed by the Kaplan-Meier procedure and log-rank test.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to William McGinnis and Scott Pletcher for Drosophila stocks. We thank Mirka Uhlirova and Willis Li for comments on the manuscript, Henri Jasper for helpful discussions, and Christine Sommers for excellent technical assistance including Drosophila transformations. This work was supported by a JP Wilmot Cancer Research Fellowship (GS) and NIH grant R03 CA123591 (DB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S, Carver LA, Posakony JW. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 2000;29:726, 728, 730, 732. doi: 10.2144/00294bm10. [DOI] [PubMed] [Google Scholar]
- Bauer JH, Poon PC, Glatt-Deeley H, Abrams JM, Helfand SL. Neuronal expression of p53 dominant-negative proteins in adult Drosophila melanogaster extends life span. Curr Biol. 2005;15:2063–2068. doi: 10.1016/j.cub.2005.10.051. [DOI] [PubMed] [Google Scholar]
- Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Bowerman B, Priess JR, Weintraub H. Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science. 1994;266:621–628. doi: 10.1126/science.7939715. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci U S A. 1996;93:13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, Hafen E, Leevers SJ, Partridge L. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- Dugan LL, Quick KL. Reactive oxygen species and aging: evolving questions. Sci Aging Knowledge Environ. 2005;2005:pe20. doi: 10.1126/sageke.2005.26.pe20. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Glintborg B, Weimann A, Kensler TW, Poulsen HE. Oltipraz chemoprevention trial in Qidong, People's Republic of China: unaltered oxidative biomarkers. Free Radic Biol Med. 2006;41:1010–1014. doi: 10.1016/j.freeradbiomed.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, Blackwell TK, Matsumoto K. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- Itoh K, Tong KI, Yamamoto M. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic Biol Med. 2004;36:1208–1213. doi: 10.1016/j.freeradbiomed.2004.02.075. [DOI] [PubMed] [Google Scholar]
- Jaiswal AK. Regulation of antioxidant response element-dependent induction of detoxifying enzyme synthesis. Methods Enzymol. 2004;378:221–238. doi: 10.1016/S0076-6879(04)78018-0. [DOI] [PubMed] [Google Scholar]
- Jasper H, Benes V, Schwager C, Sauer S, Clauder-Munster S, Ansorge W, Bohmann D. The genomic response of the Drosophila embryo to JNK signaling. Dev Cell. 2001;1:579–586. doi: 10.1016/s1534-5807(01)00045-4. [DOI] [PubMed] [Google Scholar]
- Kensler TW, Groopman JD, Sutter TR, Curphey TJ, Roebuck BD. Development of cancer chemopreventive agents: oltipraz as a paradigm. Chem Res Toxicol. 1999;12:113–126. doi: 10.1021/tx980185b. [DOI] [PubMed] [Google Scholar]
- Kobayashi M, Itoh K, Suzuki T, Osanai H, Nishikawa K, Katoh Y, Takagi Y, Yamamoto M. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- Lee JM, Li J, Johnson DA, Stein TD, Kraft AD, Calkins MJ, Jakel RJ, Johnson JA. Nrf2, a multi-organ protector? Faseb J. 2005;19:1061–1066. doi: 10.1096/fj.04-2591hyp. [DOI] [PubMed] [Google Scholar]
- Lee OH, Jain AK, Papusha V, Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J Biol Chem. 2007 doi: 10.1074/jbc.M706517200. [DOI] [PubMed] [Google Scholar]
- Lee YS, Carthew RW. Making a better RNAi vector for Drosophila: use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- McGinnis N, Ragnhildstveit E, Veraksa A, McGinnis W. A cap ‘n’ collar protein isoform contains a selective Hox repressor function. Development. 1998;125:4553–4564. doi: 10.1242/dev.125.22.4553. [DOI] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem J. 2003;374:337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr WC, Radyuk SN, Prabhudesai L, Toroser D, Benes JJ, Luchak JM, Mockett RJ, Rebrin I, Hubbard JG, Sohal RS. Overexpression of glutamate-cysteine ligase extends life span in Drosophila melanogaster. J Biol Chem. 2005;280:37331–37338. doi: 10.1074/jbc.M508272200. [DOI] [PubMed] [Google Scholar]
- Padmanabhan B, Tong KI, Ohta T, Nakamura Y, Scharlock M, Ohtsuji M, Kang MI, Kobayashi A, Yokoyama S, Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Prestera T, Zhang Y, Spencer SR, Wilczak CA, Talalay P. The electrophile counterattack response: protection against neoplasia and toxicity. Adv Enzyme Regul. 1993;33:281–296. doi: 10.1016/0065-2571(93)90024-8. [DOI] [PubMed] [Google Scholar]
- Sawicki R, Singh SP, Mondal AK, Benes H, Zimniak P. Cloning, expression and biochemical characterization of one Epsilon-class (GST-3) and ten Delta-class (GST-1) glutathione S-transferases from Drosophila melanogaster, and identification of additional nine members of the Epsilon class. Biochem J. 2003;370:661–669. doi: 10.1042/BJ20021287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. Dysfunctional KEAP1-NRF2 Interaction in Non-Small-Cell Lung Cancer. PLoS Med. 2006;3 doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal RS, Mockett RJ, Orr WC. Mechanisms of aging: an appraisal of the oxidative stress hypothesis. Free Radic Biol Med. 2002;33:575–586. doi: 10.1016/s0891-5849(02)00886-9. [DOI] [PubMed] [Google Scholar]
- Sporn MB, Liby KT. Cancer chemoprevention: scientific promise, clinical uncertainty. Nat Clin Pract Oncol. 2005;2:518–525. doi: 10.1038/ncponc0319. [DOI] [PubMed] [Google Scholar]
- Su MT, Golden K, Bodmer R. X-gal staining of Drosophila embryos compatible with antibody staining or in situ hybridization. Biotechniques. 1998;24:918–920. 922. doi: 10.2144/98246bm03. [DOI] [PubMed] [Google Scholar]
- Suh JH, Shenvi SV, Dixon BM, Liu H, Jaiswal AK, Liu RM, Hagen TM. Decline in transcriptional activity of Nrf2 causes age-related loss of glutathione synthesis, which is reversible with lipoic acid. Proc Natl Acad Sci U S A. 2004;101:3381–3386. doi: 10.1073/pnas.0400282101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi Y, Kobayashi M, Li L, Suzuki T, Nishikawa K, Yamamoto M. MafT, a new member of the small Maf protein family in zebrafish. Biochem Biophys Res Commun. 2004;320:62–69. doi: 10.1016/j.bbrc.2004.05.131. [DOI] [PubMed] [Google Scholar]
- Tu MP, Epstein D, Tatar M. The demography of slow aging in male and female Drosophila mutant for the insulin-receptor substrate homologue chico. Aging Cell. 2002;1:75–80. doi: 10.1046/j.1474-9728.2002.00010.x. [DOI] [PubMed] [Google Scholar]
- van Muiswinkel FL, Kuiperij HB. The Nrf2-ARE Signalling pathway: promising drug target to combat oxidative stress in neurodegenerative disorders. Curr Drug Targets CNS Neurol Disord. 2005;4:267–281. doi: 10.2174/1568007054038238. [DOI] [PubMed] [Google Scholar]
- Veraksa A, McGinnis N, Li X, Mohler J, McGinnis W. Cap ‘n’ collar B cooperates with a small Maf subunit to specify pharyngeal development and suppress deformed homeotic function in the Drosophila head. Development. 2000;127:4023–4037. doi: 10.1242/dev.127.18.4023. [DOI] [PubMed] [Google Scholar]
- Wakabayashi N, Itoh K, Wakabayashi J, Motohashi H, Noda S, Takahashi S, Imakado S, Kotsuji T, Otsuka F, Roop DR, et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat Genet. 2003;35:238–245. doi: 10.1038/ng1248. [DOI] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK signaling confers tolerance to oxidative stress and extends lifespan in Drosophila. Dev Cell. 2003;5:811–816. doi: 10.1016/s1534-5807(03)00323-x. [DOI] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Wu JS, Luo L. A protocol for mosaic analysis with a repressible cell marker (MARCM) in Drosophila. Nat Protoc. 2006;1:2583–2589. doi: 10.1038/nprot.2006.320. [DOI] [PubMed] [Google Scholar]
- Yu X, Kensler T. Nrf2 as a target for cancer chemoprevention. Mutat Res. 2005;591:93–102. doi: 10.1016/j.mrfmmm.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Zhang BC, Zhu YR, Wang JB, Wu Y, Zhang QN, Qian GS, Kuang SY, Li YF, Fang X, Yu LY, et al. Oltipraz chemoprevention trial in Qidong, Jiangsu Province, People's Republic of China. J Cell Biochem Suppl. 1997;28-29:166–173. [PubMed] [Google Scholar]
- Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38:769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.