Abstract
Severe structural constraints in the hepatitis A virus (HAV) capsid have been suggested as the reason for the lack of emergence of new serotypes in spite of the occurrence of complex distributions of mutants or quasispecies. Analysis of the HAV mutant spectra under immune pressure by the monoclonal antibodies (MAbs) K34C8 (immunodominant site) and H7C27 (glycophorin binding site) has revealed different evolutionary dynamics. Populations composed of complex ensembles of mutants with very low fitness or single dominant mutants with high fitness permit the acquisition of resistance to each of the MAbs, respectively. Deletion mutants were detected as components of the mutant spectra: up to 61 residues, with an average of 19, and up to 83 residues, with an average of 45, in VP3 and VP1 proteins, respectively. A clear negative selection of those replacements affecting the residues encoded by rare codons of the capsid surface has been detected through the present quasispecies analysis, confirming a certain beneficial role of such clusters. Since these clusters are located near or at the epitope regions, the need to maintain such clusters might prevent the emergence of new serotypes.
Hepatitis A virus (HAV), classified as the type species of the genus Hepatovirus within the Picornaviridae family (21), is a medically important hepatotropic virus (20). The virion capsid is composed of the structural proteins VP1, VP2, VP3, and possibly VP4, encoded in the P1 region of the genome (20, 34).
Despite some degree of nucleotide heterogeneity at the P1 genomic region (4, 11, 22, 35, 43), there is not an equivalent degree of variation at the amino acid level (20, 23, 38). It has recently been demonstrated that HAV replicates as complex dynamic mutant distributions or quasispecies (37). In this context, the high degree of conservation of the capsid amino acid sequences among independent strains and isolates of HAV would be the result of negative selection on many newly arising mutants and convergence of consensus or average sequences (14, 17). A single serotype of human HAV has been recognized (20), even though the study of the HAV evolution in cell culture has revealed the presence of some antigenic variants in the mutant spectra that were generated even in the absence of immune selection (37), as described for other viruses. Furthermore, several escape mutants, representing antigenic variants, have been selected for their resistance to different monoclonal antibodies (MAbs) (29, 31). All of these results suggest the occurrence of severe structural constraints in the HAV capsid that prevent the more extensive substitutions necessary for the emergence of a new serotype.
As stated above, several HAV escape mutants have been isolated (29, 31). However, although their frequency of isolation has not been described, it may be assumed to be very low since several neutralization cycles (an average of three to four) are required for their occurrence. Hence, the viral population dynamics during the process of selection of such escape mutants deserves further attention. In the present work, the HAV mutant spectra at different passages under the immune pressure of two MAbs (K34C8 and H7C27) have been analyzed. These MAbs represent two distinct antigenic sites: the immunodominant site (K34C8) (31, 41, 42) and the glycophorin A binding site (H7C27) (36). The immunodominant site is composed of multiple discontinuous epitopes defined by different MAbs. The glycophorin A binding site epitope defined solely by the H7C27 MAb is relevant to the antigenic structure of the capsid and also to its involvement in HAV spread in the host (36).
MATERIALS AND METHODS
Cells, viruses, and infections.
The cytopathogenic pHM175 43c strain of HAV (courtesy of T. Cromeans, Centers for Disease Control, Atlanta, GA) was plaque purified in FRhK-4 cells three times, as previously described (12), and a biological clone (pHM175 43c P0) was serially passaged 50 times in the same cell line, as previously described (7) to yield the population pHM175 43c P50. This population was further used for the analysis of the virus evolution under the selective pressure of several antibodies. The effects of two MAbs, H7C27 (generously provided by R. Decker, Abbot Laboratories, North Chicago, IL) and K34C8 (Commonwealth Serum Laboratories, Victoria, Australia), on the mutant spectra of this pHM175 43c HAV P50 population were analyzed after 6 (P6), 12 (P12), 30 (P30), and 36 (P36) passages for both MAbs and additionally after 24 passages (P24) in the case of the H7C27 MAb and after 40 passages (P40) in the case of the K34C8 MAb. At each passage, the virus was treated for 2 h at 37°C at the maximum dilution of MAbs (H7C27, 1/10,000; K34C8, 1/5,000) ensuring an infectivity neutralization of the initial P50 population higher than 95% (H7C27, 98%; K34C8, 96%), prior to its inoculation onto FrhK-4 cell monolayers. Since the initial viral numbers were around 106 50% tissue culture infective dose (TCID50) units, after neutralization the multiplicity of infection (MOI) was around 0.01 (104 TCID50 units per 106 cells), still above the minimum required to avoid bottlenecks (44). With progression of the passages, the neutralization declined and thus the MOI increased correspondingly. The postinfection media always contained the same MAbs at concentrations of 1/100,000 and 1/50,000 for H7C27 and K34C8, respectively. An unrelated ascitic antibody, generated in mice by inoculation of phosphate-buffered saline diluted 1/10 in Freund's complete adjuvant (FCA), was used as a negative control at a 1/50,000 dilution (32).
Neutralization assays.
The viral progeny of each of the studied passages for each of the three antibodies were titrated in parallel directly or after being submitted to the neutralization of the antibodies. The H7C27, K34C8, and FCA antibodies were used at the same concentrations as in the selective pressure experiments. Additionally, the populations at P36 and/or P40 were tested in reference to their neutralization by a convalescent-phase serum (HCS2) (7) as well as a polyclonal antibody generated against the pHM175 43c strain (α-HAVs) (32). The neutralization assays were carried out for 2 h at 37°C. The percentage of neutralization was calculated by comparing the titers before and after neutralization.
Genomic regions studied in the quasispecies analysis.
Two genomic regions coding for capsid proteins were studied: a fragment of the VP3-coding region (nucleotides 1470 to 1839, corresponding to amino acids 1 to 123) and a fragment of the VP1-coding region (nucleotides 2459 to 2943, corresponding to amino acids 85 to 245), which, altogether, include most of the epitopes so far described in HAV (2, 7, 29, 31, 39, 40). Furthermore, with the aim of covering a more extensive area of the capsid surface, including those regions containing clusters of rare codons (38), in some passages (P24 for the H7C27-adapting population and P36 for the K34C8-adapting population) two additional fragments were analyzed: a fragment of the VP0-coding region (nucleotides 1122 to 1353, corresponding to amino acids 107 to 183) and a second fragment of the VP3-coding region (nucleotides 1863 to 2089, corresponding to amino acids 132 to 207).
Molecular cloning and sequencing.
Viral RNA was retrotranscribed to a cDNA with the Moloney murine leukemia virus reverse transcriptase (RT; Roche), and the cDNA was copied and amplified by the thermostable Pwo polymerase (pol) from Pyrococcus woesei, which has proofreading activity (error rate of 3.2 × 10−6 substitutions/nucleotide) (28, 44). The primers used to copy and amplify the VP3-coding fragments were NH2-VP3 (5′ TCTACCTGAATGATATTTGG 3′) for the cDNA synthesis and NH2-VP3 and VP3-1431B (5′ CTTGGATCCCACTCAATGTTTTAGCTAGA 3′) for PCR amplification for one fragment and VP3-2108 (5′ AAGGAGAGGTCAATCTGTTA 3′) and VP3-1842B (5′ AAATAGGATCCAGGTAGATTA 3′) for the second VP3 fragment. The primers used to copy and amplify the VP1-coding fragment were VP1-2965 (5′ TCTGTGACAGACAGACAAATAACAAC 3′) and VP1-2428 (5′ GAGGGATCCGACATACATCAGATCATATGTC 3′). Finally, for the amplification of the VP0 fragment, the pair of primers were VP0-1355 (5′ AATTCTGACCAAACTCTAA 3′) and VP0-1121B (5′ TTGGGATCCATATGCAAGATTT 3′). The synthesis of cDNAs was performed in a final volume of 25 μl containing 8 U of Moloney murine leukemia virus RT, 0.2 mM of each nucleotide, 0.5 μM primer, 50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, and 10 mM dithiothreitol. Ten microliters of HAV RNA was denatured at 99°C for 5 min and incubated at 45°C for 1 h. DNA amplification was performed following the manufacturer's specifications in a final volume of 50 μl containing 0.5 U of the Pwo pol, 0.2 mM of each nucleotide, 0.1 μM of each primer, 10 mM Tris-HCl, 25 mM KCl, 5 mM (NH4)2SO4, 2.5 mM MgSO4 and 10 μl of the RT product and using an annealing temperature of 45°C in all reactions. Since the DNA fragments produced by Pwo pol are blunt ended and primers VP3-1431B, VP3-1842B, VP1-2428, and VP0-1121B were designed to include a BamHI restriction site at their 5′ end, the amplification products were cloned into pGEM-3Zf(+). PCR products were digested with BamHI, and the plasmid vector was digested with both BamHI and HincII. Digested DNAs were purified with the High Pure PCR product purification kit (Roche) following the directions of the manufacturer. DNA ligations were performed overnight at 16°C using T4 DNA ligase. Ligation products were transformed in Escherichia coli DH5α, and transformant clones were screened first by the standard white/blue β-galactosidase colorimetric reaction and then confirmed by colony hybridization with specific digoxigenin-labeled probes. Plasmid DNA from each clone was purified by using the Wizard Plus SV Minipreps kit (Promega). Nucleotide sequencing was carried out in an ABI PRISM 377 automated DNA sequencer, with the ABI PRISM BigDye Terminator cycle sequencing ready reaction kit (Applied Biosystems) and using vector-derived primers, as described elsewhere (5). All mutations were confirmed by sequencing both strands of DNA.
Sequence analysis.
The quasispecies complexity was analyzed by calculating the mutation frequencies and the Shannon entropy at both the nucleotide and the amino acid levels. Minimum and maximum mutation frequencies were determined as previously described (5). Normalized Shannon entropies were calculated following the formula SN = -[Σi(pi × ln pi)]/ln N, where pi is the frequency of each sequence and N is the total number of sequences in the spectrum of mutants (3). SN ranges from 0 (no diversity) to 1 (maximum diversity).
Growth competition experiments.
To determine the fitness of two isolated H7C27 MAb-resistant (MAR) mutants (D23 and D3), as well as eight K34C8 MAR mutants (Q5, F32, L1, C6, P29, G14, T10, and G16), growth competition experiments with the wild-type population grown in the presence of the nonrelated antibody at P36 (w+), were performed. The competition experiments were designed to simulate the worst case. With the H7C27 MAR clones, three different experiments were established. First, a mixture of 103 TCID50 units of the MAR viral clones and 106 TCID50 units of the w+ population were neutralized with the MAb as described above and used to infect a monolayer of 106 cells (final MAR/w+ MOI ratio of 0.001:1). In the second experiment, 106 cells were infected with a mixture of 106 TCID50 units of the MAR viral clones and 103 TCID50 units of the w+ population (MAR/w+ MOI ratio of 1:0.001) in the absence of MAb. In the third experiment, 106 cells were infected with a mixture of 106 TCID50 units of the MAR viral clones and 106 TCID50 units of the w+ population (MAR/w+ MOI ratio of 1:1), again in the absence of MAb.
With the K34C8 MAR clones, the ratios used were 104 TCID50 units of the MAR viral clones and 106 TCID50 units of the w+ population (final MAR/w+ MOI ratio of 0.01:1) in the presence of the MAb and 106 TCID50 units of the MAR viral clones and 106 TCID50 units of the w+ population (MAR/w+ MOI ratio of 1:1) in the absence of the MAb.
The mixed viral progeny of each experiment were further passed with a neutralization step with the corresponding MAb previous to the infection in the competition experiments performed in the presence of antibodies. The consensus sequences of the VP3 or VP1 fragments bearing the following marker mutations were obtained at each competition passage: position 217 of the VP1 protein in D23; position 217 of VP1; position 12 of VP3 in D3; positions 87, 123, 162, 170, and 187 of VP1 in Q5, F32, L1, C6 and P29, respectively, positions 12 and 46 of VP3 in G14 and T10, respectively; and positions 12 and 116 of VP3 in G16.
The proportion of mutant and wild-type phenotypes at each passage was inferred from the chromatogram of the consensus sequences.
RESULTS
Evolution of the mutant spectra during consecutive passages in the presence of the MAb H7C27.
The effect of MAb H7C27 on the mutant spectrum of the pHM175 43c HAV strain was analyzed. A summary of the results is shown in Table 1 and Fig. 1 and 2.
TABLE 1.
Characterization of the mutant spectrum in the presence of the antibodies used in this study
| Passage and genomic region | No. of mutations/nucleotides sequenceda | No. of mutationsb:
|
Nucleotide mutation frequencyc
|
SNd | |||||
|---|---|---|---|---|---|---|---|---|---|
| Ts | Tv | Nsyn | Syn | Indel or stop | Minimum | Maximum | |||
| P0 VP3 | 14/18,500 | 13 | 1 | 5 | 9 | 3 del | 4.3 × 10−4 | 7.6 × 10−4 | 0.20 |
| P0 VP1 | 19/24,150 | 7 | 12 | 17 | 2 | 2 del, 1 stop | 3.7 × 10−4 | 7.8 × 10−4 | 0.28 |
| FCA | |||||||||
| P6, VP3 | 8/18,500 | 7 | 1 | 3 | 5 | 0 | 3.8 × 10−4 | 4.3 × 10−4 | 0.17 |
| P12, VP3 | 9/18,500 | 5 | 4 | 2 | 7 | 3 del | 4.9 × 10−4 | 4.9 × 10−4 | 0.20 |
| P30 VP3 | 8/18,500 | 6 | 2 | 2 | 6 | 0 | 2.7 × 10−4 | 4.3 × 10−4 | 0.17 |
| P36 VP3 | 6/18,500 | 6 | 0 | 4 | 1 | 1 stop | 2.7 × 10−4 | 3.2 × 10−4 | 0.14 |
| P6 VP1 | 8/24,150 | 6 | 2 | 4 | 4 | 0 | 1.7 × 10−4 | 3.4 × 10−4 | 0.17 |
| P12 VP1 | 10/24,150 | 6 | 4 | 10 | 0 | 1 del | 8.6 × 10−5 | 4.3 × 10−4 | 0.16 |
| P30 VP1 | 7/24,150 | 6 | 1 | 1 | 6 | 2 del | 1.6 × 10−4 | 2.9 × 10−4 | 0.14 |
| P36 VP1 | 27/24,150 | 27 | 0 | 20 | 7 | 3 del | 2.1 × 10−4 | 1.1 × 10−3 | 0.28 |
| H7C27 | |||||||||
| P6 VP3 | 8/18,500 | 7 | 1 | 3 | 5 | 2 del | 4.3 × 10−4 | 4.3 × 10−4 | 0.15 |
| P12 VP3 | 7/18,500 | 2 | 5 | 3 | 4 | 1 del | 3.2 × 10−4 | 3.2 × 10−4 | 0.13 |
| P30 VP3 | 21/18,500 | 21 | 0 | 4 | 17 | 1 del | 3.8 × 10−4 | 1.1 × 10−3 | 0.31 |
| P36 VP3 | 82/18,500 | 71 | 10 | 45 | 35 | 1 stop | 6.4 × 10−4 | 3.4 × 10−3 | 0.45 |
| P6 VP1 | 8/24,150 | 3 | 6 | 7 | 1 | 3 del, 1 stop | 2.5 × 10−4 | 3.3 × 10−4 | 0.18 |
| P12 VP1 | 8/24,150 | 5 | 3 | 3 | 5 | 2 del | 3.1 × 10−4 | 3.1 × 10−4 | 0.17 |
| P24 VP1 | 18/24,150 | 14 | 4 | 14 | 4 | 0 del | 5.0 × 10−4 | 7.5 × 10−4 | 0.30 |
| P30 VP1 | 105/24,150 | 101 | 4 | 47 | 58 | 2 del | 2.1 × 10−4 | 4.3 × 10−3 | 0.26 |
| P36 VP1 | 115/24,150 | 114 | 1 | 52 | 63 | 3 del | 2.5 × 10−4 | 4.8 × 10−3 | 0.20 |
| K34C8 | |||||||||
| P6 VP3 | 5/18,500 | 4 | 1 | 2 | 3 | 0 | 2.2 × 10−4 | 2.7 × 10−4 | 0.12 |
| P12 VP3 | 3/18,500 | 0 | 3 | 2 | 1 | 0 | 1.7 × 10−4 | 1.7 × 10−4 | 0.08 |
| P30 VP3 | 33/18,500 | 32 | 1 | 10 | 23 | 3 del | 4.3 × 10−4 | 1.8 × 10−3 | 0.33 |
| P36 VP3 | 68/18,500 | 60 | 8 | 17 | 51 | 4 del | 1.2 × 10−3 | 3.7 × 10−3 | 0.57 |
| P40 VP3 | 48/18,500 | 45 | 3 | 14 | 34 | 0 del | 5.9 × 10−4 | 2.6 × 10−3 | 0.41 |
| P6 VP1 | 8/24,150 | 4 | 4 | 6 | 2 | 1 del | 2.5 × 10−4 | 3.4 × 10−4 | 0.15 |
| P12 VP1 | 7/24,150 | 5 | 2 | 1 | 6 | 4 del | 2.7 × 10−4 | 3.1 × 10−4 | 0.15 |
| P30 VP1 | 9/24,150 | 3 | 6 | 9 | 0 | 2 del | 2.9 × 10−4 | 3.7 × 10−4 | 0.19 |
| P36 VP1 | 28/24,150 | 26 | 2 | 19 | 9 | 4 del | 5.0 × 10−4 | 1.1 × 10−3 | 0.39 |
| P40 VP1 | 29/24,150 | 20 | 9 | 28 | 1 | 2 del | 3.7 × 10−4 | 1.2 × 10−3 | 0.42 |
Mutant positions are those that vary relative to the P0 consensus sequences.
Detected mutations were classified into transitions (TS) versus transversions (TV), nonsynonymous (Nsyn) versus synonymous or silent (Syn), and insertion/deletion (del) mutations.
The minimum nucleotide mutation frequency is the number of different mutations found divided by the total number of nucleotides sequenced. The maximum nucleotide mutation frequency is the total number of mutations divided by the total number of nucleotides sequenced.
The normalized Shannon entropy was calculated as SN = [Σi (pi × ln pi)]/ln N, where pi is the frequency of each nucleotide sequence and N is the total number of sequences in the spectrum of mutants.
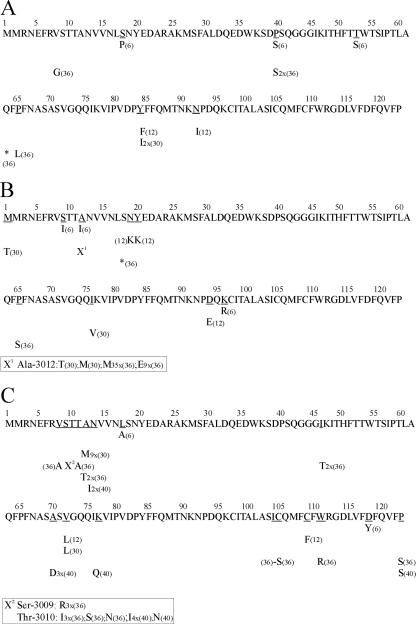
FIG. 1.
Amino acid replacements detected in a fragment of the VP3 protein through different passages of the HAV pHM175 43c strain under the selective pressure of the unrelated FCA antibody (A), H7C27 MAb (B), and K34C8 MAb (C). The replaced residues are underlined, with the new amino acid indicated below. The passage at which a given substitution occurred is indicated in parentheses. When a given substitution was present in more than one molecular clone is also indicated: i.e., “2x” means “two clones,” an asterisk represents generation of a stop codon, and “X” indicates when different replacements were detected for the same residue.
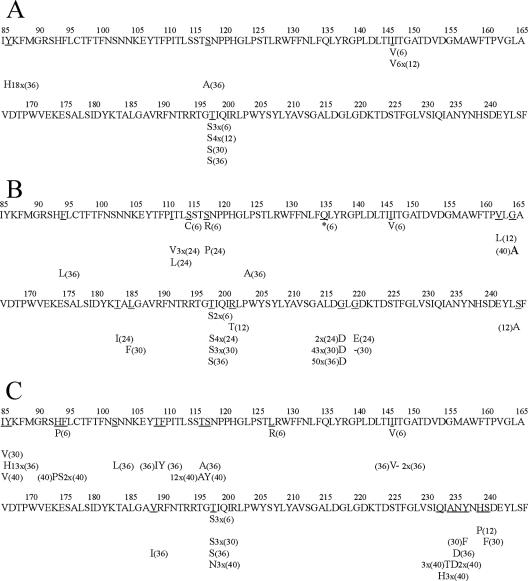
FIG. 2.
Amino acid replacements detected in a fragment of the VP1 protein through different passages of the HAV pHM175 43c strain under the selective pressure of the unrelated FCA antibody (A), H7C27 MAb (B), and K34C8 MAb (C). The replaced residues are underlined, with the new amino acid indicated below. The passage at which a given substitution occurred is indicated in parentheses. When a given substitution was present in more than one molecular clone is also indicated: i.e., “2x” indicates “two clones,” an asterisk represents generation of a stop codon, “−” represents generation of a deletion, and “X” indicates when different replacements were detected for the same residue.
No substitutions were detected in the consensus amino acid sequences of either the VP3 or VP1 regions analyzed, until P36 and P30, respectively. However, the quasispecies analysis revealed the occurrence among 50 clones of 3, 3, 4, and 45 mutations involving amino acid substitutions in the VP3 region and 7, 3, 47, and 52 mutations in the VP1 region at P6, P12, P30, and P36, respectively (Table 1). When analyzing the effect of the unrelated antibody, no substitutions were observed in the consensus amino acid sequences at any of the passages and the mutant spectra showed 3, 2, 2, and 4 substitutions in the VP3 region and 4, 10, 1, and 20 substitutions in the VP1 region (Table 1). The percentages of amino acid positions substituted at each passage of the H7C27-adapting population were 2.4, 2.4, 2.4, and 1.6 in the VP3 region (Fig. 1) and 2.5, 1.9, 1.9, and 1.9 in the VP1 region (Fig. 2). Eleven percent and 30% of these amino acid positions in the VP3 and VP1 regions, respectively, in the H7C27 spectra were the same as those substituted in the FCA spectra. Although the consensus and the dominant amino acid sequences were coincident in all cases (the dominant sequences represented around 80% of the total sequences), significant increases of the amino acid Shannon entropy were observed at P36 in the VP3 region and at P30 in the VP1 region, which proves the amino acid heterogeneity of the mutant spectra at these passages. The amino acid entropy of the mutant spectrum of VP1 of the unrelated antibody also increased at P36; however, no coincidences among the substituted residues were observed (Fig. 2). At P30 of the H7C27-adapting population, fixation of the mutation involving the substitution G→D at residue 217 of VP1, which is located in the capsid surface models (24; M. Luo, personal communication) in the immediate vicinity of K221, which defines the H7C27 binding site (Fig. 3), was almost reached, which explains the increase in entropy. This fixation definitely took place at P36, with the associated significant drop in entropy. Examination of an additional mutant spectrum of the VP1 region at P24 was undertaken to rule out the possibility of a sudden appearance of the G→D substitution at residue 217 of VP1 at P30 or, alternatively, the occurrence of a more complex quasispecies at P24. In the latter passage, 14 out of 18 mutations involved an amino acid substitution (Tables 1 and 2) and the percentage of amino acid positions substituted was 4.3 (Fig. 2), clearly indicating a higher complexity of the quasispecies than those at P12 and P30, with only 1.9% of the amino acid positions substituted in each mutant spectra. Among the seven amino acid positions changed, the fact that residue 111 was substituted in four clones and residue 217 was substituted in two clones (the latter fixed later on) was relevant. It is noteworthy that residue 111 is located on the capsid surface next to residue 114, which was previously shown to be related to the H7C27 antigenic site, its replacement inducing a lack of viral recognition by this MAb (31).
FIG. 3.
Location of the amino acid replacements detected during the passage of strain pHM175 43c under the selective pressure of the H7C27 MAb (A) or the K34C8 MAb (B) on an HAV protomer model (M. Luo, personal communication). The mutant spectrum was analyzed at P6, P12, P24, P30, and P36 (ordered vertically) in the case of the H7C27 MAb and P6, P12, P30, P36, and P40 (ordered vertically) in the case of the K34C8 MAb.
TABLE 2.
Characterization of variability at the amino acid level
| Passage and genomic region | Maximum mutation frequencya
|
Maximum indel frequency
|
SNAd | ||
|---|---|---|---|---|---|
| Nucleotide | Amino acid | Totalb | In framec | ||
| P0 VP3 | 7.6 × 10−4 | 8.1 × 10−4 | 1.6 × 10−4 | 1.6 × 10−4 | 0.10 |
| P0 VP1 | 7.8 × 10−4 | 2.2 × 10−3 | 4.1 × 10−5 | 0 | 0.14 |
| FCA | |||||
| P6 VP3 | 4.3 × 10−4 | 4.9 × 10−4 | 0 | 0 | 0.08 |
| P12 VP3 | 4.9 × 10−4 | 3.3 × 10−4 | 1.6 × 10−4 | 0 | 0.07 |
| P30 VP3 | 4.3 × 10−4 | 3.3 × 10−4 | 1.6 × 10−4 | 0 | 0.03 |
| P36 VP3 | 3.2 × 10−4 | 8.1 × 10−4 | 0 | 0 | 0.14 |
| P6 VP1 | 3.4 × 10−4 | 5.1 × 10−4 | 0 | 0 | 0.07 |
| P12 VP1 | 4.3 × 10−4 | 1.3 × 10−3 | 4.1 × 10−5 | 0 | 0.13 |
| P30 VP1 | 2.9 × 10−4 | 1.2 × 10−4 | 8.3 × 10−5 | 1.2 × 10−4 | 0.06 |
| P36 VP1 | 1.1 × 10−3 | 2.5 × 10−3 | 1.2 × 10−4 | 1.2 × 10−4 | 0.11 |
| H7C27 | |||||
| P6 VP3 | 4.3 × 10−4 | 4.9 × 10−4 | 1.1 × 10−4 | 3.3 × 10−4 | 0.12 |
| P12 VP3 | 3.2 × 10−4 | 4.9 × 10−4 | 5.4 × 10−5 | 0 | 0.10 |
| P30 VP3 | 1.1 × 10−3 | 6.5 × 10−4 | 5.4 × 10−5 | 1.6 × 10−4 | 0.13 |
| P36 VP3 | 3.4 × 10−3 | 7.6 × 10−3 | 0 | 0 | 0.23 |
| P6 VP1 | 3.3 × 10−4 | 8.7 × 10−4 | 1.2 × 10−4 | 3.6 × 10−4 | 0.16 |
| P12 VP1 | 3.1 × 10−4 | 3.7 × 10−4 | 8.3 × 10−5 | 1.2 × 10−4 | 0.11 |
| P24 VP1 | 7.5 × 10−4 | 1.7 × 10−3 | 0 | 0 | 0.27 |
| P30 VP1 | 4.3 × 10−3 | 5.8 × 10−3 | 8.3 × 10−5 | 1.2 × 10−4 | 0.24 |
| P36 VP1 | 4.8 × 10−3 | 6.5 × 10−3 | 1.2 × 10−4 | 0 | 0.05 |
| P24 VP0 | 1.2 × 10−3 | 1.1 × 10−3 | 0 | 0 | 0.10 |
| P24 VP3 bis | 1.2 × 10−3 | 2.1 × 10−3 | 0 | 0 | 0.21 |
| K34C8 | |||||
| P6 VP3 | 2.7 × 10−4 | 3.3 × 10−4 | 0 | 0 | 0.07 |
| P12 VP3 | 1.7 × 10−4 | 3.4 × 10−4 | 0 | 0 | 0.03 |
| P30 VP3 | 1.8 × 10−3 | 1.6 × 10−3 | 1.6 × 10−4 | 0 | 0.12 |
| P36 VP3 | 3.7 × 10−3 | 2.7 × 10−3 | 2.2 × 10−4 | 4.8 × 10−4 | 0.47 |
| P40 VP3 | 2.6 × 10−3 | 2 × 10−3 | 0 | 0 | 0.27 |
| P6 VP1 | 3.4 × 10−4 | 7.6 × 10−4 | 4.1 × 10−5 | 0 | 0.11 |
| P12 VP1 | 3.1 × 10−4 | 1.2 × 10−4 | 1.7 × 10−4 | 3.6 × 10−4 | 0.08 |
| P30 VP1 | 3.7 × 10−4 | 1.1 × 10−3 | 8.3 × 10−5 | 2.5 × 10−4 | 0.23 |
| P36 VP1 | 1.1 × 10−3 | 2.4 × 10−3 | 1.7 × 10−4 | 2.5 × 10−4 | 0.35 |
| P40 VP1 | 1.2 × 10−3 | 3.5 × 10−3 | 8.3 × 10−5 | 2.5 × 10−4 | 0.40 |
| P36 VP0 | 8.6 × 10−4 | 1.1 × 10−3 | 0 | 0 | 0.10 |
| P36 VP3 bis | 1.1 × 10−3 | 2.1 × 10−3 | 0 | 0 | 0.16 |
The maximum nucleotide mutation frequency is as defined in Table 1, while maximum amino acid mutation frequency is the total number of nonsynonymous mutations divided by the total number of amino acids encoded in the sequences analyzed.
The total maximum indel frequency is the total number of indels found (in frame and out of frame) divided by the total number of nucleotides sequenced.
The in-frame maximum indel generation frequency is the number of in-frame indels found divided by the total number of amino acids encoded in the sequences analyzed. These frequencies include short and long in-frame deletions.
The normalized Shannon amino acid entropy was calculated as SNA = [Σi (qi × ln qi)]/ln N, where qi is the frequency of each amino acid sequence (including those sequences with amino acid substitutions and those with in-frame deletions) and N is the total number of sequences in the spectrum of mutants. In boldface are those values higher than 0.20.
Similarly, at P36 the A→M substitution (repeated 35 times) at position 12 of VP3 was almost fixed, although the alternative A→E substitution (repeated 9 times) was also quite abundant (Fig. 1). However, this residue is harder to locate, being in the flexible amino terminus of the VP3 protein, and no previous relationship with the H7C27 binding site exists.
Evolution of the mutant spectra during consecutive passages in the presence of the MAb K34C8.
The same kind of analysis was undertaken with the MAb K34C8. The results are shown in Table 1 and Fig. 1 and 2.
No substitutions were detected in the consensus amino acid sequences of either the VP3 or VP1 region analyzed at any of the passages studied. Instead complex mutant spectra were observed at P30, P36, and even at the additional passage, P40. The numbers of mutations involving an amino acid substitution at P6, P12, P30, P36, and P40 among 50 clones were 2, 2, 10, 17, and 14 in the VP3 region and 6, 1, 9, 19, and 28 in the VP1 region (Table 1). Seven percent and 20% of the amino acid positions substituted in the VP3 and VP1 regions, respectively, in the K34C8 spectra were the same as those substituted in the FCA spectra. Although the consensus and the dominant amino acid sequences were coincident in all cases, a significant increase in the amino acid Shannon entropy at P36 in the VP3 region and P36 and P40 in the VP1 region was detected, representing the dominant sequences only around 50% of all of the sequences. Particularly, the percentages of amino acid positions substituted in each passage were of 1.6, 1.6, 1.6, 7.3, and 4.9 in the VP3 region and 2.5, 0.6, 2.5, 5.6, and 5.6 in the VP1 region. Most of the substituted residues were located near the 5X axis, in the limit between two protomers around residues 93 to 109, 127, 189 to 197, and 232 to 240 of VP1 and 8 to 17 of VP3 (Fig. 3). Additionally, a second region was located around the 3X axis affecting residues 72 and 104 to 123 of VP3 (Fig. 3).
Identification of hot spots for internal deletions.
An unexpected finding was observed regarding the occurrence of deletion mutants. A significant percentage of genomes in the populations generated in the presence of MAbs harbored long in-frame deletions, mainly at early passages in the case of the H7C27 MAb and later in the case of the K34C8 MAb. In particular, in the presence of the H7C27 MAb the mutant spectra contained, among 50 clones, 2, 0, 1, and 0 genomes with long in-frame deletions at P6, P12, P30, and P36 in the VP3 region and 3, 1, 0, and 0 genomes in the VP1 region. Similarly, in the presence of the K34C8 MAb, 0, 0, 0, 2, and 0 and 0, 3, 2, 0, and 2 genomes with long in-frame deletions were detected in the VP3 and VP1 regions, respectively at P6, P12, P30, P36, and P40. A clear tendency to accumulate in-frame deletions versus out-of-frame deletions in the populations grown in the presence of anti-HAV MAbs in contrast to those populations grown in the absence (P0) or in the presence of an unrelated antibody was observed. At P0, one of four deletion mutants (25.0%) kept the reading frame, and among a total of nine deletion mutants produced during the consecutive passages in the presence of the unrelated FCA antibody, two (22.2%) were in-frame deletions. On the contrary, 20 of 30 deleted genomes (66.7%) detected through passages with either the H7C27 MAb or the K34C8 MAb were in-frame mutants. These results confirm that the deletion mutants not only contribute to the complexity of the quasispecies to a significant degree, as can be observed comparing the frequency of amino acid mutation versus the in-frame indel frequency (Table 2), but indeed they might play a role in the adaptation of the population to the new environmental conditions. The length of the in-frame deletions in VP3 ranged from a single amino acid (14.3% of the deletion mutants) up to 61 amino acids (14.3%), with an average of 19 residues, and in VP1 from a single amino acid (21.4%) to 83 amino acids (14.3%), with an average of 45 amino acids (Fig. 4).
FIG. 4.
Genomes coding for deleted proteins among the different mutant spectra in VP3 (A) and VP1 (B).
Interestingly, though most of the long deletions mapped around the same region, mainly in VP1 (Fig. 4), only a few of them were identical, suggesting a mechanism of de novo recombination in their generation. The mean percentages of the long deletion mutants were 2.4% ± 3.0% and 4.0% ± 2.5% in the VP3 and VP1 regions, respectively. After analyzing the alignments of the recombinant genomes, several hot spots could be identified for deletions as characterized by their richness in A and U and by the occurrence of direct repetitions of identical or very similar sequences at the parting and anchoring sites (Fig. 5), as has been described previously in other picornaviruses (review in reference 2). The frequency of crossovers at these hot spots ranged from 0.3% to 0.8%. Alternatively, those deletion mutants consisting of only one or few residues may also be produced by polymerase slippage at homopolymeric tracks.
FIG. 5.
Genomes harboring nucleotide deletions detected among the different mutant spectra in VP3 (A) and VP1 (B). Direct repetitions of identical or very similar sequences at parting and anchoring sites are underlined.
Neutralization of infectivity versus antigenic variation.
Following the previously defined neutralization limits for the pHM175 HAV strain (31), which establish neutralization resistance as <30% neutralization, partial neutralization resistance as 30 to 60% neutralization, and neutralization sensitivity as >60% neutralization, it may be concluded that a significant increase in the neutralization resistance is only achieved after a considerable number of passages under the selective pressure of the MAbs (Table 3). In our particular case, however, these limits might be adjusted, bearing in mind the neutralization shown by an unrelated antibody (FCA), to <45%, 45 to 75%, and >75%, and thus it may be concluded that neutralization resistance was achieved at P30 with the H7C27 MAb (42%) and at P40 with the K34C8 MAb (27%) (Table 3). The great variability of the neutralization of the infectivity analysis is worth noting, which, apart from the inherent variability of the technique, may reflect the intrinsic nature of the quasispecies combined with the random sampling events. In fact, a positive correlation between entropy and neutralization variability could be observed, mainly at the VP1 level. Additionally, the neutralization of the viral populations at P36 and/or P40 by a convalescent-phase serum (HCS2) and a polyclonal antibody generated against the 43c strain (α-HAVs) was also tested. The population adapted to the H7C27 MAb exhibited a partial neutralization resistance to both antibodies (Table 4), while the K34C8-adapted population did not show a clear increase in resistance to neutralization (Table 4). The amino acid variability at P30 and P36 confirmed the crucial role of VP1, particularly residue 217, but in addition most probably many individual residues surrounding the glycophorin A binding epitope, as observed at P24 in the study including the two additional regions (Fig. 3A), for the acquisition of resistance to the H7C27 MAb. In contrast, for resistance to the K34C8 MAb, both VP1 and VP3 proteins are equally important, with those residues around the 5X and 3X axes being critical (Fig. 3B). However, in the latter case it was harder to define particular clue residues. Instead, larger capsid areas were the ones conferring resistance. The amino end residues of VP3, around positions 8 to 13, were substituted in the presence of either of the two antibodies.
TABLE 3.
Percentage of neutralization of infectivity by MAbs
| Passage | % of neutralization bya:
|
||
|---|---|---|---|
| FCA | H7C27 | K34C8 | |
| P0 | 29 ± 14 (21, 21, 45) | 98 ± 14 (98, 97, 98) | 95 ± 2 (96, 92, 96) |
| P6 | 37 ± 14 (45, 21, 45) | 94 ± 2 (95, 93, 93) | 93 ± 6 (87, 97, 97) |
| P12 | 23 ± 18 (34, 2, 34) | 91 ± 6 (98, 87, 87) | 96 ± 4 (91, 98, 98) |
| P18 | 27 ± 15 (19, 45, 19) | 76 ± 24 (45, 91, 91) | 86 ± 10 (78, 97, 83) |
| P24 | 30 ± 16 (40, 11, 40) | 79 ± 20 (56, 89, 92) | 73 ± 18 (54, 75, 90) |
| P30 | 23 ± 18 (2, 34, 34) | 42 ± 27 (24, 32, 72) | 68 ± 22 (42, 81, 81) |
| P36 | 27 ± 15 (19, 19, 45) | 36 ± 8 (34, 29, 45) | 52 ± 21 (75, 45, 35) |
| P40 | 27 ± 17 (11, 40, 30) | ||
FCA is an unrelated antibody, H7C27 is a MAb against the glycophorin A binding site, and K34C8 is a MAb against the immunodominant site. Values in parentheses represent the percentage of neutralization for each determination.
TABLE 4.
Percentage of neutralization of infectivity of MAb-adapted populations by polyclonal antibodies
| Antibodya | % of neutralization of infection in populationb:
|
||
|---|---|---|---|
| FCA adapted | H7C27 adapted | K34C8 adapted | |
| HCS2 | 92 ± 1 (91, 93) | 64 ± 10 (60, 67) | 73 ± 18 (60, 86) |
| α-HAVs | 95 ± 1 (94, 95) | 68 ± 6 (65, 70) | 82 ± 12 (73, 91) |
HCS2 is a human convalescent-phase serum, and α-HAVs is a polyclonal antibody generated against the pHM175 43c strain. The FCA- and H7C27-adapting populations at P36 and the K34C8-adapting-population at P40 were evaluated in front of the HCS2 convalescent-phase serum and the α-HAVs polyclonal antibody.
Values in parentheses represent the percentage of neutralization for each determination.
Rare codons and variability of the capsid surface.
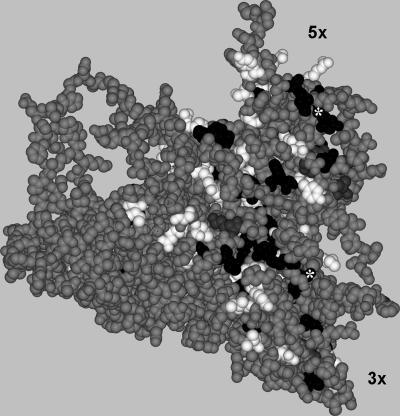
A common belief exists concerning structural constraints in the HAV capsid hampering its variability. However, the process driving the negative selection imposed by these constraints has not been clearly defined. In an attempt to explain this process to some extent, a structural analysis of the location of the substitutions detected along the studied fragments was performed. The refined version of the only existing model of the HAV protomer (24; M. Luo, personal communication) was used to visualize those surface residues substituted in the quasispecies populations. Figure 3 shows the location of the capsid surface residues substituted in some genomes of the population at each of the studied passages. While the average rates of amino acid substitutions on the capsid surface, along all of the passages studied, were 1.5%, 3.0%, and 3.4% under the pressure of the unrelated antibody (FCA), H7C27 MAb, and K34C8 MAb, respectively, these rates at the inner capsid were 1.5%, 1.0%, and 2.1%, respectively. Most of the detected substitutions (average of 84% at the surface and 76% at the inner side) occurred at or around (three residues in either direction) residues encoded by rare codons. Regarding the surface replacements, only 7.7% of these replacements occurred at the precise residues encoded by rare codons (with these residues representing 19% of the total capsid surface studied), 77% at those residues surrounding the residues encoded by rare codons (with these residues representing 49% of the capsid surface region studied), and the remaining 15.3% at residues located far away from those encoded by rare codons (with these residues representing 31% of the total capsid surface studied). However, since the regions studied did not cover all of the clusters of residues encoded by rare codons, additional analyses at those passages with the highest entropy for each MAb were performed. The quasiespecies analysis of two additional fragments, one of VP0 and another of a second region of VP3, were undertaken. These two new regions, together with the former ones, included most of the residues encoded by rare codons located at the capsid surface and represented around 60% of the total capsid surface. Nothing remarkable regarding the specific genetic data was observed in these new analyses (data not shown): particularly, no indels were detected. Particularly, only 6.2% of the replacements occurred at the precise residues encoded by rare codons (with the residues encoded by rare codons representing 20% of the total capsid surface studied), 75% at those residues surrounding the residues encoded by rare codons (with these residues around the rare-codon-encoded residues representing 53% of the capsid surface region studied), and the remaining 18.7% at residues located far away from those encoded by rare codons (with these residue representing 30% of the total capsid surface studied). These figures were clearly projected in a merged image including the residues encoded by rare codons and the residues substituted among the quasiespecies individuals, confirming the lack of coincidence of both groups of residues (Fig. 6).
FIG. 6.
HAV protomer model (M. Luo, personal communication) in which is contained a merged image of the locations of all of the substituted residues detected through all of the passages with both antibodies (residues labeled in black) and the location of the residues encoded by rare codons (residues labeled in white). Those cases in which the replaced residue was encoded by a rare codon are labeled with asterisks. Previously identified residues associated either with the H7C27 or K34C8 MAb are labeled in gray.
In an attempt to find an explanation for this fact, we analyzed all of the potential mutations in the residues encoded by such rare codons. The analysis revealed that 70% of all possible mutations would induce a significant change in rarity, while only 30% of these mutations would maintain the level of rarity of the codons. Among the actual substitutions precisely affecting the residues encoded by rare codons (Fig. 6), the one at position 1236 belonged to the latter group (UAC→UUC) and did maintain the level of rarity (26%→27%) and the other at position 3094 belonged to the first group (GAC→GAG; 19%→77%). However, while residue 1236 is encoded by a well-conserved rare codon among the different HAV strains, residue 3094 is not.
A similar pattern was observed among the biological clones isolated at those passages showing resistance to neutralization. Among 10 different mutants, 1 bore a substitution in a residue encoded by a rare codon, 6 bore substitutions in residues located near residues encoded by rare codons, and 3 bore substitutions in residues located far from residues encoded by rare codons.
This segregation of the location of the replacements was not observed at the inner capsid: to the contrary, while the residues encoded by rare codons in the inner capsid region studied represented 9% of all codons, 20.6% of the substitutions detected precisely affected them. The residues located around the ones encoded by rare codons represented 54% of all codons and 52.9% of the substitutions took place here, and finally, 26.5% of the substitutions affected the residues located far from the rare-codon-encoded residues representing 37% of the region. The observed substitutions at the inner capsid tended to decrease the level of rarity of the codons.
All of these findings suggest that the low substitution rate at the clusters of residues encoded by rare codons of the capsid surface, which are mostly located near the residues defining the antigenic sites and probably form part of such sites, may contribute to the low antigenic HAV variability.
Fittness of the MAR mutants.
One important issue is to understand the rationale of the extremely low frequency of isolation of HAV natural antigenic variants, bearing in mind that in vitro MAR mutants with a partial resistance to convalescent-phase sera may be isolated. One possibility could be that these mutants had a lower replication capacity or fitness than the wild-type viruses. To prove this premise, competition experiments between the D23 or D3 H7C27 MAR isolates as well as the Q5, F32, L1, C6, P29, G14, T10 and G16 K34C8 MAR mutants and the wild-type virus were performed. The MAR mutants isolated against the H7C27 MAb were mixed in competition with the wild-type virus population at P36 at an MAR/w+ MOI proportion of 0.001:1 and neutralized with the corresponding MAb. The progeny mixed population was again neutralized and used in a second round of infection and successively. At each passage, the consensus sequences of the VP3 and VP1 fragments containing the residues defining the resistance phenotypes (G→D at position 217 of VP1 in the D23 MAR mutant and G→D at position 217 of VP1 and A→M at position 12 of VP3 in the D3 mutant) were analyzed. As early as at the third passage, the H7C27 MAR mutant viruses were already present at a proportion of around 50%, and from the fourth passage on the MAR mutant viruses completely outcompeted the wild-type population (Fig. 7A). The second experiment was designed exactly in the other direction: i.e., in the absence of immune pressure and infection with an MAR/w+ MOI of 1:0.001. In this situation and following the same procedure stated above, even after 12 passages the wild-type virus was not able to outcompete the MAR mutants (Fig. 7B, insert). These results suggested that the wild-type virus did not present any advantage in terms of replication with respect to the mutant viruses. In order to discard the possible bottleneck imposed with an MOI of 0.001 for the wild-type virus, the experiment was repeated with an MAR/w+ MOI ratio of 1:1 (Fig. 7B). In the latter situation, again none of the viral populations could outcompete the other, indicating that the mutant viruses did not have a lower fitness in terms of replication.
FIG. 7.
Growth competition experiments. Two H7C27 MAR mutants (A and B) and eight K34C8 MAR mutants (C and D) were grown in competition with the wild-type virus. In the presence of antibodies (A and C), the MAR/w+ ratios were 1:1,000 and 1:100 for the H7C27 and K34C8 MAbs, respectively. In the absence of antibodies (B and D), the ratio was 1:1, with the exception of the insert in panel B, which had a ratio of 1,000:1.
A completely different picture was observed with the K34C8 MAR mutants since in the absence of antibody the wild-type population completely outcompetes the MAR mutants as early as at P4 while the MAR mutants were not able to completely outcompete the wild-type population even at P5 in the presence of the antibody (Fig. 7C and D), indicating a very low fitness of the mutants. This low fitness is particularly relevant, keeping in mind that the competition growth experiments in the presence of antibody of the K34C8 MAR mutants and the wild-type virus were not as adverse (ratio of 1:100) as those in the competition experiments between the H7C27 MAR mutants and the wild-type virus (ratio of 1:1,000). Special attention should be paid to MAR L1 which presented the lowest fitness and did not even appear in the consensus sequence of the mixed population at P5. The particular substitution present in this mutant (V→L at position 162 of VP1) involves a residue encoded by a rare codon which significantly changed the level of rarity from 19% to 50% (GUA→UUA).
DISCUSSION
Antigenic variation of viruses may result either from positive immune selection or from hitchhiking of substitutions at antigenic sites belonging to genomes selected by other traits or by chance events (6). In this context, it is relevant to note that antigenic variation of many viruses may occur upon virus replication in the absence of any detectable immune selection (reviewed in reference 15) and may be attributed to higher tolerance to replacements of surface residues which are relatively free of structural constraints. However, in the particular case of the HAV capsid, this flexibility is limited, and although some antigenic variants are detected among the components of the mutant spectra in the absence of immune selection, none of these genomes becomes dominant in the population (37). This result has been confirmed in the present work with the unrelated antibody (FCA)-adapting population. Another piece of evidence for the high structural constraint of the HAV external capsid surface is disclosed by the minimal differences in the average rates of amino acid substitutions at the inner and outer sides of the capsid, at least in the regions studied herein, with levels of 1.5% both inside and outside in those viral populations growing in the presence of FCA antibody. However, this difference increased under the effect of immune pressure, with average amino acid replacements inside and outside of the capsid of 1% and 3% and 2.1% and 3.4% in those populations selected against the H7C27 and K34C8 MAbs, respectively. The analysis of the mutant spectra of the viral populations growing in the presence of both antibodies revealed two different evolutionary mechanisms. On the one hand, there is the mechanism at the glycophorin A binding site with selection/fixation of the replacement at position 217 of VP1, which paralleled an increase in the neutralization resistance. On the other, there is that of the immunodominant site, which showed a complex spectrum of variants, none of them becoming dominant, but nevertheless correlating with a significant increase in neutralization resistance. In the case of the glycophorin A binding site, although other replacements indeed occurred at other capsid sites (Fig. 3) and probably at other sites not analyzed, none of them was fixed, as was concluded from the consensus sequence of the entire capsid (data not shown), and thus the resistant phenotype to the H7C27 MAb could be associated with the specific replacement G217→D since a clear association between the increase of this mutation in the population and the resistance to neutralization occurred. In contrast, the partial neutralization-resistant phenotype observed at P36 or the neutralization-resistant phenotype of P40 in the case of the population selected against the K34C8 MAb is explained by the ensemble of mutants around the antigenic site, which altogether represented 20% and 74% of the population, respectively. Different evolutionary mechanisms have also been documented in other picornaviruses, such as the antigenic diversification of site A of foot-and-mouth disease virus (26). For this virus, it has been described that even in the absence of immune pressure some antigenic variants may become dominant in the population after 20 serial passages (13) although others, involving replacements of very critical residues, only became dominant in the presence of immune selection after 10 to 20 passages (6). However, what makes HAV so special is the requirement of both immune pressure as well as a very long exposure to the antibody (more than 30 passages) in order to select antigenic variants. Additionally, while in foot-and-mouth disease virus the specific requirement of immune pressure has been interpreted as the result of a forced low-fitness rare variant selection (6), the fitness of the H7C27 MAR mutants of HAV, measured as the replication capacity in competition with the wild-type strain in the absence of the H7C27 MAb, was not decreased. At this point, again the occurrence of strong structural constraints, acting as negative selection factors, is envisaged. Thus, the balance between both factors—the escape from antibody binding and the maintenance of a proper conformation—will determine the selection of the emerging antigenic variants. A very evident role of conformation may be entailed in the case of the K34C8-adapting population, since this MAb binds to an epitope shaped only when the pentamers assemble into procapsids (42). This epitope folding could even explain the lack of dominance of a single genome, in spite of the high amino acid heterogeneity of the adapting population, provided that a replacement in one pentamer could cooperatively complement a different replacement in a second one and successively forming a mosaic-like capsid. In this sense, it is worth noting that most of the substituted residues were located near the 5X axis, in the limit between two protomers. However, although mosaic-like capsids in picornaviruses have been documented for a very long time (2), this is a premise very hard to demonstrate, particularly for HAV. Alternatively, each individual of the quasispecies could escape from the antibody binding without becoming dominant due to an equivalent relative fitness than the rest of variants of the ensemble by keeping the threshold expression level controlled by the surrounding mutant spectrum (16). In contrast, in the case of the H7C27-adapting population, although several replacements around the glycophorin A binding site were detected in the mutant spectra along the passages, only one of these replacements (G217→D) became dominant, probably reflecting a relative higher fitness of the variant harboring this replacement in comparison to the rest of the variants. The influence of relative viral fitness in the repertoire of viral mutants selected by a neutralizing antibody among the minority genomes present in a viral population has recently been proven (25). More difficult is the evaluation of the role of the deletion mutants present in the viral population in the development of the resistance to neutralization. In general, the deletion mutants selected in the presence of the H7C27 population at early passages tended to be further lost, coinciding with the increase in amino acid substitutions. In contrast in the K34C8-adapting population, the deletion mutants were never completely lost, although they showed a tendency to decrease with time. In the case of the long deletion mutants of VP1, at least 42% of the lost residues are located at the capsid surface, including some antigenic residues. In the case of VP3, 22% of the deleted residues are located on the capsid surface, also including residues involved in the antigenic structure. Deletion mutants affecting antigenic sites have been described for other picornaviruses such as poliovirus (27) and even HAV (9, 10).
The long exposure to the antibodies required to select antigenic variants may be explained by the slow quiescent kinetics of viral replication which avoids the rapid expansion of the population size necessary for the generation of a complex mutant repertoire. The slow replication of HAV is, at least in part, due to the deoptimized codon usage (33), which is conveyed in the profuse use of rare codons and its occurrence in clusters in the P1 coding region (38). It has been postulated that such clusters would decrease the translation speed by inducing a transient stop of the translational complex due to the longer time required by the rare aminoacyl-tRNA to diffuse into site A of the ribosome (8). These ribosome stallings would quantitatively impair the protein synthesis (19, 30) but qualitatively be beneficial for the proper folding of the nascent polypeptide (1, 18). However, although such a beneficial effect is hardly demonstrable, in the present work a clear negative selection of those replacements affecting the residues encoded by rare codons of the capsid surface is shown, indicating a certain beneficial role of such clusters. The need to maintain this benefit could contribute to the low variability of the HAV capsid. Many of the residues encoded by rare codons are located around the epitopes or even probably form part of them, and it is quite unlikely that a nucleotide substitution would give rise to a new codon of similar rarity and a compatible amino acid. In this context, it is worth noting that the L1 mutant, isolated as a K34C8 MAR mutant, involving the substitution of V at position 162 of VP1 by L and inducing the rare codon (GUA; 17%) substitution by a normal (UUA; 50%) codon, shows extremely low fitness with regard to the wild-type virus. Additionally, the fact that the mutations observed in the inner capsid tended to maintain a minimal frequency of rare codons confirms the previously suggested dynamics of mutation selection to preserve the balance between rate of translation and capsid protein folding (37).
Several factors contribute to the low variability of the HAV capsid. On the one hand, there is the low translation rate due to the deoptimized codon usage and the associated limited concentration of those proteins involved in RNA replication and thus low total RNA yields. On the other, there is the restricted variability of the capsid residues encoded by rare codons and located in the epitope regions. And last, but not least, there are protein structural constraints which always limit the amino acid variability, particularly keeping in mind the large interconnected immunodominant site which includes residues from VP3 and VP1 (29). Altogether, these characteristics prevent the generation of the extensive substitutions required for the emergence of a new serotype.
Acknowledgments
This work was supported in part by grants BIO2002-02625 and BIO2005-05022 from CICyT of the Ministry of Education and Science, Spain; SP22-CT-2004-502571 from the European Union; 2005SGR00966 from the Generalitat de Catalunya; and by Centre de Referència de Biotecnologia de Catalunya (CeRBa), Generalitat de Catalunya. L. Aragonès is the recipient of an FI fellowship from the Generalitat de Catalunya.
Footnotes
Published ahead of print on 5 December 2007.
REFERENCES
- 1.Adzhubei, A. A., I. A. Adzhubei, I. A. Krasheninnikov, and S. Neidle. 1996. Non-radom usage of “degenerate” codons is related to protein three-dimensional structure. FEBS Lett. 39978-82. [DOI] [PubMed] [Google Scholar]
- 2.Agol, V. I. 2002. Picornavirus genetics: an overview, p. 269-284. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 3.Airaksinen, A., N. Pariente, L. Menendez-Arias, and E. Domingo. 2003. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis. Virology 311339-349. [DOI] [PubMed] [Google Scholar]
- 4.Arauz-Ruiz, P., L. Sundqvist, Z. García, L. Taylor, K. Visoná, H. Norder, and L. O. Magnius. 2001. Presumed common source outbreaks of hepatitis A in an endemic area confirmed by limited sequencing within the VP1 region. J. Med. Virol. 65449-456. [PubMed] [Google Scholar]
- 5.Arias, A., E. Lazaro, C. Escarmis, and E. Domingo. 2001. Molecular intermediates of fitness gain of an RNA virus: characterization of a mutant spectrum by biological and molecular cloning. J. Gen. Virol. 821049-1060. [DOI] [PubMed] [Google Scholar]
- 6.Borrego, B., I. S. Novella, E. Giralt, D. Andreu, and E. Domingo. 1993. Distinct repertoire of antigenic variants of foot-and-mouth disease virus in the presence or absence of immune selection. J. Virol. 676071-6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch, A., J. F. Gonzalez-Dankaart, I. Haro, R. Gajardo, J. A. Pérez, and R. M. Pintó. 1998. A new continuous epitope of hepatitis A virus. J. Med. Virol. 5495-102. [DOI] [PubMed] [Google Scholar]
- 8.Chou, T., and G. Lakatos. 2004. Clustered bottlenecks in mRNA translation and protein synthesis. Physiol. Rev. Lett. 93198101-198104. [DOI] [PubMed] [Google Scholar]
- 9.Costa-Mattioli, M., V. Ferré, D. Casane, R. Perez-Bercoff, M. Coste-Burel, B. M. Imbert-Marcille, E. C. Andre, C. Bressollette-Bodin, S. Billaudel, and J. Cristina. 2003. Evidence of recombination in natural populations of hepatitis A. Virology 31151-59. [DOI] [PubMed] [Google Scholar]
- 10.Costa-Mattioli, M., J. Cristina, H. Romero, R. Perez-Bercof, D. Casane, R. Colina, L. Garcia, I. Vega, G. Glikman, V. Romanowsky, A. Castello, E. Nicand, M. Gassin, S. Billaudel, and V. Ferré. 2002. Molecular evolution of hepatitis A virus: a new classification based on the complete VP1 protein. J. Virol. 769516-9525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costa-Mattioli, M., A. D. Napoli, V. Ferre, S. Billaudel, R. Perez-Bercoff, and J. Cristina. 2003. Genetic variability of hepatitis A virus. J. Gen. Virol. 843191-3201. [DOI] [PubMed] [Google Scholar]
- 12.Cromeans, T., M. D. Sobsey, and H. A. Fields. 1987. Development of a plaque assay for a cytopathic, rapidly replicating isolate of hepatitis A virus. J. Med. Virol. 2245-56. [DOI] [PubMed] [Google Scholar]
- 13.Diez, J., M. G. Mateu, and E. Domingo. 1989. Selection of antigenic variants of foot-and-mouth disease virus in the absence of antibodies, as revealed by an in situ assay. J. Gen. Virol. 703281-3289. [DOI] [PubMed] [Google Scholar]
- 14.Domingo, E., C. Biebricher, M. Eigen, and J. Holland. 2001. Quasispecies and RNA virus evolution: principles and consequences. Landes Bioscience, Austin, TX.
- 15.Domingo, E., J. Diez, M. A. Martinez, J. Hernandez, A. Holguin, B. Borrego, and M. G. Mateu. 1993. New observations on antigenic diversification of RNA viruses. Antigenic variation is not dependent on immune selection. J. Gen. Virol. 742039-2045. [DOI] [PubMed] [Google Scholar]
- 16.Domingo, E., and J. Gomez. 2007. Quasispecies and its impact on viral hepatitis. Virus Res. 127131-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eigen, M., and C. K. Biebricher. 1988. Sequence space and quasispecies distribution, p. 211-245. RNA genetics. CRC Press, Boca Raton, FL.
- 18.Gavrilin, G., E. A. Cherkasova, G. Y. Lipskaya, O. M. Kew, and V. I. Agol. 2000. Evolution of circulating wild poliovirus and vaccine-derived poliovirus in an immunodeficient patient: a unifying model. J. Virol. 747381-7390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoekema, A., R. A. Kastelein, M. Vasser, and H. A. de Boer. 1987. Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol. Cell. Biol. 72914-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hollinger, F. B., and S. U. Emerson. 2001. Hepatitis A virus, p. 799-840. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams and Wilkins, Philadelphia, PA.
- 21.International Committee on Taxonomy of Viruses, M. H. V. Van Regenmortel, and International Union of Microbiological Societies, Virology Division. 2000. Virus taxonomy: classification and nomenclature of viruses: seventh report of the International committee on taxonomy of viruses: Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, CA.
- 22.Lemon, S. M., S.-F. Chao, R. W. Jansen, L. N. Binn, and J. W. LeDuc. 1987. Genomic heterogeneity among human and nonhuman strains of hepatitis A virus. J. Virol. 61735-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemon, S. M., and B. H. Robertson. 1993. Current perspectives in the virology and molecular biology of hepatitis A virus. Semin. Virol. 4285-295. [Google Scholar]
- 24.Luo, M., M. G. Rossmann, and A. C. Palmenberg. 1988. Prediction of three-dimensional models for foot-and-mouth disease virus and hepatitis A virus. Virology 166503-514. [DOI] [PubMed] [Google Scholar]
- 25.Martin, V., C. Perales, M. Davila, and E. Domingo. 2006. Viral fitness can influence the repertoire of virus variants selected by antibodies. J. Mol. Biol. 36244-54. [DOI] [PubMed] [Google Scholar]
- 26.Martinez, M. A., J. M. E. Piccone, E. L. Palma, E. Domingo, N. Knowles, and M. G. Mateu. 1991. Two mechanisms of antigenic diversification of foot-and-mouth disease virus. Virology 184695-706. [DOI] [PubMed] [Google Scholar]
- 27.Mulders, M. N., J. H. Reimerink, M. Stenvik, I. Alaeddinoglu, H. G. van der Avoort, T. Hovi, and M. P. Koopmans. 1999. A Sabin vaccine-derived field isolate of poliovirus type 1 displaying aberrant phenotypic and genetic features, including a deletion in antigenic site 1. J. Gen. Virol. 80907-916. [DOI] [PubMed] [Google Scholar]
- 28.Mullan, B., E. Kenny-Walsh, J. K. Collins, F. Shanahan, and L. J. Fanning. 2001. Inferred hepatitis C virus quasispecies diversity is influenced by choice of DNA polymerase in reverse transcriptase-polymerase chain reactions. Anal. Biochem. 289137-146. [DOI] [PubMed] [Google Scholar]
- 29.Nainan, O. V., M. A. Brinton, and H. S. Margolis. 1992. Identification of amino acids located in the antibody binding sites of human hepatitis A virus. Virology 191984-987. [DOI] [PubMed] [Google Scholar]
- 30.Parker, J. 1989. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 53273-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ping, L.-H., and S. M. Lemon. 1992. Antigenic structure of human hepatitis A virus defined by analysis of escape mutants selected against murine monoclonal antibodies. J. Virol. 662208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pintó, R. M., J. F. González-Dankaart, G. Sánchez, S. Guix, M. J. Gómara, M. García, I. Haro, and A. Bosch. 1998. Enhancement of the immunogenicity of a synthetic peptide bearing a VP3 epitope of hepatitis A virus. FEBS Lett. 438106-110. [DOI] [PubMed] [Google Scholar]
- 33.Pintó, R. M., L. Aragonès, M. I. Costafreda, E. Ribes, and A. Bosch. 2007. Codon usage and replicative strategies of hepatitis A virus. Virus Res. 127158-163. [DOI] [PubMed] [Google Scholar]
- 34.Racaniello, V. R. 2001. Picornaviridae: the viruses and their replication, p. 685-722. In B. N. Fields, D. M. Knipe, P. M. Howley, and D. E. Griffin (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 35.Robertson, B. H., R. W. Jansen, B. Khanna, A. Totsuka, O. V. Nainan, G. Siegl, A. Widell, H. S. Margolis, S. Isomura, K. Ito, T. Ishizu, Y. Moritsugu, and S. M. Lemon. 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J. Gen. Virol. 731365-1377. [DOI] [PubMed] [Google Scholar]
- 36.Sánchez, G., L. Aragonès, M. I. Costafreda, E. Ribes, A. Bosch, and R. M. Pintó. 2004. Capsid region involved in hepatitis a virus binding to glycophorin A of the erythrocyte membrane. J. Virol. 789807-9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sánchez, G., A. Bosch, G. Gomez-Mariano, E. Domingo, and R. M. Pintó. 2003. Evidence for quasispecies distributions in the human hepatitis A virus genome. Virology 31534-42. [DOI] [PubMed] [Google Scholar]
- 38.Sánchez, G., A. Bosch, and R. M. Pintó. 2003. Genome variability and capsid structural constraints of hepatitis A virus. J. Virol. 77452-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sánchez, G., R. M. Pintó, and A. Bosch. 2004. A novel CD4+ T-helper lymphocyte epitope in the VP3 protein of the hepatitis A virus. J. Med. Virol. 72525-532. [DOI] [PubMed] [Google Scholar]
- 40.Sánchez, G., R. M. Pintó, H. Vanaclocha, and A. Bosch. 2002. Molecular characterization of hepatitis A virus isolates from a transcontinental shellfish-borne outbreak. J. Clin. Microbiol. 404148-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stapleton, J. T., and S. M. Lemon. 1987. Neutralization escape mutants define a dominant immunogenic neutralization site on hepatitis A virus. J. Virol. 61491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stapleton, J. T., V. Raina, P. L. Winokur, K. Walters, D. Klinzman, E. Rosen, and J. H. McLinden. 1993. Antigenic and immunogenic properties of recombinant hepatitis A virus 14S and 70S subviral particles. J. Virol. 671080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor, M. B. 1997. Molecular epidemiology of South African strains of hepatitis A virus: 1982-1996. J. Med. Virol. 51273-279. [DOI] [PubMed] [Google Scholar]
- 44.van Nimwegen, E., J. P. Crutchfield, and M. Huynen. 1999. Neutral evolution of mutational robustness. Proc. Natl. Acad. Sci. USA 969716-9720. [DOI] [PMC free article] [PubMed] [Google Scholar]