Abstract
The symbiotic association between the roseobacter Silicibacter sp. strain TM1040 and the dinoflagellate Pfiesteria piscicida involves bacterial chemotaxis to dinoflagellate-produced dimethylsulfoniopropionate (DMSP), DMSP demethylation, and ultimately a biofilm on the surface of the host. Biofilm formation is coincident with the production of an antibiotic and a yellow-brown pigment. In this report, we demonstrate that the antibiotic is a sulfur-containing compound, tropodithietic acid (TDA). Using random transposon insertion mutagenesis, 12 genes were identified as critical for TDA biosynthesis by the bacteria, and mutation in any one of these results in a loss of antibiotic activity (Tda−) and pigment production. Unexpectedly, six of the genes, referred to as tdaA-F, could not be found on the annotated TM1040 genome and were instead located on a previously unidentified plasmid (ca. 130 kb; pSTM3) that exhibited a low frequency of spontaneous loss. Homologs of tdaA and tdaB from Silicibacter sp. strain TM1040 were identified by mutagenesis in another TDA-producing roseobacter, Phaeobacter sp. strain 27-4, which also possesses two large plasmids (ca. 60 and ca. 70 kb, respectively), and tda genes were found by DNA-DNA hybridization in 88% of a diverse collection of nine roseobacters with known antibiotic activity. These data suggest that roseobacters may use a common pathway for TDA biosynthesis that involves plasmid-encoded proteins. Using metagenomic library databases and a bioinformatics approach, differences in the biogeographical distribution between the critical TDA synthesis genes were observed. The implications of these results to roseobacter survival and the interaction between TM1040 and its dinoflagellate host are discussed.
Bacteria of the Roseobacter clade of marine Alphaproteobacteria stand out as some of the most critical players in the oceanic sulfur cycle due to the ability of several genera to degrade dimethylsulfoniopropionate (DMSP) (37, 49). Although roseobacters are widespread throughout the marine ecosystem, their abundance is significantly correlated with DMSP-producing algae, especially prymnesiophytes and dinoflagellates, such as Prorocentrum, Alexandrium, and Pfiesteria species (1, 14, 27).
Our laboratory has been studying the interaction of a roseobacter, Silicibacter sp. strain TM1040, and Pfiesteria piscicida (1, 33-36). Silicibacter sp. strain TM1040 (hereafter referred to as TM1040) was originally isolated from a laboratory microcosm culture of the heterotrophic DMSP-producing dinoflagellate P. piscicida (33). Marine algae are the major producers of DMSP in the marine environment (18), whereas bacteria, and specifically members of the Roseobacter clade, are largely responsible for DMSP catabolism (49). TM1040 degrades DMSP via a demethylation pathway producing 3-methylmercaptopropionate as a major breakdown product (33). The bacteria respond via chemotaxis to dinoflagellate homogenates and are specifically attracted to DMSP, methionine, and valine (35). Experimental evidence has shown that TM1040 motility is important in the initial phases of the symbiosis (34). Once the bacteria are in close proximity to their host, TM1040 forms a biofilm on the surface of the dinoflagellate (1, 7, 34). Thus, the symbiosis may be divided into two parts: one that involves chemotaxis and motility and a second step in which a biofilm predominates.
We have recently reported on specific phenotypes, e.g., the ability to produce antibacterial compounds and biofilm formation, that may give members of the Roseobacter clade a selective advantage and help to explain the dominance of members of this clade in association with marine algae (7). Specifically, the production of an antibiotic activity is commonly observed in roseobacters and is hypothesized to provide an advantage when colonizing phytoplanktonic hosts, such as dinoflagellates (7). The genome of TM1040 consists of a 3.2-Mb chromosome and two plasmids, pSTM1 (823 kb) and pSTM2 (131 kb) (36). A comparison between TM1040 and two other roseobacters (Silicibacter pomeroyi DSS-3 and Jannaschia sp. strain CSS-1) suggests that roseobacters have abundant and diverse transporters, complex regulatory systems, and multiple pathways for acquiring carbon and energy in seawater, with the potential to produce secondary, biologically active metabolites (36).
Biologically active metabolites, including antibacterial compounds, have been characterized from a few roseobacters. A sulfur-containing antibiotic compound, tropodithietic acid (TDA), has been isolated and chemically characterized from Phaeobacter sp. strain 27-4 (8), hereafter simply called 27-4, and Roseobacter sp. strain T5 (6). The chemical backbone of TDA (shown in Fig. 1) is a seven-member aromatic tropolone ring, which is highly significant since tropolone derivatives, notably hydroxylated forms, are widely seen as medically important sources of antibacterial, antifungal, antiviral, and antiparasitic agents (12, 38, 39). Components of the biosynthetic pathway leading to the production of thiotropocin, another tropothione derivative closely related to TDA, has been described by Cane et al. (13), who suggested that thiotropocin is synthesized from shikimate by an oxidative ring expansion of phenylacetic acid.
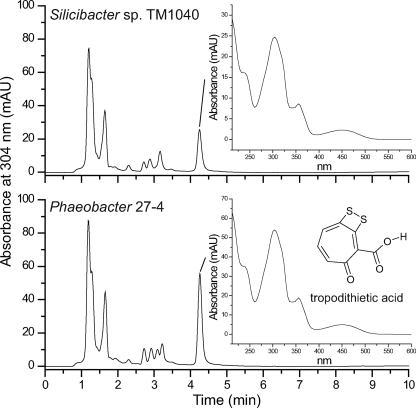
FIG. 1.
TDA. C18 reversed-phase HPLC chromatograms of ethyl acetate extracts from TM1040 and Phaeobacter sp. strain 27-4. Insets show the UV spectra of the HPLC peak corresponding to the antibiotic activity. For 27-4, the peak is TDA.
In the present study, we used both genomic and genetic techniques to uncover the genes and proteins required for TDA synthesis in TM1040 and 27-4 as models for the Roseobacter clade. In the process of locating these genes, we discovered a plasmid critical for TDA biosynthesis that is part of the TM1040 genome but escaped sequencing.
MATERIALS AND METHODS
Bacteria and media.
The strains used in the present study are listed in (Table 1). TM1040, 27-4, and Vibrio anguillarum 90-11-287 were routinely grown and maintained at 30°C in 2216 marine broth or 2216 agar as recommended by the manufacturer (BD Biosciences, Franklin Lakes, NJ). A marine basal minimal medium (MBM; 8.47g of Tris-HCl, 0.37 g of NH4Cl, 0.0022 g of K2HPO4, 11.6 g of NaCl, 6 g of MgSO4, 0.75 g of KCl, 1.47 g of CaCl2·2H2O, 2.5 mg of FeEDTA [pH 7.6], and 1 ml of RPMI 1640 vitamins [Sigma R7256] per liter) was used for determining carbon and sulfur requirements. Sole carbon sources were added at a final concentration of 1 g/liter. Escherichia coli strains were grown in Luria-Bertani broth (3) or on Luria-Bertani agar containing 1.5% Bacto agar (Becton Dickinson, Franklin Lakes, NJ). As appropriate, kanamycin was used at 120 μg per ml for Roseobacter strains and at 50 μg per ml for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype and/or phenotype | Source or reference(s) |
|---|---|---|
| Escherichia coli | ||
| DH5α | F−endA1 hsdR17 (rK− mK−) supE44 thi-1 recA1 gyrA96 relA1 φ80dlacZΔM15 | 44 |
| DH5α(λpir) | DH5Ω transduced with λpir | |
| EC100D pir+ | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ−rpsL nupG pir+(DHFR) | Epicentre |
| Roseobacters | ||
| Silicibacter sp. strain TM1040 | Wild type, antibacterial activity | 33 |
| Mutants derived from TM1040 | ||
| Silicibacter sp. strain TM1040 SM | No pigment and tda spontaneous strain | This study |
| HG1005 | paaK::EZ-Tn5,Kan | This study |
| HG1015 | tdaB::EZ-Tn5,Kan | This study |
| HG1050 | tdaF::EZ-Tn5,Kan | This study |
| HG1056 | paaJ::EZ-Tn5,Kan | This study |
| HG1080 | tdaC::EZ-Tn5,Kan | This study |
| HG1110 | tdaD::EZ-Tn5,Kan | This study |
| HG1213 | malY::EZ-Tn5,Kan | This study |
| HG1220 | cysI::EZ-Tn5,Kan | This study |
| HG1244 | tdaH::EZ-Tn5,Kan | This study |
| HG1265 | tdaE::EZ-Tn5,Kan | This study |
| HG1299 | paaI::EZ-Tn5,Kan | This study |
| HG1310 | tdaA::EZ-Tn5,Kan | This study |
| Phaeobacter sp. strain 27-4 | Wild type, antibacterial activity | 7, 21 |
| Mutants derived from 27-4 | ||
| JBB1001 | tdaB::EZ-Tn5,Kan | This study |
| JBB1003 | tdbC::EZ-Tn5,Kan | This study |
| JBB1005 | traI::EZ-Tn5,Kan | This study |
| JBB1006 | clpX::EZ-Tn5,Kan | This study |
| JBB1007 | tdbF::EZ-Tn5,Kan | This study |
| JBB1009 | tdbA::EZ-Tn5,Kan | This study |
| JBB1011 | tdbD::EZ-Tn5,Kan | This study |
| JBB1029 | tdbE::EZ-Tn5,Kan | This study |
| JBB1030 | tdaA::EZ-Tn5,Kan | This study |
| JBB1044 | metF::EZ-Tn5,Kan | This study |
| JBB1045 | tdbB::EZ-Tn5,Kan | This study |
| Other Roseobacter spp. | ||
| Roseobacter algicola 51442 | Wild type, no antibacterial activity | 7, 27 |
| Roseobacter denitrificans 33942 | Wild type, no antibacterial activity | 7, 47 |
| Roseobacter litoralis 49566 | Wild type, no antibacterial activity | 7, 47 |
| Roseobacter sp. strain TM1038 | Wild type, antibacterial activity | 7, 33 |
| Roseobacter sp. strain TM1039 | Wild type, antibacterial activity | 7, 33 |
| Roseovarius sp. strain ISM | Wild type, antibacterial activity | |
| Roseovarius sp. strain TM1035 | Wild type, antibacterial activity | 7, 33 |
| Roseovarius sp. strain TM1042 | Wild type, antibacterial activity | 7, 33 |
| Silicibacter pomeroyi strain DSS-3 | Wild type, antibacterial activity | 7, 20 |
| Sulfitobacter sp. strain 1921 | Wild type, no antibacterial activity | 7 |
| Sulfitobacter sp. strain EE36 | Wild type, antibacterial activity | 7, 9 |
| Sulfitobacter sp. strain SE62 | Wild type, no antibacterial activity | 7, 11 |
| Vibrio anguillarum 90-11-287 | Wild type, serotype O1, susceptible to TDA | 7, 48 |
| Plasmids | ||
| pSTM3 | Harboring tda genes | This study |
| pSTM3-1265 | pSTM3 carrying a Tn5 insertion in tdaE, derived from HG1265 | This study |
Characterization of antibiotic.
Bacteria were incubated in 2216 broth for 16 h at 30°C, after which the cells were removed first by centrifugation (10,000 × g) and then by filtration through a 0.22-μm-pore-size membrane (mixed-cellulose-ester membrane; Millex; Millipore, Bedford, MA), resulting in cell-free spent medium. Bacterial spent medium was either injected directly (up to 10 μl) or purified by mixed-phase anion-exchange reversed phase mini-column chromatography on Oasis MAX columns as previously described (8). TDA was analyzed by reversed-phase liquid chromatography (LC) on an Agilent 1100 high-pressure liquid chromatography (HPLC) system equipped with a diode array detector (DAD). Separation was conducted using a Phenomenex (Torrance, CA) Curosil PFP column (15 cm, 2 mm, 3 μm) using a water-acetonitrile gradient system. Both solvents contained 200 μl of trifluoroacetic acid/liter and started at 35% acetonitrile, increasing linearly to 60% in 6 min. A wavelength of 304 ± 4 nm was used for detection. LC-DAD with online high-resolution mass spectrometry using positive and negative electrospray was used for validation of the TDA detection as previously described (8).
Transposon mutagenesis and Tda− screening.
Electrocompetent roseobacter strains were prepared according to the method of Garg et al. (19) as modified by Miller and Belas (34). Random transposon insertion libraries were constructed in TM1040 and 27-4 using an EZ-Tn5<R6Kγori/KAN-2>Tnp transposome kit (Epicentre, Madison, WI). Strains were spread onto 2216 plates containing kanamycin and incubated for 1 day at 30°C. Individual kanamycin-resistant (Kanr) transposon insertion strains were transferred to 7×7 arrays on 2216 marine agar plus kanamycin to facilitate further screening. To screen for loss-of-function, antibiotic-negative (Tda−) mutants, a modification of the method described by Bruhn et al. (8) was used. Bacteria were replicated, as a 7×7 array, to a lawn of Vibrio anguillarum strain 90-11-287 (7, 8) and incubated at 20°C for 24 h, after which a zone of clearing indicative of antibiotic production was measured and compared to the parental strain (TM1040 or 27-4). For the present study, Tda− is defined as a strain lacking a detectable zone of clearing on V. anguillarum. Strains determined to be Tda− by the modified well diffusion assay were further tested by incubation at 30°C for 48 h in 2216 marine broth without shaking. Bacteria were removed by filtering through a 0.22-μm-pore-size mixed-cellulose-ester membrane, and the antibacterial activity of the supernatant was measured by using the V. anguillarum well diffusion assay as described by Bruhn et al. (7, 8).
Sole carbon and sulfur source growth.
Bacterial utilization of sole carbon sources was determined by measuring growth in MBM broth that was modified by replacing glycerol with the carbon source to be tested. The carbon compounds tested included amino acids (alanine, arginine, aspartic acid, cysteine, glutamic acid, glycine, histidine, isoleucine, leucine, methionine, phenylalanine, proline, serine, threonine, tryptophan, tyrosine, and valine), sugars (arabinose, fructose, galactose, glucose, lactose, maltose, mannose, N-acetylglucosamine, ribose, sucrose, and xylose), and tricarboxylic acid (TCA) cycle intermediates (citrate, fumurate, and succinate), as well as phenylacetic acid and sodium phenylpyruvate.
Sulfur utilization was tested by growth in modified MBM lacking sulfate and containing 10 mM DMSP, cysteine, methionine, sodium sulfate, or sodium sulfite as a sole sulfur source.
Bioinformatics analysis.
Approximately 1 μg of genomic DNA isolated from the candidate mutant was digested with NcoI, self-religated with T4 DNA ligase, and electroporated into DH5α (λpir). After selection for kanamycin resistance, Kanr colonies were picked, and the plasmid was isolated for bidirectional sequencing with transposon-specific primers as recommended by the supplier (Epicenter). The nucleotide sequence thus obtained was analyzed by BLAST analyses using DNA-DNA homology searches against the Silicibacter sp. strain TM1040 genome (accession numbers NC_008044, NC_008043, and NC_008042). The genes identified are listed in Table 2 for TM1040 and Table 3 for 27-4. Signature amino acid domains in the deduced amino acid sequence of the respective open reading frames (ORFs) were identified using BLASTP (2), Pfam (17), SMART (28), and the Conserved Domains Database (CDD; http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). Homologs in roseobacters were identified by using BLASTP analysis of Roseobase (http://www.roseobase.org) and Gordon and Betty Moore Foundation Marine Microbial Genome databases (https://research.venterinstitute.org/moore) with respective predicted protein sequence as the query sequence and a maximum E value of 1E-30. Homologs in the Global Ocean Sampling (GOS) Expedition metagenomic libraries (http://camera.calit2.net/index) (43) were identified by BLASTP analysis using a cutoff E value 1E-20.
TABLE 2.
Silicibacter sp. strain TM1040 genes and encoded proteins required for the regulation and synthesis of TDA
| Function and gene no. | GenBank accession no. | Gene | Function | Best hit ortholog/E score |
|---|---|---|---|---|
| Ring precursors, oxidation, and expansion | ||||
| TM1040_3728 | CP000376 | paaK | Phenylacetate oxidoreductase | Roseobacter sp. strain MED193 phenylacetic acid degradation oxidoreductase PaaK/8e-161 |
| TM1040_3726 | CP000376 | paaI | Phenylacetate oxygenase | Roseobacter sp. strain MED193 phenylacetic acid degradation protein PaaI/4e-110 |
| TM1040_3727 | CP000376 | paaJ | Phenylacetate oxygenase | Roseobacter sp. strain MED193 phenylacetic acid degradation protein PaaJ/2e-69 |
| EF139203 | EF139203 | tdaD | 4-Hydroxybenzoyl-CoA thioesterase | Paracoccus denitrificans PD1222 conserved hypothetical protein/2e-45 |
| EF139204 | EF139204 | tdaE | ACAD | Paracoccus denitrificans PD1222 ACAD/9e-120 |
| EF139201 | EF139201 | tdaB | β-Etherase, GST | Paracoccus denitrificans PD1222 putative β-etherase (β-aryl ether cleaving enzyme) protein/6e-56 |
| EF130202 | EF130202 | tdaC | Prephenate dehydratase | Paracoccus denitrificans PD1222 hypothetical protein/2e-45 |
| Sulfur metabolism and addition | ||||
| TM1040_2581 | CP000377 | malY | β-C-S lyase (cystathionase); amino transferase | Roseobacter sp. strain MED193 aminotransferase, classes I and II/0.0 |
| TM1040_0961 | CP000377 | tdaH | Sulfite oxidase domain protein | Sulfitobacter sp. strain NAS-14.1 hypothetical protein/7e-34 |
| TM1040_1758 | CP000377 | cysI | Sulfite reductase | Roseobacter sp. strain MED193 sulfite reductase/0.0 |
| CoA metabolism | ||||
| EF139205 | EF139205 | tdaF | Phosphopantothenoylcysteine decarboxylase | Paracoccus denitrificans PD1222 flavoprotein/2e-55 |
| Regulatory mechanism | ||||
| EF139200 | EF139200 | tdaA | LysR substrate-binding domain protein | Paracoccus denitrificans PD1222 regulatory protein, LysR:LysR, substrate binding/1e-29 |
TABLE 3.
Sole carbon source tested for TM1040 and mutants
| Gene or WT | Presence (+) or absence (-) of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Cys | Trp | Phe | Phenylacetic acid | Sodium phenylpyruvate | Sodium phenylbutyrate | 2216 | Other amino acid | |
| WT | + | + | + | + | + | + | + | + |
| paaI | + | - | - | - | - | - | + | + |
| paaJ | + | - | - | - | - | - | + | + |
| paaK | + | - | - | - | - | - | + | + |
| tdaA | + | + | + | + | + | + | + | + |
| tdaB | + | + | + | + | + | + | + | + |
| tdaC | + | + | + | + | + | + | + | + |
| tdaD | + | + | + | + | + | + | + | + |
| tdaE | + | + | + | + | + | + | + | + |
| tdaF | + | + | + | + | + | + | + | + |
| cysI | + | - | - | - | - | - | + | - |
| malY | + | + | + | + | + | + | + | + |
| tdaH | + | + | + | + | + | + | + | + |
DNA extraction and separation.
Chromosomal DNA was extracted from bacterial cells by routine methods (3) or by using a DNeasy blood and tissue kit (Qiagen, Valencia, CA). Plasmid DNA was prepared by the alkaline lysis method (3) and digested with NcoI (New England Biolabs, Beverly, MA), and the resulting restriction fragments were separated by agarose gel electrophoresis in Tris-acetate-EDTA buffer.
Pulsed-field gel electrophoresis (PFGE) was performed by using a CHEF DR-III clamped homogeneous electric field system (Bio-Rad, Richmond, CA) with a 1% agarose gel, a 3- to 15-s pulse ramp, an electrophoresis rate of 6.0 V/cm with an included angle of 120° at a constant temperature of 14°C, and a run time of 26 h. Gels were stained with ethidium bromide and visualized with a Typhoon 9410 (Amersham Biosciences, Piscataway, NJ).
PCR amplification.
Multiplex PCR amplification was used to screen for the presence of tda genes in Tda− mutants. A 716-bp sequence internal to tdaE was amplified by using the primers 5′-CAGATGATGGTGCCAAAGGACTAT-3′ and 5′-GGTCAGTTTCTTCTGCACATACTGG-3′, while (in the same reaction) an internal 401-bp fragment of flaA (accession number CP000377, locus tag TM1040_2952) was also amplified by using the primers 5′-TTGCAGTATCCAATGGTCGTG-3′ and 5′-TGAATTGCGTCAGAGTTTGCC-3′ as a control. The standard PCR amplification conditions were 100 μM concentrations of each deoxynucleoside triphosphate, 0.2 μM concentrations of each primer, and 1 U of Taq DNA polymerase (New England Biolabs) in 1× reaction buffer (New England Biolabs) with an initial denaturing step at 94°C for 3 min, followed by 30 cycles of 94°C for 1 min each, annealing at 55°C for 30 s, and an elongation at 72°C for 1 min.
To detect the tdaA-E locus, PCR amplification was conducted with a forward primer complementary to tdaA (5′-CGCTTTCCGGAACTGGAGAT-3′) and a reverse primer complementary to tdaE (5′-GGCTGCCGTATAGTTTCAGCA-3′) using the Expand Long Template PCR system (Roche Applied Science, Indianapolis, IN), and the PCR program conditions and cycle parameters were as described by the supplier.
DNA hybridization.
DNA-DNA hybridization by Southern slot blot (3) was used to detect the presence of tda genes in other roseobacters. The roseobacter strains used were Phaeobacter sp. strain 27-4, Roseobacter algicola ATCC 51442, Roseobacter denitrificans ATCC 33942, Roseobacter litoralis ATCC 49566, Roseobacter sp. strain TM1038, Roseobacter sp. strain TM1039, Roseovarius sp. strain TM1035, Roseovarius sp. strain TM1042, Roseovarius sp. strain ISM, Silicibacter pomeroyi DSS-3, Silicibacter sp. strain TM1040, Sulfitobacter sp. strain EE36, Sulfitobacter sp. strain 1921, Sulfitobacter sp. strain SE62, and Vibrio anguillarum 90-11-287. After extraction, 100 ng of total genomic DNA purified from each strain was spotted onto a positively charged nylon membrane (Roche). The DNA was cross-linked to the membrane with UV light by using a Stratalinker UV cross-linker (Stratagene, La Jolla, CA), followed by prehybridization of the membrane at 25°C for 30 min, using a DIG High Prime DNA Labeling and Detection Starter Kit II (Roche) as described by the manufacturer. The membrane was incubated at 25°C overnight with a double-stranded DNA probe prepared by HindIII digestion of a plasmid bearing tdaA cloned from strain HG1310 that was labeled with digoxigenin-dUTP using random priming as recommended by the manufactures (Roche). Unbound labeled DNA was removed from the membrane by two 5-min treatments in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate, followed by two 15-min treatments in 0.2× SSC-0.1% sodium dodecyl sulfate (3). In the Southern blot, the membrane was prehybridized for 30 min in the same buffer, to which was added a tdaE gene probe, and the probe was allowed to hybridize overnight at 42°C. The blots were washed under high-stringency conditions according to the manufacturer's protocol (Roche) and exposed to Lumi-Film chemiluminescent detection film (Roche) for subsequent detection of the hybridization signal.
RESULTS
TM1040 produces the sulfur-containing antibiotic TDA.
In a previous report (7), we showed that TM1040 produces an extracellular broad spectrum antibacterial compound capable of inhibiting or killing many bacteria. In continuing these studies, we found that greater antibacterial activity occurred when the bacteria were grown in a nutrient broth culture under static conditions. The clearing zone used to assess antibiotic production was 11 mm greater in samples tested from cultures that were not shaken compared to those that were. Under static conditions, TM1040 cells attached to one another forming rosettes and produced a distinct yellow-brown pigment. These phenotypes are consistent with those described for Phaeobacter sp. strain 27-4 (8) and other roseobacters (7). Nonpigmented colonies were sometimes seen after TM1040 was incubated on nutrient agar, and subsequent analysis revealed that these white spontaneous mutants also had lost antibacterial activity.
TM1040 produces an antibiotic and shares phenotypic traits with other roseobacters, notably 27-4, whose antibiotic has been identified as TDA (8). To compare the antibacterial compound produced by TM1040 to TDA, cell-free supernatants from TM1040 and 27-4 were analyzed by HPLC using previously described methods (8). A peak from TM1040 had the same retention time as the TDA peak from 27-4 (Fig. 1, 4.2 min). The UV spectra corresponding to both peaks were the same as the published spectrum of TDA (Fig. 1, insets) (8, 29). Mass spectroscopy analysis of this compound from TM1040 was also consistent with the conclusion that TDA is the antibacterial metabolite produced by TM1040 (data not shown).
Identification of genes involved in the synthesis of TDA.
With the exception of some genes involved in shikimate and phenylacetate metabolism (36), analysis of the genome sequence of TM1040 does not suggest genes likely to participate in the biosynthesis and regulation of TDA. To detect such genes, a random-insertion transposon bank of 11,284 Kanr colonies was generated in TM1040 and screened for the Tda− phenotype that indicates the loss of antibiotic production. Approximately 0.7% of the transposon mutants (81 of 11,284) were defective in both TDA synthesis and pigment formation.
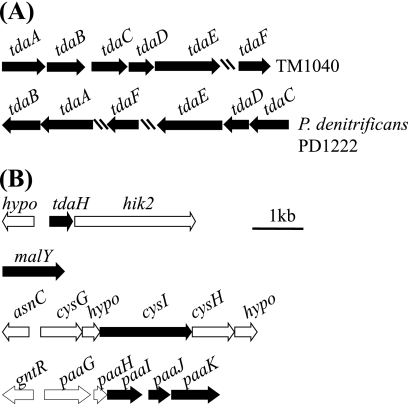
To help identify the genetic basis of the phenotype, TM1040 DNA adjacent to each side of the transposon was sequenced in all 81 of the Tda− mutants. Surprisingly, the transposon-associated sequences from 32 or nearly 40% of the Tda− mutants did not match DNA sequence in the annotated TM1040 genome (36). However, the newly identified sequences overlapped and were assembled into one large contiguous DNA fragment of 4.5 kb harboring at least six ORFs, designated tdaA to tdaF (Table 2 and Fig. 2A). These genes are not part of the original annotation of the genome, suggesting that this DNA may have been lost from the sequenced variant of TM1040. Below we present a thorough analysis of these “orphan” genes that were later found to be involved in TDA biosynthesis and to reside on a 130-kb plasmid.
FIG. 2.
Genes required for synthesis of TDA in TM1040. The black boxes indicate the ORF interrupted by the transposon. Arrows indicate ORFs transcriptional orientations, hatch marks indicate a break in the region, and the relative distance is indicated by the 1-kb marker. (A) tdaA∼tdaF genes reside on a plasmid, with their closest homologs found on the chromosome of P. denitrificans PD1222. An intergenic space of 54 bp separates TM1040 tdaA and tdaB, and 345 bp separate tdaB from tdaC-E, which overlap each other by one bp, and >10 kb separate tdaF from tdaE. (B) The remainder of the genes involved in TDA biosynthesis are located either on the chromosome (tdaH, malY, and cysI) or, in the case of the genes involved in phenylacetate catabolism (paaIJK), on plasmid pSTM1.
DNA adjacent to the transposons in 49 Tda− mutants matched the available genome sequence. In these strains we assessed the presence of tdaE with PCR and could not detect an amplification product in 43 of the 49 mutants. The loss of plasmid-borne tdaA-F might cause their Tda− phenotype, and this type of loss might also account for low-frequency spontaneous loss of TDA synthesis (estimated at <10−5 cells). These mutants were not investigated further.
In three of the six mutants that retained tdaE, the transposons disrupted putative genes encoding phenylacetate catabolism, paaI, paaJ, and paaK (Fig. 2B). Their deduced amino acid sequences were similar to homologs in other roseobacters (Table 2). In other bacteria, paaGHIJK encodes a ring-hydroxylating complex of proteins that is responsible for the first step in the aerobic catabolism of phenylacetate involving coenzyme A (CoA) activation (31, 40), producing 1, 2-dihydro-phenylacetate-CoA (16, 23). The loss of TDA synthesis from disruption of the paa genes supports the biochemical evidence of phenylacetate metabolism in thiotropocin synthesis published in 1992 by Cane et al. (13).
Mutants with defects in phenylacetate metabolism were also unable to grow on phenylalanine, phenylacetic acid, tryptophan, sodium phenylpyruvate, or phenylbutyrate as a sole carbon source (Table 3). This result is consistent with the hypothesis that paaIJK of TM1040 function in the phenylacetate catabolism pathway similarly to other bacteria (36).
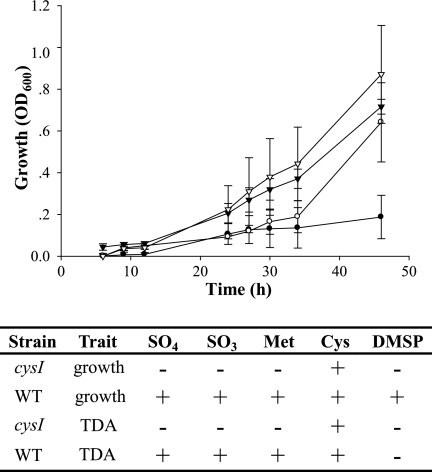
TDA is a disulfide-modified tropolone compound, indicating that sulfur metabolism must be involved in TDA synthesis. This idea is supported by the identification of three Tda− mutants (Table 2), each with a transposon inserted in a gene whose product is involved in sulfur metabolism: cysI, malY, and an ORF (tdaH) with homology to sulfite oxidase (Table 2). The identification of these genes suggests that sulfur from reductive sulfur pathways is used and incorporated into TDA, which was tested by observing the growth of the sulfur metabolism mutants on a minimal medium containing a sole sulfur source (see Materials and Methods). The results are shown in Fig. 3. The cysI mutant grew when provided complex sulfur sources or cysteine and was unable to utilize DMSP, SO32−, SO42−, or methionine. The addition of cysteine to the medium resulted in enhanced growth of the cysI mutant, as well as increased synthesis of TDA (Fig. 3).
FIG. 3.
Growth and TDA synthesis are affected by mutations in cysI. TM1040 (inverted triangles) and a cysI mutant (HG1220; circles) were grown in minimal medium lacking sulfate and containing either methionine (closed symbols) or cysteine (open symbols), and growth was measured optically at 600 nm. Unlike the wild-type, the CysI− mutant cannot grow on methionine, but does utilize cysteine. Measurement of antibiotic activity indicates that the cysI defect also affects TDA synthesis, which is corrected by the addition of cysteine to the medium but not by the addition of methionine, DMSP, sulfite, or sulfate.
TDA biosynthesis genes resided on a 130-kb plasmid.
A bioinformatic analysis was done on TdaA-F to help elucidate the potential function of these proteins (Table 2). Interestingly, these proteins share their strongest homology with a similar set of proteins encoded by chromosome 1 of Paracoccus denitrificans PD1222 (accession no. NC_008686), a nonmotile alphaproteobacterium first isolated from soil by Beijerinck (4). TdaA (Table 2) has homology with LysR regulatory proteins, possessing a helix-turn-helix DNA-binding domain and a LysR substrate-binding domain (15, 45). TdaA was the only regulatory protein detected in the present study, perhaps indicating that it is the sole regulator of TDA synthesis. The remaining ORFs encode putative enzymes. TdaB contains a glutathione S-transferase (GST) domain and belongs to the bacterial GST protein family (Table 1). TdaC has an amino acid domain with homology to prephenate dehydratase (PheA), an enzyme involved in the conversion of chorismate to prephenate, a step in the pathway leading to phenylacetate synthesis (50).
The involvement of CoA metabolism, addition, or modification is evident from the functional domains on TdaD and TdaE. TdaD is predicted to be a member of the thioesterase superfamily of acyl-CoA thioesterases (Table 2) (5), TdaE encodes a putative acyl-CoA dehydrogenase (ACAD) (24), and TdaF has homology to aldehyde dehydrogenase (26).
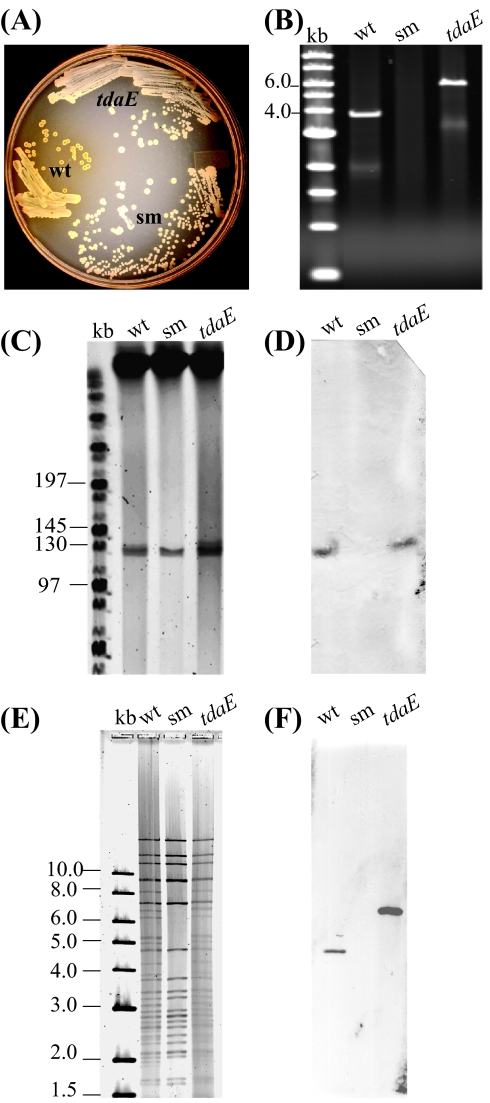
The secondary evidence suggests that tdaA-F reside on a “cryptic” plasmid that may be spontaneously lost. To develop a means to test the hypothesis, we used three strains, TM1040, a spontaneous Tda− nonpigmented strain of TM1040 (TM1040SM), and HG1265 (tdaE:Tn) (Fig. 4A and Table 1), along with a PCR amplification using primers for tdaA-E, predicted to generate a 3.8-kp product from wild-type DNA. As shown in Fig. 4B, PCR amplification of wild-type DNA gave the predicted 3.8-kb band, a 5.7-kp product when tdaE:Tn DNA was used as a template, and no product when the DNA from the SM strain was amplified, indicating that the SM strain had lost the tdaA-E locus.
FIG. 4.
TM1040 tda genes reside on a plasmid that undergoes a low frequency spontaneous loss. (A) Pigment synthesis. TM1040 (wt) produces a yellow-brown extracellular pigment that is correlated with TDA synthesis. In contrast, a tdaE:Tn mutant (strain HG1265) and a spontaneous mutant (sm; TM1040SM) are nonpigmented and have lost the ability to produce both TDA and pigment. (B) Spontaneous loss of pigment and antibiotic activity results from a loss of tda genes. PCR amplification of tdaE results in a band from wild-type (wt) and tdaE:Tn DNA, respectively, with the additional 2 kb in size of the tdaE:Tn product resulting from insertion of the transposon. No product was amplified from the spontaneous nonpigmented mutant (sm). (C) PFGE separation of total DNA obtained from TM1040 (wt), the spontaneous nonpigmented mutant (sm), and the tdaE:Tn mutant. (D) Southern blot hybridization of the PFGE gel to labeled tdaE DNA. (E) NcoI digestion of plasmid DNA isolated from TM1040 (wt), the spontaneous nonpigmented mutant (sm), and HG1265 (tdaE:Tn), respectively. The resulting patterns of DNA bands were compared to each other and to an in silico NcoI digestion of pSTM2 (see Fig. S1 in the supplemental material). (F) Southern blot hybridization of NcoI-digested plasmid DNA to tdaE.
Total DNA from TM1040, TM1040SM, and HG1265 (tdaE:Tn) was separated by PFGE. As observed in Fig. 4C, all three strains had high-molecular-weight DNA, presumably a mixture of chromosomal and pSTM1, and a band or bands at ca. 130 kb, corresponding to the size of pSTM2 (132 kb) (36). Close inspection of this region and comparison between the SM DNA lane (middle, Fig. 4C) and either the TM1040 or tdaE:Tn DNA (left and right lanes, respectively) shows that the SM band is thinner than either TM1040 or tdaE:Tn, hinting that SM DNA is missing a DNA species in this size range that overlaps with pSTM2. Repeated attempts to change PFGE conditions failed to resolve this region further. To overcome this limitation, a Southern blot (Fig. 4D) using a tdaE DNA probe was performed on the gel shown in Fig. 4C, and the results confirmed that the SM DNA, while possessing a 130-kb band, fails to hybridize to tdaE. In contrast, both wild-type DNA and tdaE:Tn DNA hybridizes to the expected band (ca. 130 kb). This confirms the loss of tda DNA in SM and adds evidence supporting the idea that the missing tda DNA is on a plasmid. It does not rule out the (unlikely) possibility that tda genes reside on pSTM2 and are somehow deleted from that known molecule.
To resolve the issue, we isolated plasmids from each of the three strains (TM1040, TM1040SM, and HG1265) and subjected each mixture to NcoI digestion (Fig. 4E), chosen because an in silico NcoI digestion of pSTM2 provided a recognizable pattern of DNA fragments. As can be seen in Fig. 4E, the TM1040SM DNA digest had much fewer bands than wild-type DNA or DNA from tdaE:Tn. This would be expected if the TM1040SM strain lost a large plasmid. Consistent with the hypothesis, Southern blotting showed that a tdaE probe hybridized to a 4.5-kb fragment in wild-type plasmid DNA and to a 6.4-kb fragment from plasmids isolated from the tdaE:Tn strain (Fig. 4E).
The EZ:Tn transposon contains a kanamycin resistance gene, as well as the oriR6K origin of replication, permitting replication in permissive hosts carrying the pir gene (25). Thus, the plasmid from tdaE:Tn was used to transform E. coli EC100D (Table 1) with a subsequent selection for kanamycin resistance (see Materials and Methods). This transformation was successful despite a very low transformation efficiency, resulting in 7 CFU per μg of mixed plasmid DNA, and provides strong evidence for the existence of an ∼130-kb plasmid harboring tda genes. This new plasmid is called pSTM3.
Twelve random colonies were chosen from the transformation with pSTM3, and the NcoI digestion pattern of each was compared. Four common restriction digestion patterns emerged from this analysis (see Fig. S2 in the supplemental material). Although each plasmid was PCR positive for the tda genes (data not shown) and the set of four shared many common bands, they had remarkably different patterns, indicating that deletion and/or rearrangements had occurred during or after the transfer of pSTM3 to E. coli. The reason and molecular mechanism underlying these band pattern differences is not known; however, the sum of the results indicates that TM1040 harbors an ∼130-kb plasmid, pSTM3, that is essential for TDA and pigment biosynthesis and that may be spontaneously lost in laboratory culture.
Distribution of tda genes in other Roseobacter spp.
We used EZ:Tn to construct a 6,321-member library in 27-4 and screened these mutants for the Tda− phenotype. A total of 37 Tda− mutants were found, 12 of which were analyzed further. Two of the twelve ORFs mutated were similar to TdaA (identity 38%) and TdaB (identity 55%) from TM1040 (Table 4), suggesting that these two roseobacter types share a common TDA biosynthesis and regulation scheme. The remaining nine genes were not identified as important to TDA synthesis in TM1040 and had various degrees of homology to genes in the annotated TM1040 genome but, unlike TM1040, were not part of the phenylacetate or reductive sulfur pathways. The one exception was 27-4 metF (Table 4), which may possibly be involved in sulfur metabolism (46).
TABLE 4.
Phaeobacter sp. strain 27-4 genes and encoded proteins required for the regulation and synthesis of TDA
| Function and mutant no. | GenBank accession no. | Gene | Function | Best hit ortholog/E score |
|---|---|---|---|---|
| Ring precursors, oxidation, and expansion | ||||
| JBB1001/JBB1030 | EF139212 | tdaB | β-Etherase, GST | Sinorhizobium meliloti putative β-etherase (β-aryl ether cleaving enzyme/4e-52 |
| Sulfur metabolism and addition | ||||
| JBB1044 | EF139218 | metF | 5-Methyltetrahydrofolate-homocysteine S-methyltransferase | Silicibacter sp. strain TM1040 MetF protein/2e-77 |
| CoA metabolism | ||||
| JBB1009 | EF139215 | tdbA | d-β-Hydroxybutyrate dehydrogenase | Roseovarius sp. strain 217 d-β-hydroxybutyrate dehydrogenase/2e-32 |
| JBB1045 | EF139216 | tdbB | Phosphate acetyltransferase | Roseobacter sp. strain MED193 phosphate acetyltransferase/8e-81 |
| Transport: import and export | ||||
| JBB1003 | EF139213 | tdbC | Lytic transglycosylase, peptidase C14 | Roseobacter sp. strain MED193 hypothetical protein/6e-85 |
| JBB1005 | EF139221 | traI | TraI, type IV (Vir-like) secretion | Rhodobacter sphaeroides 2.4.1 TraI/5e-58 |
| JBB1011 | EF139222 | tdbD | Type I secretion target repeat protein | Roseobacter sp. strain MED193 type I secretion target repeat protein/8e-54 |
| JBB1029 | EF139216 | tdbE | Oligopeptide/dipeptide ABC transporter | Silicibacter sp. strain TM1040 binding-protein-dependent transport systems inner membrane component/6e-124 |
| Regulatory mechanism | ||||
| JBB1006 | EF139220 | clpX | ATP-dependent Clp protease | Silicibacter sp. strain TM1040 ATP-binding subunit ClpX/1e-47 |
| JBB1007 | EF139214 | tdbF | RNase D | Roseobacter sp. strain MED193 RNase D/6e-49 |
| JBB1030 | EF139217 | tdaA | LysR substrate-binding domain protein | Paracoccus denitrificans PD1222 regulatory protein, LysR:LysR, substrate-binding/3e-51 |
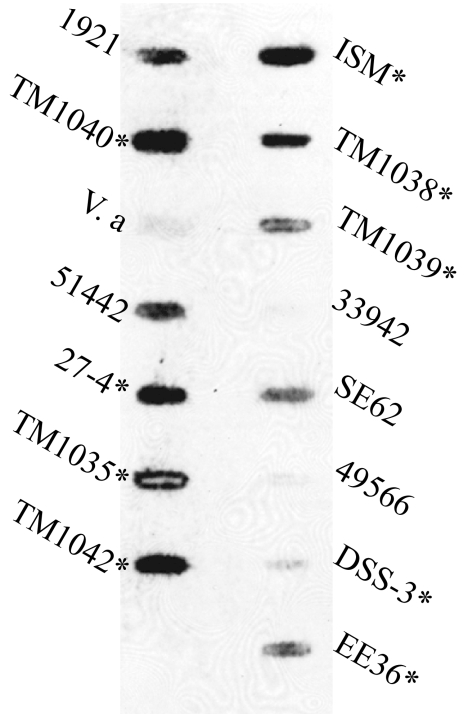
We also used DNA-DNA hybridization to measure the hybridization of a tdaA-F gene probe to DNA from 14 Roseobacter clade species (Fig. 5). The tda probe hybridized to eight of the nine roseobacter spp. that have been reported to produce antibacterial activity (Fig. 5), with the ninth, Silicibacter pomeroyi DSS-3, showing a low amount of hybridization. Three of six non-antibiotic-producing roseobacters also positively hybridized to the tda DNA. This may have resulted from a strain that has very low tda expression and antibiotic activity below the detection limits of the well diffusion assay or from spurious hybridization to non-tda DNA. The tda probe did not hybridize with DNA from V. anguillarum, implying that the second possibility is the more likely scenario.
FIG. 5.
DNA from other Roseobacter species hybridizes to tda DNA. Total DNA was extracted from 13 roseobacters, TM1040, and a nonroseobacter control species (V. anguillarum) and used in a slot blot hybridization with labeled tda DNA. Positive hybridization was strongly correlated with measurable antibiotic activity (indicated by an asterisk). The strains used were as follows: ISM, Roseovarius strain ISM; TM1038, Roseobacter sp. strain TM1038; TM1039, Roseobacter sp. strain TM1039; 33942, Roseobacter denitrificans ATCC 33942; SE62, Sulfitobacter strain SE62; 49566, Roseobacter litoralis ATCC 49566; DSS-3, Silicibacter pomeroyi DSS-3; EE36, Sulfitobacter strain EE36; 1921, Sulfitobacter strain 1921; TM1040, Silicibacter sp. strain TM1040; V.a, Vibrio anguillarum; 51442, Roseobacter algicola ATCC 51442; 27-4, Phaeobacter 27-4; TM1035, Roseovarius sp. strain TM1035; and TM1042, Roseovarius sp. strain TM1042.
Distribution of tda genes in the environment.
The marine genome and metagenomic databases were searched for sequences with homology to one of the twelve genes (Table 2) required for TDA synthesis by TM1040. Although homologs to the proteins involved in phenylacetate and reductive sulfur metabolism were quite commonly found within the 14 selected roseobacter genomes (Jannaschia sp. strain CCS1, Silicibacter pomeroyi DSS-3, Sulfitobacter sp. strain EE-36, Sulfitobacter sp. strain NAS-14.1, Sagittula stellata E-37, Rhodobacterales bacterium HTCC2654, Roseobacter sp. strain MED193, Roseovarius nubinhibens ISM, Loktanella vestfoldensis SKA53, Oceanicola batsensis HTCC2597, Oceanicola granulosus HTCC2516, Roseovarius sp. strain 217, Roseovarius sp. strain HTCC2601, and Roseovarius sp. strain TM1035) in Roseobase (http://www.roseobase.org) and the Gordon and Betty Moore Foundation Marine Microbial Genome databases (https://research.venterinstitute.org/moore), close homologs of TdaA-F were absent (at a BLASTP E value cutoff of 1E-30). Although the reason for the absence of homologs is not known, it is possible, although unlikely, that all 14 roseobacters do not produce TDA, produce an antibacterial activity that involves another compound, or lost their tda plasmid. The last possibility is most likely to have resulted from laboratory culturing; therefore, we searched for Tda homologs in the GOS metagenomic database (http://camera.calit2.net) (43) that should contain abundant uncultivated roseobacter DNA.
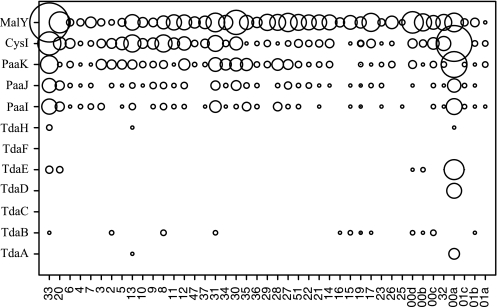
The data gathered from searching the GOS data set database are shown graphically in Fig. 6, where a circle and its relative size indicates the presence and abundance (respectively) of a given protein. As observed with the roseobacter genomes, phenylacetate and reductive sulfur metabolism proteins were readily found at numerous sites, with the greatest abundance of PaaIJK and CysI at site GS00a, a Sargasso Sea sample (31°32′6"N, 63°35′42"W). Positive Tda protein “hits” were also recorded in a hypersaline pond sample (GS033) and a sample obtained from Lake Gatun, Panama Canal (Fig. 6). In no sample did we find hits to all 12 proteins involved in TDA biosynthesis.
FIG. 6.
Presence and relative abundance of each of the Tda proteins identified in TM1040 (rows) in the GOS metagenomic database. Relative abundance is indicated by the size of the circle. GOS sample numbers are indicated on the horizontal axis.
DISCUSSION
It is not surprising members of the Roseobacter clade, whose genomes reveal a great potential for the synthesis of bioactive molecules (10, 32), produce TDA. Many marine bacteria produce an antibiotic activity (30, 36), and there are numerous reports of antibacterial activity from roseobacters, including a compound that produces a probiotic effect on scallop larvae (41, 42) and is antagonistic to gammaproteobacterium strains (41), as well as a compound that is antagonistic to fish larval bacterial pathogens (21, 22). From our data, it is likely that much of the antibiotic activity seen in roseobacters may be due to plasmid-borne tda genes.
TDA activity and biosynthesis depends on culture conditions and the physiology of TM1040. Bruhn et al. (7) have shown that TDA activity is significantly enhanced when TM1040 is cultured in a static nutrient broth, a condition that accentuates rosette and biofilm formation, as well as the synthesis of TDA and pigment. We have divided the symbiosis into two phases: the motile phase in which TM1040 cells actively respond to dinoflagellate-derived molecules by swimming toward the host, and the sessile phase, whereupon having located the zoospore, the bacteria cease motility and form rosettes and a biofilm on the surface of the dinoflagellate (1, 34, 35). Thus, there is a direct correlation between rosette and biofilm formation, pigment production, and TDA biosynthesis, all of which may affect the symbiosis.
There is a direct link between the spontaneous appearance of nonpigmented Tda− colonies and the loss of pSTM3 of TM1040. As we have reported, more than 40 of the mutants initially screened as Tda− were ultimately found to have lost pSTM3. This suggests that pSTM3 is lost at a relatively low frequency during laboratory cultivation of TM1040. Instability of the Tda+ phenotype is not unique to TM1040. The appearance of spontaneous nonpigmented Tda− mutants or variants has been observed in other roseobacters, including Phaeobacter sp. strain 27-4 (7) and Roseobacter gallaeciensis T5 (6). The simplest explanation for the cause of these spontaneous mutants is a loss of a plasmid carrying one or more critical genes required for TDA synthesis. Indeed, 27-4 possesses at least two plasmids of ca. 60 and 70 kb, respectively (data not shown). We speculate that one or both of these plasmids may be involved in the TDA biosynthesis of 27-4, and tdaA and tdaB, identified by transposon insertion mutagenesis in 27-4 Tda− mutants, reside on one of these plasmids (data not shown). It is also worth noting that the transformation of E. coli with pSTM3 resulted in instability of the plasmid and the apparent loss or rearrangement of plasmid DNA sequences when in the foreign host (see Fig. S2 in the supplemental material). The nucleotide sequence of pSTM3 is currently under way in our laboratory. Preliminary data indicate that pSTM3 harbors a repC that is distinct from the repC found on pSTM2, which further supports the existence of pSTM3 as a discrete DNA, separate from pSTM2.
One of the unexpected results from our study is the paucity of Tda homologs in the genomes of other sequenced roseobacters. There are several possible explanations why Tda homologs may be difficult to find, but the strongest lines of evidence support the idea that tda genes and Tda proteins have poorly conserved sequences, which is highlighted when TdaA (38% identity) and TdaB (55% identity) from 27-4 are compared to the same proteins from TM1040. Evidence of poorly conserved gene sequences is also apparent in other data in the present study. The Southern slot blot shown in Fig. 5 was done under low-stringency hybridization and produced several weak positive signals. These weak positives are likely caused by poor DNA-DNA homology, further supporting the idea that the tda gene (and Tda protein) sequences are poorly conserved among the roseobacters. The choice of algorithm parameters used in BLAST searches that may also preclude finding genes or proteins with poor sequence conservation to the Tda target. For example, the amino acid sequence divergence between Tda proteins of TM1040 and other roseobacters could result in BLASTP E values greater than our chosen cutoff (1E-20 or less). Indeed, when higher E values are used, more Tda homologs are found in the roseobacter genome database (data not shown). Despite the difficulties in finding Tda homologs, many of the roseobacters used in the present study have been shown by Bruhn et al. (7) to produce an antibiotic activity that is correlated with rosette and biofilm formation and coincides with the production of a yellow-brown pigment, phenotypes associated with TDA synthesis in both TM1040 and 27-4.
Tda homologs were differentially distributed in the GOS metagenomics data set. The two metagenomic samples that showed relatively good Tda homolog hits were from a site in the Sargasso Sea and a hypersaline pond, respectively. It is interesting that DMSP is thought to be used by algae as an osmolyte that protects the cells against changes in salinity (51). Although our results suggest that DMSP is not used as a sole sulfur source in the biosynthesis of TDA, the correlation between salinity, DMSP, and the presence of Tda homologs makes for a tantalizing hypothesis. However, the apparent differential distribution of tda genes in the GOS metagenomic data set cannot be confirmed unless the sequencing coverage at each site is also considered. Further, if the genes are indeed distributed differentially, the selection may be on other characteristics of the organisms that carry them, not necessarily these genes.
To the best of our knowledge, this is the first report describing the genes and proteins required for TDA synthesis by roseobacters and highlighting the occurrence of tda genes on a previously unknown plasmid (pSTM3) of TM1040. Although this report answers numerous questions about TDA genetics, it has also opened new and exciting avenues for discovery. For example, underscoring and extending earlier biochemical studies (13), our data, specifically the identification of paaIJK and tdaC (prephenate dehydratase), indicate that TDA biosynthesis originates from the shikimate pathway and proceeds through phenylacetate. The results also emphasize a role for phenylacetate-CoA and CoA metabolism as vital to TDA production and suggest that the reductive sulfur pathway moving through CysH and CysI is critical for TDA activity.
The biosynthesis of TDA is predicted to have several beneficial effects on TM1040-dinoflagellate symbiosis. TDA may benefit the dinoflagellate by acting as a probiotic with antibacterial activity, whose action prevents the growth and colonization of bacteria on the surface of the dinoflagellate that could potentially harm the zoospore. In turn, the antibacterial activity of TDA may enhance the growth of TM1040 cells attached to the zoospore by warding off other biofilm-forming bacteria that compete with TM1040 for space on the surface of and nutrients from P. piscicida. Although DMSP appears not to be a primary source of the sulfur atoms of TDA, it is probable that one or more non-DMSP sulfur-containing metabolites produced by the dinoflagellate are used by TM1040 in the biosynthesis of TDA. Studies are currently under way in our laboratory to investigate how TDA biosynthesis affects TM1040-dinoflagellate symbiosis.
Supplementary Material
Acknowledgments
We gratefully acknowledge the comments, advice, and encouragement given by the members of the Belas laboratory and Russell Hill for advice and use of the PFGE.
This study was supported by a grant from the National Science Foundation (MCB0446001).
Footnotes
Published ahead of print on 11 January 2008.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alavi, M., T. Miller, K. Erlandson, R. Schneider, and R. Belas. 2001. Bacterial community associated with Pfiesteria-like dinoflagellate cultures. Environ. Microbiol. 3:380-396. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S., W. Gish, W. Miller, E. Myers, and D. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M. 2001. Current protocols in molecular biology. J. Wiley, New York, NY.
- 4.Baker, S. C., S. J. Ferguson, B. Ludwig, M. D. Page, O. M. Richter, and R. J. van Spanning. 1998. Molecular genetics of the genus Paracoccus: metabolically versatile bacteria with bioenergetic flexibility. Microbiol. Mol. Biol. Rev. 62:1046-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benning, M. M., G. Wesenberg, R. Liu, K. L. Taylor, D. Dunaway-Mariano, and H. M. Holden. 1998. The three-dimensional structure of 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. strain CBS-3. J. Biol. Chem. 273:33572-33579. [DOI] [PubMed] [Google Scholar]
- 6.Brinkhoff, T., G. Bach, T. Heidorn, L. Liang, A. Schlingloff, and M. Simon. 2004. Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl. Environ. Microbiol. 70:2560-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruhn, J. B., L. Gram, and R. Belas. 2007. Production of antibacterial compounds and biofilm formation by roseobacter species are influenced by culture conditions. Appl. Environ. Microbiol. 73:442-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruhn, J. B., K. F. Nielsen, M. Hjelm, M. Hansen, J. Bresciani, S. Schulz, and L. Gram. 2005. Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the roseobacter clade. Appl. Environ. Microbiol. 71:7263-7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchan, A., L. S. Collier, E. L. Neidle, and M. A. Moran. 2000. Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine roseobacter lineage. Appl. Environ. Microbiol. 66:4662-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchan, A., J. M. Gonzalez, and M. A. Moran. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchan, A., E. L. Neidle, and M. A. Moran. 2001. Diversity of the ring-cleaving dioxygenase gene pcaH in a salt marsh bacterial community. Appl. Environ. Microbiol. 67:5801-5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Budihas, S. R., I. Gorshkova, S. Gaidamakov, A. Wamiru, M. K. Bona, M. A. Parniak, R. J. Crouch, J. B. McMahon, J. A. Beutler, and S. F. Le Grice. 2005. Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res. 33:1249-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cane, D. E., Z. Wu, and J. E. Van Epp. 1992. Thiotropocin biosynthesis. Shikimate origin of a sulfur-containing tropolone derivative. J. Am. Chem. Soc. 114:8479-8483. [Google Scholar]
- 14.Dantzer, W. R., and R. E. Levin. 1997. Bacterial influence on the production of paralytic shellfish toxins by dinoflagellate algae. J. Appl. Microbiol. 83:464-469. [DOI] [PubMed] [Google Scholar]
- 15.Ezezika, O. C., S. Haddad, T. J. Clark, E. L. Neidle, and C. Momany. 2007. Distinct effector-binding sites enable synergistic transcriptional activation by BenM, a LysR-type regulator. J. Mol. Biol. 367:616-629. [DOI] [PubMed] [Google Scholar]
- 16.Ferrandez, A., B. Minambres, B. Garcia, E. R. Olivera, J. M. Luengo, J. L. Garcia, and E. Diaz. 1998. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 273:25974-25986. [DOI] [PubMed] [Google Scholar]
- 17.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gage, D. A., D. Rhodes, K. D. Nolte, W. A. Hicks, T. Leustek, A. J. Cooper, and A. D. Hanson. 1997. A new route for synthesis of dimethylsulphoniopropionate in marine algae. Nature 387:891-894. [DOI] [PubMed] [Google Scholar]
- 19.Garg, B., R. C. Dogra, and P. K. Sharma. 1999. High-efficiency transformation of Rhizobium leguminosarum by electroporation. Appl. Environ. Microbiol. 65:2802-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez, J. M., J. S. Covert, W. B. Whitman, J. Henriksen, F. Mayer, B. Scharf, R. Schmitt, A. Buchan, J. Fuhrman, R. P. Kiene, and M. A. Moran. 2003. Silicibacter pomeroyi sp. nov. and Roseovarius nubinhibens sp. nov., DMSP demethylating bacteria from marine environments. Int. J. Syst. Evol. Microbiol. 53:1261-1269. [DOI] [PubMed] [Google Scholar]
- 21.Hjelm, M., O. Bergh, A. Riaza, J. Nielsen, J. Melchiorsen, S. Jensen, H. Duncan, P. Ahrens, H. Birkbech, and L. Gram. 2004. Selection and identification of autochthonous potential probiotic bacteria from turbot larvae (Scophthalmus maximus) rearing units. Syst. Appl. Microbiol. 27:360-371. [DOI] [PubMed] [Google Scholar]
- 22.Hjelm, M., A. Riaza, F. Formoso, J. Melchiorsen, and L. Gram. 2004. Seasonal incidence of autochthonous antagonistic Roseobacter spp. and Vibrionaceae strains in a turbot larva (Scophthalmus maximus) rearing system. Appl. Environ. Microbiol. 70:7288-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ismail, W., M. El-Said Mohamed, B. L. Wanner, K. A. Datsenko, W. Eisenreich, F. Rohdich, A. Bacher, and G. Fuchs. 2003. Functional genomics by NMR spectroscopy: phenylacetate catabolism in Escherichia coli. Eur. J. Biochem. 270:3047-3054. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J. J., M. Wang, and R. Paschke. 1993. Crystal structures of medium-chain acyl-CoA dehydrogenase from pig liver mitochondria with or without substrate. Proc. Natl. Acad. Sci. USA 90:7523-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolter, R., M. Inuzuka, and D. R. Helinski. 1978. Transcomplementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15:1199-1208. [DOI] [PubMed] [Google Scholar]
- 26.Kupke, T., M. Uebele, D. Schmid, G. Jung, M. Blaesse, and S. Steinbacher. 2000. Molecular characterization of lantibiotic-synthesizing enzyme EpiD reveals a function for bacterial Dfp proteins in coenzyme A biosynthesis. J. Biol. Chem. 275:31838-31846. [DOI] [PubMed] [Google Scholar]
- 27.Lafay, B., R. Ruimy, C. Rausch de Traubenberg, V. Breittmayer, M. J. Gauthier, and R. Christen. 1995. Roseobacter algicola sp. nov., a new marine bacterium isolated from the phycosphere of the toxin-producing dinoflagellate Prorocentrum lima. Int. J. Syst. Bacteriol. 45:290-296. [DOI] [PubMed] [Google Scholar]
- 28.Letunic, I., R. R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34:D257-D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, L. 2003. Investigation of secondary metabolites of North Sea bacteria: fermentation, isolation, structure elucidation, and bioactivity. Ph.D. dissertation. Georg-August-Universität zu Göttingen, Gottingen, Sweden.
- 30.Long, R., and F. Azam. 2001. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol. 67:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luengo, J. M., J. L. Garcia, and E. R. Olivera. 2001. The phenylacetyl-CoA catabolon: a complex catabolic unit with broad biotechnological applications. Mol. Microbiol. 39:1434-1442. [DOI] [PubMed] [Google Scholar]
- 32.Martens, T., L. Gram, H. P. Grossart, D. Kessler, R. Muller, M. Simon, S. C. Wenzel, and T. Brinkhoff. 2007. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb. Ecol. 54:31-42. [DOI] [PubMed] [Google Scholar]
- 33.Miller, T. R., and R. Belas. 2004. Dimethylsulfoniopropionate (DMSP) metabolism by Pfiesteria-associated Roseobacter spp. Appl. Environ. Microbiol. 70:3383-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, T. R., and R. Belas. 2006. Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ. Microbiol. 8:1648-1659. [DOI] [PubMed] [Google Scholar]
- 35.Miller, T. R., K. Hnilicka, A. Dziedzic, P. Desplats, and R. Belas. 2004. Chemotaxis of Silicibacter sp. TM1040 toward dinoflagellate products. Appl. Environ. Microbiol. 70:4692-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moran, M. A., R. Belas, M. A. Schell, J. M. Gonzalez, F. Sun, S. Sun, B. J. Binder, J. Edmonds, W. Ye, B. Orcutt, E. C. Howard, C. Meile, W. Palefsky, A. Goesmann, Q. Ren, I. Paulsen, L. E. Ulrich, L. S. Thompson, E. Saunders, and A. Buchan. 2007. Ecological genomics of marine roseobacters. Appl. Environ. Microbiol. 73:4559-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moran, M. A., J. M. Gonzalez, and R. P. Kiene. 2003. Linking a bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiol. J. 20:375-388. [Google Scholar]
- 38.Morita, Y., E. Matsumura, T. Okabe, M. Shibata, M. Sugiura, T. Ohe, H. Tsujibo, N. Ishida, and Y. Inamori. 2003. Biological activity of tropolone. Biol. Pharm. Bull. 26:1487-1490. [DOI] [PubMed] [Google Scholar]
- 39.Morita, Y., E. Matsumura, H. Tsujibo, M. Yasuda, T. Okabe, Y. Sakagami, Y. Kumeda, N. Ishida, and Y. Inamor. 2002. Biological activity of 4-acetyltropolone, the minor component of Thujopsis dolabrata SIeb. et Zucc. hondai Mak. Biol. Pharm. Bull. 25:981-985. [DOI] [PubMed] [Google Scholar]
- 40.Olivera, E. R., B. Minambres, B. Garcia, C. Muniz, M. A. Moreno, A. Ferrandez, E. Diaz, J. L. Garcia, and J. M. Luengo. 1998. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: the phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. USA 95:6419-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz-Ponte, C., V. Cilia, C. Lambert, and J. Nicolas. 1998. Roseobacter gallaeciensis sp. nov., a new marine bacterium isolated from rearings and collectors of the scallop Pecten maximus. Int. J. Syst. Evol. Microbiol. 48:537-542. [DOI] [PubMed] [Google Scholar]
- 42.Ruiz-Ponte, C., J. F. Samain, J. L. Sanchez, and J. L. Nicholas. 1999. The benefit of a Roseobacter species on the survival of scallop larvae. Mar. Biotechnol. 1:52-59. [DOI] [PubMed] [Google Scholar]
- 43.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y. H. Rogers, L. I. Falcon, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 46.Sekowska, A., H. F. Kung, and A. Danchin. 2000. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J. Mol. Microbiol. Biotechnol. 2:145-177. [PubMed] [Google Scholar]
- 47.Shiba, T. 1991. Roseobacter litoralis gen. nov., sp. nov., and Roseobacter denitrificans sp. nov., aerobic pink-pigmented bacteria which contain bacteriochlorophyll a. Syst. Appl. Microbiol. 14:140-145. [Google Scholar]
- 48.Skov, M., K. Pedersen, and J. L. Larsen. 1995. Comparison of pulsed-field gel-electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar 01. Appl. Environ. Microbiol. 61:1540-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner-Dobler, I., and H. Biebl. 2006. Environmental biology of the marine roseobacter lineage. Annu. Rev. Microbiol. 60:255-280. [DOI] [PubMed] [Google Scholar]
- 50.Xue, Y., and W. N. Lipscomb. 1995. Location of the active site of allosteric chorismate mutase from Saccharomyces cerevisiae, and comments on the catalytic and regulatory mechanisms. Proc. Natl. Acad. Sci. USA 92:10595-10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoch, D. C. 2002. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl. Environ. Microbiol. 68:5804-5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.