Abstract
An antibody generated to an α-keto amide containing hapten 1 catalyzes the cis-trans isomerization of peptidyl-prolyl amide bonds in peptides and in the protein RNase T1. The antibody-catalyzed peptide isomerization reaction showed saturation kinetics for the cis-substrate, Suc-Ala-Ala-Pro-Phe-pNA, with a kcat/Km value of 883 s−1⋅M−1; the reaction was inhibited by the hapten analog 13 (Ki = 3.0 ± 0.4 μM). Refolding of denatured RNase T1 to its native conformation also was catalyzed by the antibody, with the antibody-catalyzed folding reaction inhibitable both by the hapten 1 and hapten analog 13. These results demonstrate that antibodies can catalyze conformational changes in protein structure, a transformation involved in many cellular processes.
The rate-limiting step in the slow folding reactions of many proteins involves cis-trans isomerization about peptidyl-prolyl amide bonds (1). The assistance of peptidyl-prolyl cis-trans isomerization catalysts (rotamases) in the folding of several proteins, including type III collagen (2), mouse Ig light chain (3), RNase T1 (4), RNase A (5), staphylococcal nuclease (6), creatine kinase (7), and cytochrome c (8), has been reported along with several in vivo examples (9, 10). Rotamases also have been proposed to play important physiological roles in signal transduction. The isomerases cyclophilin (cyclosporin-binding proteins) (11) and FKBP (FK506/rapamycin-binding proteins) (12) suppress T cell activation by inhibiting calcineurin (13); the recently identified mitotic Pin1 (14) regulates mitotic progression by catalyzing sequence-specific and phosphorylation-dependent proline isomerization (15). The biological importance of peptidyl-prolyl isomerization, as well as other protein conformational changes associated with protein folding and signal transduction, have led us to develop antibody catalysts for this class of transformations. Such catalysts might provide insights into the mechanisms of these novel enzymatic reactions as well as tools for controlling protein function in vitro and in vivo.
The cis-trans isomerization of the prolyl amide bond can occur either by noncovalent stabilization of a twisted amide (16, 17) or by tetrahedral adduct formation involving an active site nucleophile (18, 19). In the case of Pin1, the participation of an active site cysteine residue during peptide bond isomerization is supported by crystallographic, pH titration, and mutagenesis studies (20). In contrast, structural and mutagenesis studies of cyclophilins and FKBP have ruled out mechanisms involving covalent catalysis (21). The crystal structures of FK506 and the FK506-FKBP complex show that the N,N-disubstituted α-keto amide group in FK506 exists in a nonplanar conformation. This observation has led to the suggestion that the α-keto amide is a surrogate for the twisted amide bond of a bound peptide substrate, consistent with a mechanism involving substrate strain (22, 23).
In principle, an antibody generated against an α-keto amide containing hapten might catalyze proline isomerization by either of the above mechanisms. The α-keto amide group of hapten 1, like that in FK506, presumably exists in an orthogonal conformation, because of allylic [A(1,3)] strain (22, 23). In addition, the α-keto amide functionality in 1 contains an electrophilic carbonyl group that might elicit an active site nucleophile in the antibody combining site capable of forming a tetrahedral adduct. The electrophilicity of the ketone carbonyl moiety in α-keto amides, acids, and esters is well precedented (24); a crystal structure of trypsin complexed with the ketoacid (4-amidinophenyl)pyruvate revealed that the active site residue Ser-195 adds into the ketone carbonyl group to form a tetrahedral adduct (25). Indeed, it has been shown recently by Yli-Kauhaluoma et al. (26) that antibodies elicited against a keto amide containing tripeptide catalyze the cis-trans prolyl isomerization of the corresponding substrate with kcat/Km values of 170–1,100 s−1⋅M−1. Here we report that antibodies elicited against hapten 1 not only catalyze prolyl isomerization in a peptide but also the proline isomerization-dependent folding of RNase T1.
MATERIALS AND METHODS
Hapten Synthesis.
(4S,5R)-N-tert-butyloxycarbonyl)-4-methyl-5-carboxy-2-oxazolidinone (4). Dimethylaminopyridine (DMAP; 4.7 g, 3.8 mmol), di-tert-butyl pyrocarbonate (3.6 g, 3 mmol), and triethylamine (3.2 ml, 2.3 mmol) were added to a solution of (4S,5R)-(−)-4-methyl-5-phenyl-2-oxazolidinone 3 (3.4 g, 19.2 mmol) in 35 ml of tetrahydrofuran (THF). The mixture was stirred at room temperature for 1 h after which time more di-tert-butyl pyrocarbonate (4.5 g, 3.8 mmol) was added. Stirring was continued for 2 h until analysis by TLC indicated complete conversion of the starting material. The solvent was evaporated, and the resulting oily residue was redissolved in methylene chloride (150 ml) and washed with 1.0 N aqueous HCl (3 × 80 ml) and brine (80 ml). The organic layer was concentrated and dried to afford 5.18 g (97%) of a white solid: Rf = 0.65 [50% (vol/vol) EtOAc in hexane]; mp 97–98°C. The Boc-protected oxazolidinone was dissolved without further purification in 40 ml of MeCN, 40 ml of CCl4, and 60 ml of water. Sodium periodate (64.16 g, 0.3 mol) was added to this solution followed by 83 mg of ruthenium (III) chloride hydrate (RuCl3⋅3H2O). The mixture was stirred vigorously at room temperature for 24 h, after which time another 2.5 equivalents of periodate (10.7 g, 0.05 mol) and 1 mol% RuCl3⋅3H2O (41.5 mg, 0.2 mmol) were added. After an additional 48 h, the solvent was evaporated, and the residue was partitioned between EtOAc (250 ml) and brine (50 ml). The organic layer was dried and concentrated to afford 6.4 g of a black oil. The crude material was applied to a silica column and eluted with 10% EtOAc in hexane. The fractions containing pure product were combined, concentrated, and dried under high vacuum overnight to afford 2.31 g (48%) of a white solid: Rf = 0.12 (EtOAc); mp 117°C (dec); 1H NMR (400 MHz, CDCl3) δ 1.41 ppm (d, 3H, J = 6.5 Hz), 1.56 (s, 9H), 4.55 (q, 1H, J = 6.5 Hz), 5.05 (d, 1H, J = 7.3 Hz), 6.10 (br s, 1H); 13C NMR (125.7 MHz, CDCl3) δ 14.91 ppm, 27.95, 52.72, 73.76, 84.74, 148.76, 150.49, 168.65; mass spectrum (FAB+) m/z 246 (MH+). Exact mass calculated for C10H15NO6 was 246.0978; found was 246.0977.
(2R,3S)-methyl-N-(tert-butyloxycarbonyl)-2-hydroxy-3-aminobutanoate (6).
Carboxylic acid 4 was converted quantitatively to its methyl ester (5) by using diazomethane. Cesium carbonate (0.66 g, 2.0 mmol) was added to a solution of ester 5 (2.40 g, 9.28 mmol) in MeOH (150 ml). The reaction mixture was stirred at room temperature for 3 h, neutralized with citric acid, and concentrated in vacuo. The resulting residue was dissolved in EtOAc (250 ml) and washed with brine (125 ml). The aqueous layer was extracted further with EtOAc (2 × 80 ml), and the organic layers were combined, dried over MgSO4, and concentrated in vacuo to afford 1.9 g of a white solid. The crude material was purified by flash chromatography on silica gel and eluted with a gradient of 6:1 to 3:1 (vol/vol) hexane/EtOAc. The fractions containing pure product were combined, concentrated, and dried to afford the methyl aminobutanoate (1.18 g of a white solid), which was saponified with 1.0 N NaOH to provide aminobutanoic acid 6 (0.99 g, 91%) as a white solid: Rf = 0.09 (1:2 hexane/EtOAc); mp 104°C (dec); 1H NMR (400 MHz, CD3OD) δ 1.06 ppm (d, 3H, J = 6.9 Hz), 1.43 (s, 9H), 3.96–4.01 (m, 1H), 4.18–4.21 (m, 1H); 13C NMR (100.6 MHz, CD3OD) δ 14.71 ppm, 28.73, 49.95, 73.86, 80.16, 157.37, 175.87; mass spectrum (FAB+) m/z 220 (MH+). Exact mass calculated for C9H18NO5 was 220.1185; found was 220.1187.
l-prolyl-l-phenylalanyl-p-nitroanilide, trifluoroacetate salt (8).
N-(tert-butyloxycarbonyl)-l-proline (0.86 g, 4.0 mmol), l-phenylalanine-p-nitroanilide (1.13 g, 4 mmol), and 1-hydroxybenzotriazole hydrate (HOBT, 0.56 g, 4.2 mmol) were dissolved in dimethylformamide (DMF, 20 ml). The reaction mixture was cooled to 4°C before 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC, 1.30 g, 6.7 mmol) was added in three batches. The mixture was left overnight to warm to room temperature and was concentrated in vacuo. The residue was redissolved in methylene chloride (75 ml), successively washed with saturated NaHCO3 (2 × 30 ml), water (30 ml), 1.0 M citrate (pH 4.0, 2 × 30 ml), and brine (30 ml). The organic phase was dried over MgSO4 and concentrated in vacuo. Purification by flash chromatography on silica gel using a gradient of 6:1 to 1:4 (vol/vol) hexane/EtOAc afforded 1.72 g (89%) of Boc-Pro-Phe-pNA as a white foam. Deprotection of Boc-Pro-Phe-pNA with trifluoroacetic acid (TFA) gave 1.73 g (98%) of the desired salt (8) as a white solid: Rf = 0.07 (1:2 hexane/EtOAc); mp 226°C (dec.); 1H NMR (400 MHz, CD3OD) δ 1.94–2.13 ppm (m, 3H), 2.40–2.49 (m, 1H), 3.04 (dd, 1H, J = 13.7 Hz, 8.6 Hz), 3.19 (dd, 1H, J = 13.7 Hz, 6.7 Hz), 3.25–3.40 (m, 2H), 4.27 (dd, 1H, J = 8.6 Hz, 6.7 Hz), 4.78 (dd, 1H, J = 8.6 Hz, 6.7 Hz), 7.16–7.30 (m, 5H), 7.73 (d, 2H, J = 9.3 Hz), 8.16 (d, 2H, J = 9.3 Hz); 13C NMR (100.6 MHz, CD3OD) δ 24.93 ppm, 31.03, 38.94, 47.44, 57.46, 60.87, 120.52, 125.68, 128.05, 129.57, 130.26, 137.70, 144.77, 145.59, 169.76, 172.02; mass spectrum (FAB+) m/z 383 (MH+). Exact mass calculated for C25H23N4O4 was 383.1719; found was 383.1720.
N-tert-butyloxycarbonyl)-l-α-hydroxyalanyl-l-prolyl-l-phenylalanyl-p-nitroanilide (9).
Compound 9 was prepared by the procedure described for the preparation of Boc-Pro-Phe-pNA using carboxylic acid 6 (0.615, 2.81 mmol), amine 8 (1.46, 2.95 mmol), HOBT (0.40 g, 2.95 mmol), diisopropylethylamine (DIEA, 0.51 ml, 2.95 mmol), and EDC (0.646 g, 3.37 mmol) in DMF (10 ml) to afford 1.59 g (97%) of a white solid: Rf = 0.19 (1:2 hexane/EtOAc); mp 144°C; 1H NMR (400 MHz, CDCl3) δ 0.80 ppm (d, 3H, J = 6.9 Hz), 1.43 (s, 9H), 1.95–2.04 (m, 3H), 2.14–2.21 (m, 1H), 3.27 (d, 2H, J = 13.7 Hz, 6.9 Hz), 3.43 (d, 1H, J = 7.3 Hz), 3.71–3.77 (m, 3H), 4.39 (dd, 1H, J = 7.2 Hz, 3.2 Hz), 4.43–4.47 (m, 1H), 4.78–4.86 (m, 2H), 6.80 (d, 1H, J = 7.3 Hz), 7.18–7.33 (m, 5H), 7.77 (d, 2H, J = 9.1 Hz), 8.17 (d, 2H, J = 9.1 Hz), 8.68 (br s, 1H); 13C NMR (100.6 MHz, CDCl3) δ 13.75 ppm, 25.35, 28.33, 28.56, 36.64, 47.72, 48.20, 54.97, 61.73, 71.82, 79.96, 119.43, 124.83, 127.30, 128.90, 129.07, 136.36, 143.56, 143.62, 155.38, 169.34, 171.18, 173.04; mass spectrum (FAB+) m/z 584 (MH+). Exact mass calculated for C29H38N5O8 was 584.2720; found was 584.2725.
N-(tert-butyloxycarbonyl)-l-alanyl-l-α-hydroxyalanyl-l-prolyl-l-phenylalanyl-p-nitroanilide (10).
Boc-peptide 9 (0.60 g, 3.16 mmol) was deprotected and subsequently coupled as described above with N-(tert-butyloxycarbonyl)-l-alanine (1.63 g, 2.72 mmol), HOBT (0.43 g, 3.16 mmol), DIEA (0.55 ml, 3.16 mmol), and EDC (0.66 g, 3.44 mmol) in DMF (10 ml). Flash chromatography on silica gel using a gradient of 5:1 hexane/EtOAc to EtOAc afforded peptide 10 (0.98 g, 55%) as an off-white foam: mp 124°C (dec.); 1H NMR (400 MHz, CDCl3) δ 0.91 ppm (d, 3H, J = 6.7 Hz), 1.33 (d, 3H, J = 7.0 Hz), 1.44 (s, 9H), 1.90–1.99 (m, 3H), 2.09–2.15 (m, 1H), 3.17–3.35 (m, 2H), 3.60–3.80 (m, 3H), 4.00–4.50 (m, 2H), 4.34–4.46 (m, 4H), 4.74–4.84 (m, 1H), 5.12 (br s, 1H), 6.87 (br s, 1H), 7.14–7.30 (m, 6H), 7.78 (d, 2H, J = 8.2 Hz), 8.14 (d, 2H, J = 8.6 Hz), 9.03 (br s, 1H); 13C NMR (100.6 MHz, CDCl3) δ 13.84 ppm, 17.99, 25.23, 28.24, 28.67, 36.71, 47.46, 47.75, 55.17, 60.37, 61.64, 71.42, 80.64, 119.43, 124.82, 127.22, 128.82, 129.12, 143.55, 143.72, 155.62, 169.59, 171.22, 172.97, 173.61; mass spectrum (FAB+) m/z 655 (MH+). Exact mass calculated for C32H43N6O9 was 655.3092; found was 655.3106.
N-succinyl-l-alanyl-l-α-hydroxyalanyl-l-prolyl-l-phenylalanyl-p-nitroanilide (11).
Peptide 10 (0.32 g, 0.48 mmol) was deprotected by TFA and then coupled to succinic anhydride (56.0 mg, 0.54 mmol), using DIEA (1.80 ml, 1.0 mmol) in DMF (5 ml) to afford 0.28 g of crude product. The material was purified by reversed-phase preparative HPLC (Dynamax Chromosorb C18) using a gradient of 20–100% MeCN in aqueous 0.1% TFA over 80 min at a flow rate of 9 ml/min. The desired product was eluted as a single peak (detected at 320 nm) with a retention time of 30.2 min. The fractions were collected and lyophilized to afford 173.2 mg (51%) of a white solid: mp 122°C (dec.); 1H NMR (400 MHz, CD3OD) δ 1.12 ppm (d, 3H, J = 6.9 Hz), 1.33 (d, 3H, J = 7.2 Hz), 1.74–1.83 (m, 1H), 1.85–1.99 (m, 2H), 2.09–2.17 (m, 1H), 2.41–2.69 (m, 4H), 3.11 (dd, 1H, J = 13.7 Hz, 8.6 Hz), 3.22 (dd, 1H, J = 13.7 Hz, 6.4 Hz), 3.66–3.80 (m, 2H), 4.13–4.18 (m, 1H), 4.28–4.40 (m, 3H), 4.69 (dd, 1H, J = 8.6 Hz, 6.4 Hz), 7.15–7.25 (m, 5H), 7.86 (d, 2H, J = 9.2 Hz), 8.17 (d, 2H, J = 9.2 Hz); 13C NMR (100.6 MHz, CD3OD) δ 14.33 ppm, 14.56, 17.84, 17.88, 26.07, 29.99, 30.07, 30.25, 31.22, 31.34, 38.20, 50.59, 50.64, 57.03, 57.07, 62.24, 62.33, 72.66, 120.72, 125.61, 127.85, 129.50, 129.61, 130.22, 130.30, 138.29, 138.32, 144.70, 145.57, 172.13, 172.16, 173.40, 173.58, 174.22, 174.41, 174.66, 174.78, 175.19, 176.49; mass spectrum (FAB+) m/z 655 (MH+). Exact mass calculated for C31H39N6O10 was 655.2728; found was 655.2730.
N-succinyl-l-alanyl-l-α-ketoalanyl-l-prolyl-l-phenylalanyl-p-nitroanilide (1).
To a solution of oxalyl chloride (0.35 ml, 0.40 mmol) in methylene chloride (5.0 ml) at −78°C was added dropwise a solution of freshly redistilled dimethyl sulfoxide (DMSO) (0.43 ml, 6.0 mmol) in methylene chloride (1.2 ml). After 10 min, a solution of α-hydroxy peptide 11 (132 mg, 0.20 mmol) in MeCN (2.0 ml, plus a 1-ml rinse) was added via cannula under an atmosphere of nitrogen at a rate to maintain the internal temperature below −65°C. After 2.5 h, triethylamine (1.2 ml) was added, allowing the mixture to warm up to −20°C. The reaction was quenched by the addition of cold aqueous ammonium chloride solution. The aqueous layer was extracted with methylene chloride (3 × 30 ml), and the organic phases were combined and evaporated to afford 141.2 mg of crude product. The material was further purified by HPLC as for 11. The desired product was eluted with a retention time of 26.4 min. The fractions were collected and lyophilized to afford 89.5 mg (68%) of a white powder: mp 163°C; variable temperature (20–110°C) 1H NMR experiments in DMF-d7 showed initial broadening and finally the coalescing of peaks, suggesting the existence of rotamers. 1H NMR (400 MHz, DMF-d7, 25°C, major rotamer) δ 1.25 ppm (d, 1.5H, J = 7.1 Hz), 1.29 (d, 1.5H, J = 7.2 Hz), 1.31–1.34 (m, 1.5H), 1.42 (d, 1.5H, J = 7.1 Hz), 1.68–1.75 (m, 0.5H), 1.79–1.95 (m, 2H), 1.98–2.45 (m, 1H), 2.45–2.63 (m, 3.5H), 3.05–3.19 (m, 1H), 3.28–3.38 (m, 1H), 3.43–3.55 (m, 1.5H), 3.59–3.71 (m, 1.5H), 4.30–4.40 (m, 0.5H), 4.41–4.53 (m, 1H), 4.60–4.69 (m, 0.5H), 4.75–4.87 (m, 1.5 H), 4.89–4.93 (m, 0.5H), 7.19–7.4 (m, 6H), 7.90–8.09 (m, 2H), 8.11 (dd, 1H, J = 17.7 Hz, 7.0 Hz), 8.25 (t, 2H, J = 8.7 Hz), 8.35–8.38 (m, 1H); 13C NMR (125.7 MHz, DMF-d7, 25°C, major rotamer) δ 14.42 ppm, 14.60, 18.01, 18.22, 26.45, 29.91, 30.27, 30.86, 31.31, 31.61, 38.20, 50.55, 50.79, 57.31, 57.52, 62.43, 62.81, 120.21, 125.63, 128.05, 129.58, 129.34, 130.30, 130.39, 138.27, 138.46, 144.77, 145.55, 172.11, 172.26, 173.44, 173.68, 174.21, 174.53, 174.86, 174.99, 175.14, 176.52, 206.35; mass spectrum (FAB+) m/z 653 (MH+). Exact mass calculated for C31H37N6O10 was 653.2571; found was 653.2571.
Hapten Analog (Inhibitor) Synthesis.
l-deoxo-phenylalanyl-p-nitroanilide (14). A solution of borane in THF (1.0 M, 24.8 ml) was cooled to 4°C and l-phenylalanine-p-nitroanilide (0.7 g, 2.5 mmol) was added in several batches. The resulting yellow solution was heated to reflux for 2 h, then cooled to 4°C and quenched by slow addition of a mixture of 50% MeOH (vol/vol) in THF until the solution stopped bubbling. The solvent was evaporated, aqueous 1 N HCl (14 ml) and MeOH (14 ml) were added, and the mixture was heated gently at 50°C. After 1 h, the reaction mixture was neutralized with 1 N NaOH, and then concentrated partially in vacuo. The aqueous layer was extracted with EtOAc (4 × 200 ml), and the organic layers were combined, dried, and concentrated to give 0.64 g (96%) of a yellow oil: 1H NMR (400 MHz, DMSO-d6) δ 2.57 ppm (dd, 1H, J = 13.8 Hz, 7.2 Hz), 2.77 (dd, 1H, J = 13.8 Hz, 5.2 Hz), 2.99–3.05 (m, 1H), 3.08–3.16 (m, 2H), 6.62 (d, 2H, J = 9.3 Hz), 7.16–7.34 (m, 6H), 7.96 (d, 2H, J = 9.2 Hz); 13C NMR (100.6 MHz, DMSO-d6) δ 41.03 ppm, 48.57, 51.77, 110.85, 126.07, 126.18, 128.26, 129.21, 135.58, 139.14, 154.70; mass spectrum (FAB+) m/z 272 (MH+). Exact mass calculated for C15H18N3O2 was 272.1399; found was 272.1396.
N-(tert-butyloxycarbonyl)-l-alanyl-l-α-hydroxyalanyl-l-prolyl-l-deoxo-phenylalanyl-p-nitroanilide (16).
Compound 14 (0.62 g, 2.3 mmol) was coupled with N-(tert-butyloxycarbonyl)-l-proline (0.52g, 2.40 mmol), HOBT (0.32 g, 2.4 mmol), and EDC (0.52, 2.7 mmol) in DMF (10 ml). The resultant product was purified by flash chromatography on silica gel to afford 1.0 g of a yellow solid, which was treated with TFA to give 0.98 g of the desired salt (15). Compound 15 (0.52 g, 1.1 mmol) then was reacted with 6 (0.23 g, 1.0 mmol), HOBT (0.15 g, 1.1 mmol), DIEA (0.19 ml, 1.1 mmol), and EDC (0.24 g, 1.2 mmol) in DMF (5 ml). After silica gel chromatography in 1:3 hexane/EtOAc, 0.53 g (91%) of a yellow foam was obtained: Rf = 0.10 (1:2 hexane/EtOAc); 1H NMR (400 MHz, CDCl3, major rotamer at 25°C) δ 1.02 ppm (d, 2.16H, J = 6.8 Hz), 1.07 (d, 0.85H, J = 6.8 Hz), 1.41, 1.44 (2s, 9H), 1.66–1.76 (m, 1H), 1.84–2.15 (m, 3H), 2.90–2.94 (m, 2H), 3.24–3.40 (m, 2H), 3.72–3.75 (m, 2H), 3.85–3.95 (m, 1H), 4.19–4.32 (m, 2H), 4.39 (d, 1H, J = 3.8 Hz), 6.59–6.64 (m, 2H), 7.18–7.29 (m, 5H), 7.97–8.02 (m, 2H); 13C NMR (125.7 MHz, CDCl3) δ 14.14 ppm, 25.30, 28.16, 28.33, 31.37, 36.41, 38.42, 46.28, 47.64, 48.17, 50.57, 61.46, 71.69, 79.76, 111.08, 126.30, 126.94, 128.74, 129.97, 137.08, 137.73, 153,47, 155.37, 162.50, 171.49, 172.51; mass spectrum (FAB+) m/z 570 (MH+). Exact mass calculated for C29H40N5O7 was 570.2928; found was 570.2920.
N-Acetyl-l-alanyl-l-α-ketoalanyl-l-prolyl-l-deoxo-phenylalanyl-p-nitroanilide (13).
Boc-protected 16 (0.51 g, 0.9 mmol) was treated with TFA to give 0.38 g (92%) of the trifluoroacetate salt as a yellow foam, which was coupled with N-acetyl-l-alanine (143.0 mg, 1.1 mmol), HOBT (148.0 mg, 1.1 mmol), triethylamine (TEA, 0.25 ml), and EDC (230 mg, 1.2 mmol) to afford 0.5 g of crude product as a yellow solid. The material was purified by preparative HPLC to give 336 mg (53%) of a yellow powder. Freshly distilled pyridine (50 μL) and Dess-Martin periodinane (170 mg, 0.40 mmol) were added to a solution of this material (182 mg, 0.31 mmol) in methylene chloride (4 ml). After 1.5 h at room temperature, the reaction was cooled to 0°C and stopped by the addition of saturated sodium sulfite (5 ml). After being stirred for 5 min at 0°C, the reaction mixture was extracted with methylene chloride (4 × 40 ml). The organic layers were combined, dried, and evaporated in vacuo. The residue was purified by preparative HPLC to give 149 mg (82%) of the desired product as a yellow powder: mp 163–165°C; UV (MeOH) 210 nm (ɛ 10, 500), 382 (12,000); 1H NMR (400 MHz, DMSO-d6, major rotamer at 25°C) δ 1.09–1.18 ppm (m, 4.5H), 1.32 (d, 1.5H, J = 7.1 Hz), 1.57–1.98 (m, 4H), 1.78 (s, 1.5H), 1.80 (s, 1.5H), 2.71–2.86 (m, 2H), 3.15–3.24 (m, 2H), 3.25–3.60 (m, 2H), 4.02–4.35 (m, 2H), 4.42–4.55 (m, 1H), 4.57–4.80 (m, 1H), 6.63–6.67 (m, 2H), 7.15–7.30 (m, 5H), 7.98–8.03 (m, 2H); mass spectrum (FAB+) m/z 583 (MH+). Exact mass calculated for C29H39N6O7 was 583.2881; found was 583.2881.
Protein-Hapten Conjugates.
Protein-hapten conjugates were prepared as previously described (27). The epitope densities were determined by comparing the UV difference spectra of the conjugated and the unconjugated proteins at 320 nm (BSA conjugate, ɛhapten = 10,560) and 316 nm [keyhole limpet hemocyanin (KLH) conjugate, ɛhapten = 10,440]. Epitope densities of 8.1 and 6.7 were measured for the BSA and KLH conjugates, respectively.
Peptide Isomerization Assays.
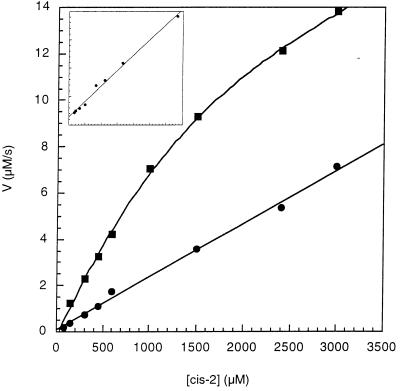
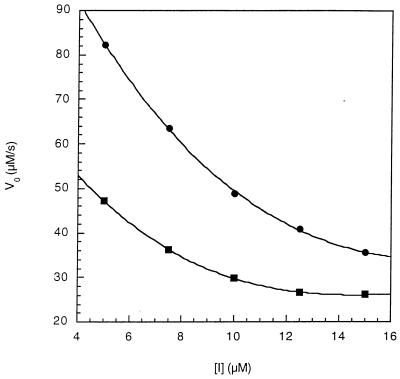
These assays were performed by the described method (28) at 0°C in 50 mM Hepes/50 mM NaCl, pH 7.8 buffer containing 2% trifluoroethanol (TFE) in the presence of antibody 47 Mar.1 (7 μM) or absence of antibody. A buffer solution containing antibody and α-chymotrypsin (40 μl of 60 mg/ml solution in 1 mM HCl) was incubated for 10 min at 0°C in a 0.4-ml cuvette. Substrate (8 μl of 6.25–250 mM stock solutions in anhydrous TFE containing 470 mM LiCl; cis/trans = 60:40) was rapidly added, and the reaction progress curves [with cis-(2) in the range of 0.125–3 mM] were recorded spectrophotometrically by measuring the absorbance increase at 460 nm (ɛ 270) as a function of time (over 10 min, 150 data points/min). The absorbance data points then were transferred to a Macintosh computer, converted to [time (min), OD460] pairs, and saved as a raw data file by using kaleidagraph. These data were fitted to an integrated rate equation (including terms for both the catalyzed and uncatalyzed isomerization rates) derived by Kofron et al. (28) for the calculation of kcat and Km values, using the dynafit software package (ref. 29; for more information about dynafit, see: http://www.biokin.com) running on a power Macintosh. See Fig. 2 for initial reaction velocities calculated from this program versus substrate concentrations. Inhibition experiments were measured at two substrate concentrations (0.6 mM and 0.3 mM of cis-2) with inhibitor (13) concentrations in the range of 5–15 μM (see Fig. 4). The Ki value (3.0 ± 0.4 μM) was calculated by using nonlinear least-squares optimization to a competitive equation as described (28), using dynafit.
Figure 2.
Plot of reaction velocity versus substrate concentration for the cis to trans isomerization reaction of 2. Initial velocities were calculated from progress curves measured at 0°C in 50 mM Hepes/50 mM NaCl, pH 7.8 buffer containing 2% TFE, in the presence of 7.0 μM antibody 47 Mar.1 (▪) and in the absence of a catalyst (•). (Inset) The corresponding Lineweaver–Burk plot for the antibody-catalyzed isomerization reaction.
Figure 4.
V0 vs. [I] plot for antibody 47 Mar.1 catalyzed cis to trans isomerization reaction of 2 in the presence of inhibitor 13. Antibody concentration was 7.0 μM. Initial velocities were obtained by the same methods as described in Fig. 2 at two concentrations of substrate 2: 1 mM [cis-(2) = 0.6 mM] (•) and 0.5 mM [cis-(2) = 0.3 mM] (▪).
Protein Folding Assays.
RNase T1 from Aspergillus oryzae was purchased from Boehringer Mannheim, purified by ion exchange chromatography (Mono Q), and dialysed into distilled water at 4°C. The protein concentration was determined by absorbance (A280 = 1.9 OD for 1 mg/ml of RNase T1) (30), and the sample solution then was quantitatively divided into small vials for lyophilization to dryness. RNase T1 was denatured by incubation in 8 M urea, 0.1 M Tris⋅HCl, 1 mM EDTA, pH 8.0 at 25°C for 2 h. Refolding was initiated by diluting 10 μl of unfolded protein with 390 μl of 0.1 M Tris⋅HCl, 1 mM EDTA, pH 8.0, containing the appropriate concentrations of the antibody, hapten 1, and hapten-analog 13 in a 0.4-ml fluorimeter cell at 10°C. The final concentrations for refolding reactions were 3.0 μM RNase T1 in 0.2 M urea, 0.1 M Tris⋅HCl, 1 mM EDTA, pH 8.0 at 10 ± 0.1°C. The refolding reaction was monitored by the increase in fluorescence at 320 nm (5 nm slit) after excitation at 268 nm (1 nm slit) using a Hitachi F-4500 fluorescence spectrophotometer. The excitation shutter was closed between the individual measurements to minimize radiation damage during the kinetic experiments (31). All data were reproducible with deviations of less than 3.5%.
RESULTS AND DISCUSSION
Hapten Synthesis and Antibody Production.
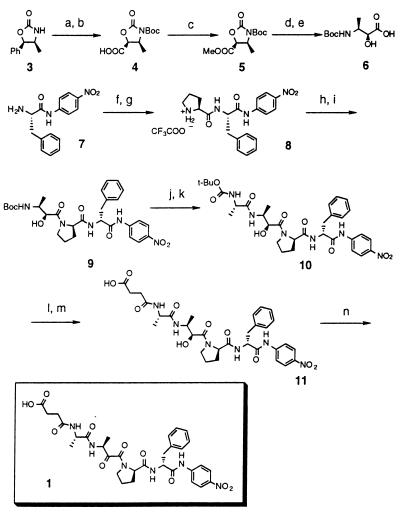
To facilitate kinetic comparisons to the natural enzymes, hapten 1 (Fig. 1) was designed to mimic the peptide succinyl-l-alanyl-l-alanyl-l-prolyl-l-phenylalanyl-p-nitroaniline (Suc-Ala-Ala-Pro-Phe-pNA) (2), a commercially available substrate for kinetic and mechanistic studies of cyclophilins and FKBP. The peptide Suc-Ala-Ala-Pro-Phe-pNA undergoes spontaneous cis to trans isomerization at 37°C with a first-order rate constant of approximately 0.15 s−1 (18), a relatively low background rate for small peptides. Synthesis of hapten 1 was carried out from the hydroxy-acid precursor (6) by standard peptide coupling protocols (32) as shown in Fig. 1. Hydroxy-acid 6 was prepared in five steps from oxazolidinone 3. Treatment of (4S,5R)-(−)-4-methyl-5-phenyl-2-oxazolidinone with Boc-anhydride and triethylamine afforded the N-Boc oxazolidinone (97%), which subsequently was oxidized with ruthenium trichloride (RuCl3⋅3H2O) and sodium periodate to give acid 4 (48%). Quantitative conversion of the acid to its methyl ester (5) was accomplished by treatment with diazomethane. The resultant oxazolidinone (5) was selectively ring-opened with powdered cesium carbonate in methanol to give the methyl ester (54%), which was saponified to provide the desired hydroxy-acid (6) in 91% yield. Carbodiimide-mediated coupling of N-Boc-l-proline with l-Phe-p-nitroaniline (7) in DMF gave the Boc-protected dipeptide (89%). Acidolytic deprotection of the Boc group using TFA provided amine 8 in 98% yield as a trifluoroacetate salt. Tripeptide 9 was generated by coupling amine 8 with hydroxy-acid 6 in DMF (97%). Treatment with TFA afforded the amine-TFA salt, which was coupled with N-Boc-alanine as described above, to give the Boc-tetrapeptide (10) in 55% yield. Again, the N-terminal Boc group was deprotected by using TFA, and the resultant amine-TFA salt was acylated with succinic anhydride in the presence of DIEA. The hydroxy-peptide (11) was isolated in 51% yield after purification by reversed-phase HPLC. Swern oxidation of the alcohol moiety followed by further HPLC separation provided hapten 1 in 68% yield.
Figure 1.
Synthesis of hapten 1. Conditions: (a) (Boc)2O, triethylamine,97%; (b) RuCl3/NaIO4, 48%; (c) CH2N2, 100%; (d) Cs2CO3, 54%; (e) 1N NaOH, 91%; (f) Boc-Pro/DIEA/HOBT, EDC, 0°C → room temperature (RT), 89%; (g) TFA, 0°C, 98%; (h) DIEA/HOBT; (i) + 6, EDC, 0°C → RT, 97%; (j) TFA, 0°C, 100%; (k) Boc-Ala/DIEA/HOBT, EDC, 0°C → RT, 55%; (l) TFA, 0°C, 100%; (m) Succinic anhydride/DIEA, 51%; (n) (COCl)2, DMSO, −78°C → −20°C, 68%.
Antibodies specific for hapten 1 were produced by standard methods using BALB/c mice immunized with the keyhole limpet hemocyanin conjugate of 1 (27). ELISA assays with the BSA conjugate of 1 identified more than 100 monoclonal cell lines expressing antibody specific for 1. Ascites fluid generated from the cell lines was purified by protein G affinity chromatography. A total of 67 antibodies were found by ELISA assays to bind both free hapten 1 and substrate 2. Antibodies were judged to be greater than 95% pure by SDS/PAGE.
Catalytic Assays and Kinetic Characterization.
The most widely used method to assay peptidyl-prolyl cis-trans isomerization is an enzyme-coupled assay that depends on the inability of α-chymotrypsin to hydrolyze Suc-Ala-Xaa-Pro-Phe-pNA when the Xaa-Pro bond is in the cis conformation (28). Although a direct fluorimetric assay (33) has been developed, the substrate scope is narrow, and the low substrate concentrations used in the fluorimetric assay may limit its application to antibody-catalyzed reactions. Reliable methods for analyzing the kinetics by NMR spectroscopy recently have been reported (34, 35); however, these methods are not convenient for screening large numbers of antibodies. By using the enzyme-coupled assay, three antibodies were identified that catalyzed the isomerization reaction of substrate 2. One catalyst (47 Mar.1) was further purified by ion exchange (Mono S) chromatography and characterized in detail.
The apparent first-order rate constants and kinetic parameters for both the uncatalyzed and antibody-catalyzed reactions were derived by analysis of the progress curves by using the computer program dynafit (29). The background rate of cis to trans isomerization of substrate 2 at 0°C was measured to be 0.0024 s−1 in 50 mM Hepes/50 mM NaCl, pH 7.8 buffer containing 2% TFE and 9.4 mM LiCl (LiCl-TFE mixture results in increased concentration of the cis substrate, simplifying kinetic analysis). Antibody 47 Mar.1 exhibited Michaelis-Menten behavior with kcat = 1.58 s−1, Km = 1.79 mM, and kcat/Km = 883 s−1⋅M−1 for the cis substrate (cis-2) (Fig. 2). For comparison, the cis to trans isomerization reactions of substrate 2 catalyzed by the natural enzymes cyclophilin (from calf thymus, Sigma) and FKBP (from recombinant human, Sigma) have kcat/Km values of 1.2 × 107 s−1⋅M−1, and 6.4 × 105 s−1⋅M−1, respectively. Under similar assay conditions, these kinetic data and the background rate of 2 are in good accordance with previously determined values (28).
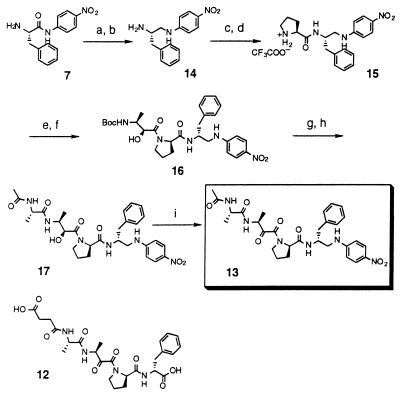
To verify that catalysis is occurring in the antibody combining site, hapten inhibition studies were carried out. However, the results were difficult to interpret because of hydrolysis of hapten 1 by chymotrypsin. Attempts to inhibit catalysis using a portion of the hapten epitope (12) were unsuccessful. A nonhydrolyzable analog (13) of the hapten (1) therefore was synthesized that contains an aminomethylene isostere in place of the susceptible p-nitroaniline bond (Fig. 3). The amide bond of l-Phe-p-nitroaniline was selectively reduced by using borane-THF; acidic work-up with aqueous HCl led to the isolation of diamine 14 in 96% yield. Coupling of this amine with Boc-proline followed by acidolytic deprotection with TFA afforded dipeptide 15 as a TFA salt in 95% yield. Further coupling of 15 with hydroxy-acid 6 gave a 91% yield of product 16. Another round of TFA deprotection (92%) and coupling (53%) with N-acetyl-l-alanine afforded 17, which was oxidized by Dess-Martin periodinane. After final purification by preparative HPLC, the desired hapten analog (13) was obtained in 82% yield.
Figure 3.
Synthesis of hapten analog (inhibitor) 13 and the structure of 12. Conditions: (a) BH3⋅THF; (b) aqueous HCl, 96% in two steps; (c) Boc-Pro/EDC/HOBT, 0°C → room temperature (RT), 94%; (d) TFA, 0°C, 96%; (e) DIEA/HOBT, 0°C; (f) + 6, EDC, 0°C, 91% in two steps; (g) TFA, 0°C, 92%; (h) N-acetyl-Ala/DIEA/HOBT, EDC, 0°C → RT, 53%; (i) Dess-Martin periodinane, −78°C, 82%.
Hapten analog 13 was found to inhibit the catalytic activity of antibody 47 Mar.1 (Fig. 4). Least-squares Dixon analysis with two sets of substrate concentrations (0.6 mM and 0.3 mM of cis-2) afforded a Ki value of 3.0 ± 0.4 μM. This result indicates that catalysis is associated with the antibody combining site. Not surprisingly, hapten analog 13 is also a good inhibitor for the cis-trans isomerization reactions of 2 catalyzed by cyclophilin (Ki = 14 μM) and FKBP (Ki = 11 μM). The rate acceleration of this catalytic antibody (kcat/kuncat = 658) is very close to the value of Km/Ki (746), consistent with preferential stabilization of the transition state by the antibody. The antibody-catalyzed reaction is insensitive to pH variations, and no solvent deuterium isotope effect is observed (28). These findings, as with the natural rotamases, suggest the lack of involvement of catalytic groups in the antibody-substrate complex. Therefore it is likely the α-keto amide functional group in hapten 1 is acting as a mimic of a conformationally distorted transition state rather than as an electrophile to elicit a nucleophile in the antibody binding site.
Antibody-Catalyzed Protein Folding.
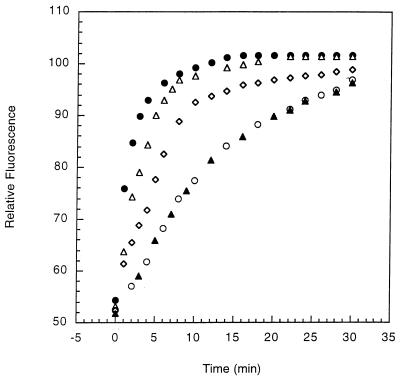
The natural protein, RNase T1, then was examined as a possible substrate for the antibody. RNase T1 is a small protein (104 amino acids) with four prolyl peptide bonds (36) that has been widely used as a model protein for folding studies of the natural rotamases, cyclophilins (37), and FKBP (38). The unfolding of RNase T1 by urea or guanidine hydrochloride is a completely reversible reaction, and the refolding experiments can be carried out by monitoring the fluorescence enhancement of the sole tryptophan residue, which is completely buried in the folded protein (37). In the absence of an isomerase, the refolding reaction is approximately first-order at early time and complete after 65 min at 10°C, consistent with the model that refolding is roughly a sum of two phases, an intermediate phase with a time constant, τ ≈ 400 s, and a very slow phase with τ ≈ 3,000 s (4). Antibody 47 Mar.1 (6.4 μM) leads to a 4- to 5-fold increase in the folding rate of RNase T1 (Fig. 5). Furthermore, the antibody-catalyzed reaction is completely inhibited by the addition of 9.1 μM hapten 1 or by 10.6 μM hapten analog 13, again demonstrating that the folding reaction is associated with the antibody combining site. Additionally, several other antibodies elicited against hapten 1 have been examined, and none of them catalyze the folding reaction. For comparison, FKBP (0.14 μM) increases the rate of RNase T1 folding 7- to 8-fold over the background reaction under the same assay conditions. However, unlike the antibody, catalysis by FKBP is not inhibited by hapten 1 or analog 13 at concentrations up to 150 μM.
Figure 5.
Catalysis of RNase T1 refolding reaction by antibody 47 Mar.1. The change in fluorescence at 320 nm is shown as a function of the time. The final conditions for refolding were 3.0 μM RNase T1 in 0.2 M urea, 0.1 M Tris⋅HCl, 1 mM EDTA, pH 8.0 at 10 ± 0.1°C. Uncatalyzed folding reaction in the absence of a catalyst (▴); folding reactions in the presence of 6.4 μM antibody 47 Mar.1 (▵), 6.4 μM antibody, and an additional hapten 1: 3.7 μM (◊) and 9.1 μM (○), and 0.14 μM FKBP (•).
The refolding of RNase T1 catalyzed by the antibody is more complicated and less efficient than isomerization of the relative small, unstructured peptide. Several exponential phases and intermediates (4) are involved during the folding pathway from the denatured to the native state, making it difficult to characterize in detail the peptidyl-prolyl bond isomerization reaction(s). Spontaneous refolding of RNase T1 chains (with four prolyl residues: Ser-37–Tyr-38–Pro-39–His-40, Ser-53–Ser-54–Pro-55–Tyr-56, Glu-58–Trp-59–Pro-60–Ile-61, and Gly-71–Ser-72–Pro-73–Gly-74) presumably is determined by the slow trans [→ cis isomerization of the Tyr-38–Pro-39 and Ser-54–Pro-55 peptide bonds, which are both cis in the native state, but are predominantly (80–90%) trans in the unfolded protein (4)]. However, catalysis of isomerization at Pro-55 by natural rotamases may be the main contributor for the acceleration of RNase T1 folding because this proline, unlike Pro-39, which is buried in the native protein and also presumably in folding intermediates, is solvent-exposed (39). The ability of antibody 47 Mar.1 to catalyze folding of RNase T1 suggests that the antibody has relatively broad substrate specificity. This may result from the uncharged hydrophobic nature of the hapten. The reduced rate of the antibody-catalyzed reaction compared with FKBP and cyclophilins suggests that the antibody may distort the amide bond but may not effectively catalyze the bond rotation reaction. In this regard, spirocyclic ring systems may be more efficient analogs than α-keto amide moieties for mimicking an orthogonal transition state. Nevertheless, this study and that of Yli-Kauhaluoma et al. (26) suggest that the use of distorted substrate analogs is a reproducible means for generating prolyl isomerization catalysts. Similar approaches have been used to generate antibodies that catalyze the isomerization of carbon-carbon sigma bonds in bridged biphenyls (40). Finally, the demonstration that antibodies can catalyze conformational changes involving the protein backbone raises the possibility that antibodies also can be generated that catalyze other conformational changes such as those involved in signal transduction processes, either in vitro or in vivo.
Acknowledgments
We thank Elizabeth Sweet and Yoko Oei for assistance with antibody production. This work was supported by the National Institutes of Health, predoctoral fellowships from the National Science Foundation and American Chemical Society (L.C.H.-W.), and a postdoctoral fellowship from the Cancer Research Fund of the Damon-Runyon Walter Winchell Foundation (L.M.). P.G.S. is a Howard Hughes Medical Institute Investigator and a W. M. Keck Foundation Investigator.
ABBREVIATIONS
- FKBP
FK506/rapamycin-binding proteins
- THF
tetrahydrofuran
- HOBT
1-hydroxybenzotriazole hydrate
- DMF
dimethylformamide
- EDC
1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride
- TFA
trifluoroacetic acid
- DIEA
diisopropylethylamine
- DMSO
dimethyl sulfoxide
- TFE
trifluoroethanol
References
- 1.Nall B T. In: Mechanisms of Protein Folding. Pain R H, editor. Oxford: IRL; 1994. pp. 80–103. [Google Scholar]
- 2.Bächinger H P. J Biol Chem. 1987;262:17144–17148. [PubMed] [Google Scholar]
- 3.Lang K, Schmid F X, Fischer G. Nature (London) 1987;329:268–270. doi: 10.1038/329268a0. [DOI] [PubMed] [Google Scholar]
- 4.Schmid F X. Annu Rev Biophys Biomol Struct. 1993;22:123–143. doi: 10.1146/annurev.bb.22.060193.001011. [DOI] [PubMed] [Google Scholar]
- 5.Lang K, Schmid F X. Nature (London) 1988;331:453–455. doi: 10.1038/331453a0. [DOI] [PubMed] [Google Scholar]
- 6.Veeraraghavan S, Nall B T, Fink A L. Biochemistry. 1997;36:15134–15139. doi: 10.1021/bi971357r. [DOI] [PubMed] [Google Scholar]
- 7.Yang H, Zhong H, Zhou H. Biochim Biophys Acta. 1997;1338:147–150. doi: 10.1016/s0167-4838(97)00026-5. [DOI] [PubMed] [Google Scholar]
- 8.Lin L N, Hasumi H, Brandts J F. Biochim Biophys Acta. 1988;956:256–266. doi: 10.1016/0167-4838(88)90142-2. [DOI] [PubMed] [Google Scholar]
- 9.Steinmann B, Bruckner P, Superti-Furga A. J Biol Chem. 1991;266:1299–1303. [PubMed] [Google Scholar]
- 10.Lodish H F, Kong N. J Biol Chem. 1991;266:14835–14838. [PubMed] [Google Scholar]
- 11.Fischer G. Angew Chem Int Ed Engl. 1994;33:1415–1436. [Google Scholar]
- 12.Galat A. Eur J Biochem. 1993;216:687–707. doi: 10.1111/j.1432-1033.1993.tb18189.x. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber S L, Albers M W, Brown E J. Acc Chem Res. 1993;26:412–420. [Google Scholar]
- 14.Lu K P, Hanes S D, Hunter T. Nature (London) 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 15.Yaffe M B, Schutkowski M, Shen M, Zhou X Z, Stukenberg P T, Rohfeld J-U, Xu J, Kuang J, Kirschner M W, Fisher G, et al. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 16.Fischer G, Berger E, Bang H. FEBS Lett. 1989;250:267–270. doi: 10.1016/0014-5793(89)80735-5. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Ke H. Biochemistry. 1996;35:7362–7368. doi: 10.1021/bi960278x. [DOI] [PubMed] [Google Scholar]
- 18.Harrison R K, Stein R L. Biochemistry. 1990;29:1684–1689. doi: 10.1021/bi00459a003. [DOI] [PubMed] [Google Scholar]
- 19.Rosen M K, Standaert R F, Galat A, Nakatsuka M, Schreiber S L. Science. 1990;248:863–866. doi: 10.1126/science.1693013. [DOI] [PubMed] [Google Scholar]
- 20.Ranganathan R, Lu K P, Hunter T, Noel J P. Cell. 1997;89:875–886. doi: 10.1016/s0092-8674(00)80273-1. [DOI] [PubMed] [Google Scholar]
- 21.Van Duyne G D, Standaert R F, Karplus P A, Schreiber S L, Clardy J. Science. 1991;252:839–842. doi: 10.1126/science.1709302. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Albers M W, Chen C M, Schreiber S L, Walsh C T. Proc Natl Acad Sci USA. 1990;87:2304–2308. doi: 10.1073/pnas.87.6.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albers M W, Walsh C T, Schreiber S L. J Org Chem. 1990;55:4984–4986. [Google Scholar]
- 24.Li Z Z, Patil G S, Golubski Z E, Hori H, Tehrani K, Foreman J E, Eveleth D D, Bartus R T, Powers J C. J Med Chem. 1993;36:3472–3480. doi: 10.1021/jm00074a031. [DOI] [PubMed] [Google Scholar]
- 25.Walter J, Bode W. Hoppe-Seylers Z Physiol Chem. 1983;364:949–959. doi: 10.1515/bchm2.1983.364.2.949. [DOI] [PubMed] [Google Scholar]
- 26.Yli-Kauhaluoma T J, Ashley J A, Lo C H L, Coakley J, Wirsching P, Janda K D. J Am Chem Soc. 1996;118:5496–5497. [Google Scholar]
- 27.Jacobsen J, Schultz P G. Proc Natl Acad Sci USA. 1994;91:5888–5892. doi: 10.1073/pnas.91.13.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kofron J L, Kuzmic P, Kishore V, Colón-Bonilla E, Rich D H. Biochemistry. 1991;30:6127–6134. doi: 10.1021/bi00239a007. [DOI] [PubMed] [Google Scholar]
- 29.Kuzmic P. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 30.Egami F, Takahashi K, Uchida T. Prog Nucleic Acids Res Mol Biol. 1964;3:59–101. doi: 10.1016/s0079-6603(08)60739-4. [DOI] [PubMed] [Google Scholar]
- 31.Schönbrunner E R, Schmid F X. Proc Natl Acad Sci USA. 1992;89:4501–4513. doi: 10.1073/pnas.89.10.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodanszky M, Bodanszky A. The Practice of Peptide Synthesis. Berlin: Springer; 1984. pp. 1–180. [Google Scholar]
- 33.Garcia-Echeverria C, Kofron J L, Kuzmic P, Kishore V, Rich D H. J Am Chem Soc. 1992;114:2758–2259. [Google Scholar]
- 34.Videen J S, Stamnes M A, Hsu V L, Goodman M. Biopolymers. 1994;34:171–175. doi: 10.1002/bip.360340203. [DOI] [PubMed] [Google Scholar]
- 35.Kern D, Kern G, Scherer G, Fischer G, Drakenberg T. Biochemistry. 1995;34:13594–13602. doi: 10.1021/bi00041a039. [DOI] [PubMed] [Google Scholar]
- 36.Pace C N, Heinemann U, Hahn U, Saenger W. Angew Chem Int Ed Engl. 1991;30:343–360. [Google Scholar]
- 37.Kiefhaber T, Quaas R, Hahn U, Schmid F X. Biochemistry. 1990;29:3061–3070. doi: 10.1021/bi00464a024. [DOI] [PubMed] [Google Scholar]
- 38.Tropschug M, Wachter E, Mayer S, Schönbrunner E R, Schmid F X. Nature (London) 1990;346:674–677. doi: 10.1038/346674a0. [DOI] [PubMed] [Google Scholar]
- 39.Schmid F X, Frech C, Scholz C, Walter S. Biol Chem. 1996;377:417–424. [PubMed] [Google Scholar]
- 40.Uno T, Ku J, Prudent J R, Huang A, Schultz P G. J Am Chem Soc. 1996;118:3811–3817. [Google Scholar]