Abstract
Epstein-Barr virus (EBV) latent infection, and its associated oncogenic potential, is dependent on genome maintenance functions of EBV nuclear antigen 1 (EBNA-1), one of six EBNAs expressed from a common promoter (Wp and then Cp) upon infection of naive B cells. Subsequent host-mediated silencing, however, necessitates the expression of EBNA-1 from the EBNA-1-specific promoter Qp to ensure against genome loss during cell division, including EBV-associated malignancy. Here we addressed the mechanism by which EBNA-1 represses Qp through binding downstream of the transcription start site and the role of this autoregulatory function in EBV latency. Our results revealed that EBNA-1 does not inhibit transcription from Qp, as previously predicted, but acts post- or cotranscriptionally to block the processing of primary transcripts. This does not, however, require the RGG motifs responsible for strong but nonspecific RNA binding by EBNA-1. Within isogenic B-cell lines using either Cp/Wp or Qp, EBNA-1 occupancy of Qp is equivalent, suggesting that autoregulation occurs, albeit to different degrees, during full and restricted EBV latency programs. Finally, in cell lines using Cp or Wp for EBNA expression, unprocessed transcripts from Qp are detectable in the absence of corresponding mRNAs, providing further evidence that this novel mechanism of EBNA-1 action functions during latency. This posttranscriptional mechanism of regulation would provide an efficient means to monitor and regulate EBNA-1 expression from Qp, ensuring levels adequate for genome maintenance but, perhaps more importantly, below an immunogenic threshold above which latently infected cells may be at risk for elimination by EBNA-1-specific cytotoxic T cells.
Among the Epstein-Barr virus (EBV) latency gene products, EBV nuclear antigen 1 (EBNA-1) is the only protein required for maintenance of the episomal EBV genome (29, 61), a process essential for EBV persistence and thus its oncogenic potential (24). The genome maintenance functions of EBNA-1 require sequence-specific binding to two functionally distinct elements of the latent infection origin of plasmid DNA replication, oriP: the family of repeats (FR), consisting of approximately 20 30-bp repeats, each containing an EBNA-1 binding site (collectively, the region I EBNA-1 binding sites), and following ∼800 bp of intervening DNA, a region of dyad symmetry containing four additional (region II) EBNA-1 binding sites (19, 36, 38). EBNA-1 bound to the region of dyad symmetry orchestrates the assembly of the cellular DNA replication machinery at oriP and the initiation of DNA synthesis (6, 11, 12, 46), whereas FR-bound EBNA-1 serves to partition EBV genomes to daughter cells by tethering its viral DNA cargo to host metaphase chromosomes via its interaction with a cellular chromosome-associated protein (20, 48) or direct interaction with host DNA through an AT hook mechanism (47). Additionally, EBNA-1 can function as a transcriptional activator when bound to multiple sites within FR, activating promoters of latency-associated genes located within 10 kbp on either side of oriP, including Cp capable of driving the expression of the six-member family of EBNA proteins during the latency III or growth program of latency gene expression (Fig. 1) (1, 13, 29, 37, 52, 60).
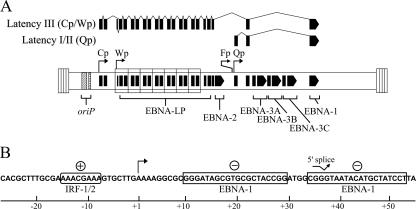
FIG. 1.
EBNA-1 promoter usage and regulation. (A) Exon structures of EBNA-1 mRNAs expressed from Cp or Wp during the latency III program of EBV or from Qp during the latency I and II programs are shown relative to the EBV genome in its linear configuration bounded by short terminal repeats (not to scale). The fourth known EBNA-1 promoter, Fp, is located approximately 200 bp upstream of Qp and is active during the EBV lytic cycle. EBNA exons are bracketed; the large internal repeat 1 elements that encode the majority of EBNA-LP are boxed. Whereas all EBNAs can be expressed from Wp or Cp, Qp is used exclusively for the expression of EBNA-1. (B) Positive (+) and negative (−) regulatory elements of Qp relevant to this work are indicated; numbering is relative to the major site of transcription initiation, +1 (bent arrow). Note that the first two exons (Q and U in the text) of the EBNA-1 mRNA from Qp are noncoding.
A third and lower-affinity set of EBNA-1 binding sites (region III) is located ∼43 kbp downstream of oriP (2, 19). These two sites lie in tandem at position +10 of the promoter Qp, which drives the exclusive expression of EBNA-1 during the restricted programs of EBV latency gene expression (latency I and latency II; Fig. 1). The principal significance of this EBNA-1-only pattern of EBNA transcription, brought about by epigenetic silencing of the common EBNA promoters Cp and Wp (5, 17, 35, 40, 41, 45, 56), is that it enables the essential expression of EBNA-1 in the absence of the remaining five EBNA proteins that, though important for the establishment of a persistent infection, would ultimately subject the infected cell to elimination by the host cytotoxic T-cell response directed against epitopes within either the EBNAs themselves or viral proteins (e.g., LMP-2A) whose expression these EBNAs (as transcription factors) regulate (39). By contrast, sustained expression of EBNA-1 is possible in part due to properties of its glycine-alanine repeat (GAr) domain that prevent major histocompatibility complex (MHC) class I-restricted presentation of EBNA-1 epitopes by the infected cell (26, 27, 54, 55, 62). Qp is also responsible for EBNA-1 expression within EBV-associated tumors, e.g., Burkitt lymphoma (BL) and nasopharyngeal carcinoma (33, 45, 50). Thus, appropriate expression of EBNA-1 through Qp is pivotal to the normal biology of EBV, as well as to its oncogenic potential.
Whereas EBNA-1 bound to oriP can activate transcription, the effect of EBNA-1 on Qp-driven gene expression is a potent repression (42, 45, 53). This apparent autoregulatory function presumably prevents overexpression of EBNA-1 during restricted latency (Qp active) and may contribute to repression of Qp during latency III (Cp/Wp active). In reporter-based assays, repression of Qp is evident at the mRNA as well as the protein level, suggesting that EBNA-1 regulates Qp through a transcriptional mechanism (42, 45). However, this does not necessarily exclude a posttranscriptional mode of action. For example, the EBNA-1 binding sites could serve as a platform to promote local interaction of EBNA-1 with its nascent transcripts through the strong RNA-binding capability of its N terminus (28, 51), potentially blocking their processing and/or transport to the cytoplasm. Here we directly addressed the mechanism of this EBNA-1 function and demonstrate that repression is indeed mediated through a posttranscriptional, or more likely cotranscriptional, inhibition of pre-mRNA processing. This novel mode of action, however, appears not to require EBNA-1's strong RNA-binding capability. Furthermore, we provide evidence that this posttranscriptional regulation by EBNA-1 occurs in the context of latent EBV infection and that it may be the primary determinant of Qp inactivity during latency III. Most importantly, autoregulation likely contributes to immune evasion by preventing unnecessary new synthesis, thus limiting the production of EBNA-1 peptides derived from defective ribosomal products believed to be the primary source of EBNA-1 epitopes presented in association with MHC class I antigens (55, 58).
MATERIALS AND METHODS
Cell lines and culture.
Cells were maintained in RPMI 1640 medium containing 2 mM l-glutamine and 10% defined fetal bovine serum (HyClone). Louckes and BJAB are EBV-negative BL and B-lymphoma cell lines, respectively. Paired BL lines (i) KemI and KemIII and (ii) MutuI and MutuIII (15) maintain EBV latency I and latency III, respectively. BL lines Oku and Sal maintain the Wp-restricted program of EBV latency (21). LHF and TN11/10 are EBV-transformed lymphoblastoid cell lines (LCLs) that maintain latency III.
Plasmids and reporter assay.
Plasmids pOGH.006 and pOGH.059 contained EBV DNA from −681 to +75 and −143 to +5068, respectively, relative to the Qp transcription start site (+1) upstream of the promoterless human growth hormone (hGH) gene in pOGH (32). Transient expression of EBNA-1 or the EBNA-1 mutant NΔ450-641 (22) (gift from B. Sugden) was achieved from vector pSG5 (Stratagene). Cells (8 × 106) in fresh growth medium were electroporated in a cuvette with a 0.4-cm gap in a Bio-Rad Gene Pulser (250 V, 960 μF) with 10 μg reporter plasmid and 5 μg EBNA-1 expression plasmid or pSG5. If cells were not used for RNA analysis or nuclear run-on assay, 1 μg of β-galactosidase expression plasmid (pCMV-βgal) was included to normalize for differences in transfection efficiency. The amount of hGH in culture medium was determined in duplicate by radioimmunoassay (Nichols Institute) and normalized to the β-galactosidase activity (adjusted for total protein assayed) present within the cell extract.
Nuclear run-on assay of transcription.
Two protocols were used, one using purified nuclei and the other using cells permeabilized with lysolecithin. Purified nuclei were prepared from 4 × 107 viable cells (pooled from several transfections) by the alternate protocol for isolation of nuclei by sucrose gradient centrifugation and were kept until use in glycerol storage buffer at 2 × 107 to 3 × 107 nuclei per 200 μl (one labeling reaction mixture) in liquid nitrogen, all exactly as previously described (14). Nascent RNA was labeled by incorporation of [α-32P]UTP at 30°C for 30 min, followed by treatment with DNase I and proteinase K as previously described (14). Nuclei were solubilized in 7 ml RNA-BEE (Tel-Test), and following addition of 100 μg yeast tRNA, 32P-labeled RNA was purified from 1-ml aliquots according to the manufacturer's instructions, precipitated in ethanol, and dissolved in H2O (100 μl total). Labeling of RNA within cells (8 × 106 per reaction mixture) permeabilized in 26% lysolecithin (Sigma-Aldrich) (8) was performed as previously described (4, 8) at 37°C for 30 min. Cells were then treated with DNase I and proteinase K, and RNA was purified as described above. Equal numbers of counts per minute of 32P-labeled RNA from each sample (with or without EBNA-1) were heat denatured and hybridized to slot blots at 65°C for 40 h in 1.5 ml of 500 mM Na2HPO4-7% sodium dodecyl sulfate (SDS)-1 mM EDTA (pH 8.0). Blots were then washed in decreasing concentrations (2.0× to 0.5×) of SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) in 1% SDS at 65°C, dried, and processed by phosphorimage analysis and autoradiography. Slot blots contained 5 μg M13 phage single-stranded DNA containing the sense or antisense strand of hGH, 5 μg λ phage DNA (control for nonspecific hybridization), and 2.5 μg of the DNA fragment(s) encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and/or 28S rRNA (normalization controls).
RNA (Northern) blot and reverse transcription (RT)-PCR analyses.
Total RNA was isolated with RNA-BEE (Tel-Test), and residual DNA was removed by digestion with RQ1 DNase (Promega). For RNA blot analysis, 10 μg of RNA was fractionated by electrophoresis through a 1.2% agarose-2.2 M formaldehyde gel and analyzed by Northern blot hybridization with 32P-labeled hGH cDNA by standard techniques. Blots were reprobed for GAPDH mRNA to monitor RNA loading.
For detection of Qp-derived transcripts by either standard or quantitative (real-time) RT-PCR, cDNA synthesis was primed with an RT primer that would anneal within either the first intron or a downstream exon to amplify primary or mRNA transcripts, respectively. The cDNA was then amplified by PCR with a 5′ forward (Fw) primer that would anneal near the beginning of the first exon (common to the primary transcript and mRNA) and the appropriate 3′ reverse (Rv) primer that would anneal near the beginning of the first intron to detect primary transcripts or within the second exon to detect mRNAs (see Fig. 3). Cellular GAPDH mRNA was amplified in parallel as a general positive control. All of the primers used in this study are described in Table 1, as are the TaqMan probes and GAPDH primers purchased from Applied Biosystems (GAPDH Control Reagents, ABI 402869). First-strand cDNA was synthesized from 1 μg total RNA with 2 pmol RT primer in a 20-μl reaction mixture with SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. For standard PCR, 1 μl cDNA was amplified with the Expand High Fidelity PCR system (Roche) in the presence of 250 nM each Fw and Rv PCR primer in a 25-μl reaction mixture. As controls for DNA contamination, amplification was performed on equal aliquots of primary transcript cDNA synthesis reaction mixtures that did not contain reverse transcriptase (−RT) and also with each primer set in the absence of template cDNA. Thermal cycling conditions were as follows: 94°C for 2 min and then 35 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 45 s, followed by 72°C for 7 min. To quantitatively measure cDNAs representing transcripts originating from Qp within various B-cell lines (see Fig. 7), a real-time PCR was performed in an ABI Prism 7900HT sequence detector system with Qp TaqMan probes (both from Applied Biosystems) labeled with a 5′ 6-carboxyfluorescein reporter dye and a 3′ 6-carboxytetramethylrhodamine quencher dye. Amplification of 1 μl cDNA (synthesized as described above) was performed in triplicate within 50-μl reaction mixtures containing 500 nM each Fw and Rv primers and TaqMan probe in 1× TaqMan Universal PCR Mastermix. Thermal cycling conditions were as follows: 95°C for 10 min and then 40 cycles of 95°C for 15 s and 60°C for 1 min. A standard curve of threshold cycle (CT) values was generated from serial fivefold dilutions of Qp primary transcript cDNA generated from MutuI cell RNA (Qp active for EBNA-1 expression). The CT values obtained from test samples (each amplified in triplicate) from three independent cDNA synthesis reaction mixtures were plotted on this standard curve, and the relative quantity was calculated with the Sequence Detector software, version 2.1 (Applied Biosystems). To arrive at an accurate estimation of Qp-specific transcripts, it was necessary to exclude those initiating from the lytic-cycle EBNA-1 promoter Fp (∼200 bp upstream of Qp; Fig. 1), as well as transcripts originating further upstream, e.g., Cp. To do this, levels were determined for cDNA amplified with a Fw primer (SP2-Fw) that would anneal between the Fp and Qp transcription start sites, and these values were subtracted from those obtained with the 5′ primer Qp-Fw, which would anneal to all cDNAs originating either at Qp or upstream.
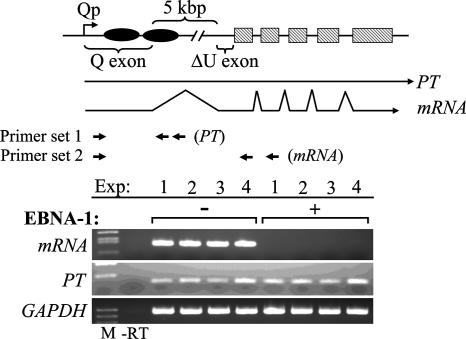
FIG. 3.
EBNA-1 blocks mRNA processing. Primary transcript (PT) and mRNA expressed from a Qp-driven hGH reporter gene (pOGH.059; top) in the absence or presence of EBNA-1 in transiently transfected EBV-negative BL cells was assessed by RT-PCR and agarose gel electrophoresis. Cells were transfected with 10 μg pOGH.059 (Qp coordinates −143 to +5068) and 5 μg pSG5 or an equal amount of pSG5-EBNA-1 expression vector. RT-PCR was performed with total RNA isolated from cells at 24 h posttransfection; RT and PCR primers used to amplify either PT or mRNA are shown relative to the structures of the unspliced (PT) and spliced (mRNA) transcripts originating from the Qp-hGH reporter construct. Primer sets 1 and 2 were used to amplify PT and mRNA cDNAs, respectively; the rightmost arrow in each set represents the RT primer. Note that the Rv PCR primer used to amplify mRNA (set 2) actually spanned the junction of the second and third exons of the reporter construct. Amplification of GAPDH cDNA was included as a control for RNA integrity. −RT, RT-PCR carried out in the absence of reverse transcriptase (a representative result is shown; all were negative). The primers used are described in Table 1. The results shown are from four independent experiments (Exp). M, DNA size standards.
TABLE 1.
Primers and TaqMan probes used in this study
| Application (figure[s]) and primer or probe | Location | Sequence (5′ to 3′) | EBV genome coordinates |
|---|---|---|---|
| RT-PCR, PT vs mRNA (3, 5) | |||
| Qp-Fw | Q exon | AAGGCGCGGGATAGC | 62425-62439 |
| Qp-Rv | Q/U intron | TGCCAAAATGTAAGGATAGC | 62487-62468 |
| hGH-Rv | 1st and 2nd exons of hGH | GAGCCTGTAGCCATTGC | NAa |
| Q intron-RT | Q/U intron | ACACCCCAGTTGGTGCAT | 62871-62854 |
| hGH-RT | 2nd exon of hGH | CGTTGTCAAAAAGCCTGGAT | NA |
| GAPDH-Fw | 2nd exon | GAAGGTGAAGGTCGGAGTC | NA |
| GAPDH-Rv | 4th exon | GAAGATGGTGATGGGATTTC | NA |
| GAPDH-RT | 5th exon | TTGATTTTGGAGGGATCTCG | NA |
| PT mapping (7) | |||
| SP2-Fw | Between Fp and Qp start sites | CCTGTCACCACCTCCCTGAT | 62289-62308 |
| Qp-Fw | Q exon | AAGGCGCGGGATAGC | 62425-62439 |
| Qp-Rv | Q/U intron | TGCCAAAATGTAAGGATAGC | 62487-62468 |
| Q intron-RT | Q/U intron | ACACCCCAGTTGGTGCAT | 62871-62854 |
| K-Rv | K exon | CTCTATGTCTTGGCCCT | 108151-108135 |
| K-RT | K exon | CTTTGCAGCCAATGCAA | 108199-108183 |
| ChIP (6) | |||
| ChIP Qp-Fw | Qp (−87 to −64) | GACCACTGAGGGAGTGTTCCACAG | 62336-62359 |
| ChIP Qp-Rv | Q/U intron (+116 to +96) | ACACCGTGCGAAAAGAAGCAC | 62538-62518 |
| BamHI-D-Fw | BamHI-D region | TTGGGTGTGGATACCCATGT | 134101-134120 |
| BamHI-D-Rv | BamHI-D region | GTGGTCAGGACCGATGAGAT | 134331-134312 |
| TaqMan probes (6, 7) | |||
| Qp | Q exon (+40 to +19) | TTACCCGCCATCCGGTAGCGCAb | 62462-62441 |
| GAPDH | 4th exon | CAAGCTTCCCGTTCTCAGCC | NA |
NA, not applicable.
Ends with first four nucleotides of the Q/U intron, which are identical to first four nucleotides of the second exon, U.
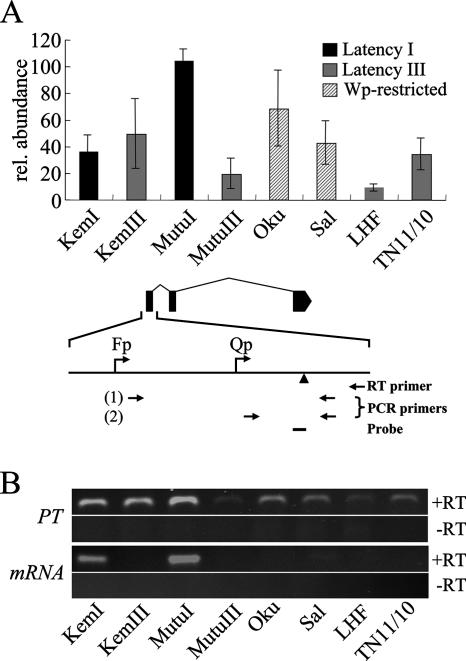
FIG. 7.
Evidence of posttranscriptional regulation in latently infected B cells. (A) Detection by quantitative RT-PCR of unspliced transcripts from Qp in B-cell lines maintaining either latency I, latency III, or Wp-restricted latency. Data on the vertical axis represent the means of three independent RT reaction mixtures in which each cDNA sample was amplified in triplicate and CT values were normalized to the level of GAPDH cDNA amplified from the same sample. Values obtained with RNA from MutuI cells were arbitrarily set at 100; error bars indicate standard errors of the means. Positions of RT and PCR primers used are shown relative to the Qp-specific EBNA-1 mRNA (not to scale). (1) Primer set (SP2-Fw and Qp-Rv; Table 1) used to detect transcripts initiating at Fp and upstream. (2) Primer set (Qp-Fw and Qp-Rv) used to detect transcripts initiating at Qp and upstream. The amount of Qp-specific transcript was determined by subtraction of the value obtained with primer set 1 from that obtained with primer set 2. rel., relative. (B) Representative results obtained by standard (nonquantitative) RT-PCR demonstrating the presence of unspliced transcript (PT) and absence of Qp-specific EBNA-1 mRNA in RNA samples analyzed in panel A. PT was amplified as described above, with primer set 2; to amplify EBNA-1 mRNA, RT was primed within the 5′ end of the EBNA-1 coding exon (K) and amplified with a nested Rv primer (K-Rv; Table 1) and Qp-Fw. Lack of a product in the −RT control for detection of PT confirms that the data in panel A are not derived from contaminating viral DNA. The absence of detectable Qp-specific EBNA-1 mRNA in cells that use Cp or Wp but express unspliced transcripts from Qp as shown in panel A supports a posttranscriptional mechanism of regulating EBNA-1 expression from Qp during latency III and Wp-restricted latency. Note that because the PT cDNA levels amplified here are not corrected for transcripts initiating upstream of Qp, there is not complete correlation between the PT results in panels A and B.
Chromatin immunoprecipitation (ChIP).
EBNA-1 occupancy of Qp within KemI and KemIII cells was determined with the ChIP-IT kit (Active Motif). Briefly, 1.25 × 106 cells were fixed in 1% paraformaldehyde, washed, lysed, and sonicated. The soluble fraction was precleared with salmon sperm DNA-protein A-agarose (Upstate) incubated overnight with 10 μl rabbit antiserum to EBNA-1 (gift from J. Hearing) or 4 μg control immunoglobulin G (IgG). Immune complexes from these precipitations and a mock precipitation were precipitated with protein A-agarose, eluted in 50 mM NaHCO3-1% SDS, and incubated at 65°C for 4 h to overnight to reverse cross-linking. Samples were digested with proteinase K, and DNA was column purified and dissolved in 100 μl H2O. For quantification of Qp DNA obtained by ChIP, 1 μl of sample (equivalent to the material obtained from 1.25 × 104 cells) was subjected to real-time PCR (thermal cycling conditions as for a quantitative RT-PCR) in triplicate with primers that would amplify a 203-bp DNA fragment of Qp (−87 to +116) and a TaqMan probe representing the sequence between +19 and +40 (Table 1). The standard curve was generated by amplification of Qp DNA from fivefold serial dilutions of cleared KemI cell lysate that was subjected to immunoprecipitation. As an additional control for specificity, semiquantitative amplification of a 231-bp DNA fragment from the EBV BamHI-D region lacking EBNA-1 binding sites was performed on material obtained by immunoprecipitation with EBNA-1 antiserum or IgG control antibody and by mock immunoprecipitation; here, the level of BamHI-D DNA recovered with EBNA-1 antiserum was at or below that obtained with IgG or in the mock precipitation (data not shown), confirming the specificity of the EBNA-1 immunoprecipitation.
RESULTS
Repression by EBNA-1 is posttranscriptional.
The close proximity (+10) of the EBNA-1 binding sites to the transcription start site suggests that EBNA-1 represses Qp activity by interfering with transcription, either by inhibiting recruitment of transcription factors to the promoter or by acting as a blockade to the advancing RNA polymerase II complex. However, two observations raised the possibility that EBNA-1 might not regulate Qp transcriptionally. First, EBNA-1 no longer has an effect on reporter expression when binding sites are placed at +50 of a heterologous promoter (45) or are relocated to a downstream intron within a Qp-driven hGH reporter gene (J.T.S., unpublished observation), arguing against a transcriptional blockade. Second, we have found by electrophoretic mobility shift assay (data not shown) that EBNA-1 is unable to interfere with the binding of IRF-2, the principal transactivator of Qp (31, 43), to its response element 8 bp upstream of the transcription start site (Fig. 1).
To directly address the nature of EBNA-1 repression, we performed nuclear run-on assays of Qp-directed transcription in the presence and absence of EBNA-1. Two different Qp-hGH reporter constructs were used, both of which support Qp-specific transcription (32). The first (pOGH.006) contained DNA from −681 to +75, in which the first exon (Q) and 39 bp of the adjacent intron were fused to the 5′ exon of the five-exon hGH gene. The second construct (pOGH.059) extended from −143 through the entire 5,018-bp first intron and 14 bp of the second exon (U), which was fused to the 5′ exon of hGH. We reasoned that the latter construct, which includes all known regulatory elements of Qp, would be more representative of the genomic EBNA-1 transcription unit. Further, there could be no transcription from the adjacent Fp, which initiates approximately 200 bp upstream of the Qp start site, albeit under conditions that support the virus replication cycle (23, 30, 33, 44). Upon transient transfection of EBV-negative Louckes BL cells, EBNA-1 repression was monitored by assay of hGH in the culture supernatant, while transcription was assessed by nuclear run-on assay. As demonstrated by the results in Fig. 2, whereas EBNA-1 repression of hGH expression was typically greater than 90%, a parallel effect on hGH transcription was not observed. In all, this experiment was performed five times (four times with pOGH.006 and once with pOGH.059) with either of two run-on assays, one with isolated nuclei (14) and the other with permeabilized cells (8). When data were normalized to either 28S RNA (all five experiments) or GAPDH (three experiments), hGH transcription observed in the presence of EBNA-1 was 143.8% (standard error [SE], 26.3%) or 158.3% (SE, 55.8%), respectively, of that observed in the absence of EBNA-1. Although these data suggest that EBNA-1 may have a slight positive influence on transcription, they clearly indicate that the dominant negative effect of EBNA-1 on Qp-driven gene expression is posttranscriptional in nature.
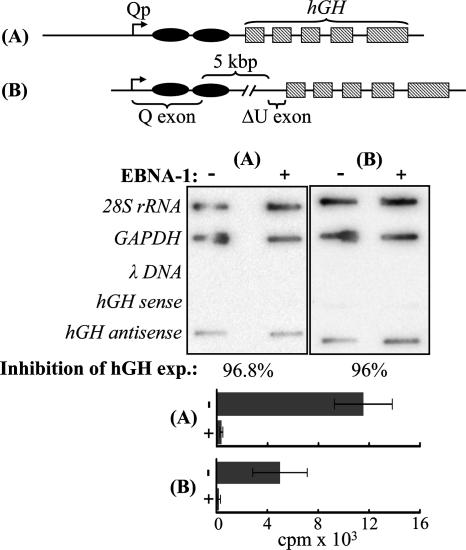
FIG. 2.
EBNA-1 repression is posttranscriptional. The effect of EBNA-1 on transcription from Qp was assessed by nuclear run-on assay with EBV-negative BL cells transiently transfected with either of two Qp-driven hGH reporter constructs and pSG5 or an equal amount of a pSG5-based EBNA-1 expression vector. (Top) Reporter constructs (not to scale). (A) pOGH.006, Qp coordinates −681 to +75. (B) pOGH.059, Qp coordinates −143 to +5068, containing all known regulatory elements of Qp, the entire 5,018-bp first intron, and 14 bp of the second and noncoding exon (U). Ovals depict the EBNA-1 binding sites within the first exon (Q), and shaded boxes represent hGH exons. (Middle) Results of run-on assay of transcription from pOGH.006 (A) and pOGH.059 (B) in the absence (−) and presence (+) of EBNA-1. The signal resulting from hGH-specific transcription is detected with single-stranded DNA representing hGH antisense (coding) strand DNA on the slot blot. Results are representative of five independent experiments in which the mean value for transcription in the presence of EBNA-1 was 143.8% (SE, 26.3%) or 158.3% (SE, 55.8%) of that during its absence when normalized to 28S RNA (all five experiments) or GAPDH (three experiments), respectively. Inhibition of hGH (protein) expression by EBNA-1 was greater than 93% in four experiments and 72% in one experiment. (Bottom) Parallel analysis of hGH protein expression in either the absence (−) or the presence (+) of EBNA-1 by radioimmunoassay revealed at least 96% inhibition by EBNA-1 when hGH assays were performed on individual transfected-cell cultures prior to pooling them for their respective nuclear run-on assay.
EBNA-1 inhibits pre-mRNA processing.
We next assessed whether EBNA-1 might interfere with the processing of primary transcripts, which would be consistent with the lack of an effect on transcription and our previous finding that EBNA-1 inhibits the accumulation of mRNA (42, 45). We therefore compared, by RT-PCR, the levels of primary (unspliced) transcript in the presence and absence of EBNA-1 expression. As shown in Fig. 3, whereas mRNA levels were dramatically reduced by EBNA-1, as expected, primary transcript levels (i.e., those from which the first intron had not been excised) were largely unaffected. When RT-PCR was performed under quantitative (real-time) PCR conditions, we consistently observed a small (1.17-fold) increase in primary transcript levels in the presence of EBNA-1 (data not shown). Thus, EBNA-1 is able to block the splicing of at least the cap-proximal exon of a Qp-derived transcript, with relatively little degradation of affected transcripts as a result. Because splicing is one of three integrated pre-mRNA processing events (5′ capping, splicing, and 3′-end processing) that occur as the nascent transcript emerges from the advancing polymerase II complex (65), we conclude that EBNA-1 represses Qp-driven gene expression by inhibition of one or more aspects of cotranscriptional processing of the pre-mRNA.
Repression by EBNA-1 is through its C-terminal domain.
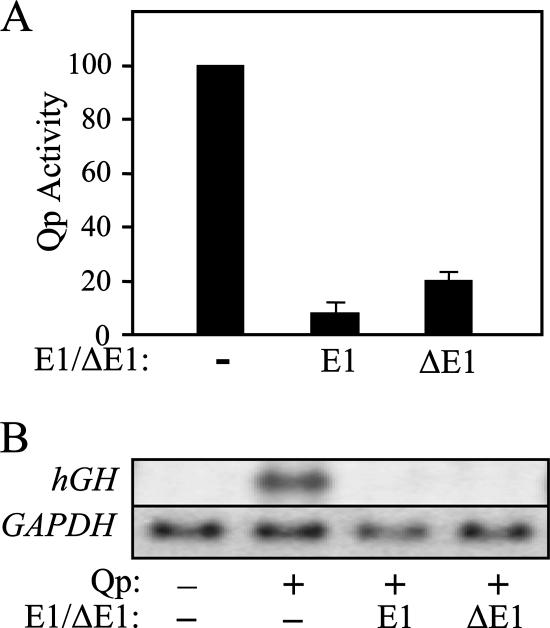
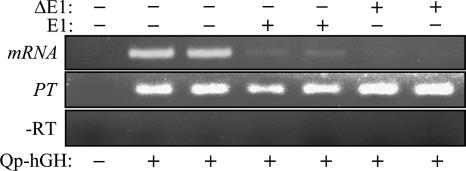
Deletion or mutation of the EBNA-1 binding sites within Qp results in loss of EBNA-1 responsiveness (42, 45, 53), indicating that specificity of repression by EBNA-1 is dependent on its sequence-specific DNA-binding property (although the context of the EBNA-1 response element within the gene is critical, as noted above). Given the strong but largely nonspecific RNA-binding capability of EBNA-1 (28, 51), we next explored the possibility that EBNA-1, when positioned downstream of the transcription start site as in Qp, might mediate repression through interaction with its nascent transcript. RNA binding by EBNA-1 is attributed to three RGG RNA-binding motifs within the N-terminal half of the protein (28). To determine whether these or other elements within the N-terminal domain of EBNA-1 are required for repression, we assessed the ability of a deletion mutant form of EBNA-1, NΔ450-641, to repress Qp-mediated reporter expression. NΔ450-641, referred to here as ΔE1, lacks amino acids (aa) 1 to 378 and 387 to 450 of the 641-aa EBNA-1 sequence but contains the nuclear localization signal (aa 379 to 386) and the C-terminal 191 aa spanning the DNA-binding and dimerization domains (aa 461 to 604) of EBNA-1 (22). When we compared it to full-length EBNA-1 (E1), ΔE1 appeared as effective in the repression of reporter (hGH) protein and mRNA expression (Fig. 4A and B). Further, analysis by RT-PCR indicated that ΔE1 too is able to suppress splicing of the cap-proximal exon (Fig. 5). Thus, the C-terminal domain of EBNA-1 is sufficient to mediate the posttranscriptional repression observed with full-length EBNA-1 (Fig. 3), without a contribution from RNA-binding or other domains of the N terminus of EBNA-1.
FIG. 4.
The C terminus of EBNA-1 is sufficient to repress Qp-mediated gene expression. The relative effects of full-length and NΔ450-641 EBNA-1 (E1 and ΔE1, respectively) on Qp-driven hGH expression were assessed in transiently transfected, EBV-negative BL cells. (A) Effects of E1 and ΔE1 on hGH protein expression. Cells were transfected with 10 μg pOGH.006 and either 5 μg pSG5 or an equal amount of pSG5-E1 or pSG5-ΔE1. Transfections were done in triplicate, and hGH values were normalized for transfection efficiency; error bars indicate the standard deviations of the means of duplicate hGH determinations for each transfection. (B) Northern blot analysis of the effects of E1 and ΔE1 on hGH mRNA in EBV-negative BL cells; transfections as in panel A. The first lane contained RNA from cells transfected with the promoterless hGH reporter plasmid pOGH; the blot was reprobed for GAPDH mRNA to monitor the loading of RNA.
FIG. 5.
The N terminus of EBNA-1 is not required for posttranscriptional repression. Following transfection, the primary transcript (PT) and mRNA expressed from pOGH.059 (Qp coordinates −143 to +5068) in the absence or presence of E1 or ΔE1 were amplified by RT-PCR (as in Fig. 3) and cDNA was detected by agarose gel electrophoresis. −RT, RT-PCR carried out in the absence of reverse transcriptase in the respective PT reaction mixtures. Lane pairs 2 and 3, 4 and 5, and 6 and 7 represent results from two independent transfections for each of the hGH reporter and E1/ΔE1 expression construct combinations indicated.
EBNA-1 autoregulation within latently infected cells.
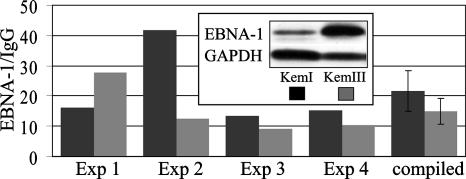
We next sought evidence for autoregulation in the context of latent EBV infection within B-cell lines that either express the six-member family of EBNAs from either Cp or Wp (latency III) or use Qp for the exclusive expression of EBNA-1 (latency I). Because usage of Qp might reflect a difference in binding by EBNA-1, we first examined the EBNA-1 occupancy of Qp during latency I and III infections by ChIP. EBNA-1 binding to Qp within B-cell lines has been noted previously (6, 10). However, due to the different origins of the B-cell lines examined previously, e.g., LCL versus BL, as well as their unknown viral genome copy number and corresponding level of EBNA-1 expression, it is not possible to conclude from earlier reports whether there are latency program-specific differences in EBNA-1 binding to Qp. We therefore examined EBNA-1 occupancy of Qp within two BL cell lines, KemI and KemIII (gift from A. Rickinson), that originated from the same tumor but which stably maintain latency I or III, respectively. These cell lines are, in theory, isogenic, and thus any difference in EBNA-1 binding to Qp within them is more likely related to the latency program instead of the genetic background (cellular or viral) or B-cell origin.
Analysis of EBNA-1 occupancy by ChIP and quantitative PCR yielded similar recoveries of Qp DNA from KemI and KemIII cells, although we achieved greater recovery of Qp from immunoprecipitates of EBNA-1 from KemI lysates in three of four experiments (Fig. 6). Quantification of the EBV DNA content within these lines indicated that the genome copy number in KemI is approximately twice that of KemIII (data not shown), yet EBNA-1 levels were consistently higher in KemIII cells, which use Cp for EBNA-1 expression (Fig. 6, insert). Although these and additional variables inherent to ChIP made it difficult to precisely determine relative occupancy, our results suggest that there is not a substantial difference in EBNA-1 binding to Qp between closely matched cells that differ primarily in the EBV latency program that they maintain.
FIG. 6.
EBNA-1 is bound to Qp during latency I and latency III. Detection of EBNA-1 occupancy of Qp by ChIP during latency I and latency III in KemI and KemIII BL cells, respectively. Data were acquired by quantitative (real-time) PCR amplification of Qp DNA (−87 to +116) immunoprecipitated with antiserum to EBNA-1 and are expressed relative to values obtained by parallel immunoprecipitation with a non-EBNA-1-specific IgG antibody; amplification of a 231-bp region of EBV BamHI-D DNA, lacking EBNA-1 binding sites, following EBNA-1 immunoprecipitation consistently yielded values below the background (data not shown). Results from four independent ChIP assays are shown; a compilation of these data, including the standard errors of means, is shown on the right. See Table 1 for descriptions of the primers and TaqMan probes used. (Insert) Immunoblot analysis of EBNA-1 levels relative to GAPDH in KemI and KemIII BL cells. Exp, experiment.
We next sought more direct evidence of posttranscriptional repression of Qp in vivo by searching for the presence of unprocessed EBNA-1 transcripts in a panel of BL cell lines and LCLs that do not use Qp. RNAs from KemI and MutuI BL cells maintaining latency I (EBNA-1 expression from Qp) were included as positive controls for detection of primary transcripts. A quantitative RT-PCR assay was used, as levels of endogenous primary transcripts are too low to be detected by nuclease protection assay, if they are indeed present. To exclude overlapping transcripts originating from Cp or Wp during latency III, or Fp in a low percentage of lytically infected cells, we used a 5′ PCR primer that would anneal either immediately downstream of the Qp transcription start site (to detect transcripts from Qp and upstream promoters) or one that would anneal upstream of this site but downstream of Fp to permit detection of transcripts originating from Fp or further upstream (Fig. 7A). The difference between the results obtained with these two 5′ primers, in conjunction with a common 3′ primer specific for the first intron, should represent transcripts initiating from Qp. As shown in Fig. 7A, not only were we able to detect unspliced transcripts originating from Qp in BL cells maintaining latency I (KemI and MutuI), we detected them as well in BL cell lines and LCLs maintaining latency III. This was true also for two variant BL cell lines, Oku and Sal, that use Wp instead of either Cp or Qp for the expression of EBNA-1, as well as for the EBNA-3 proteins (the EBNA-2 open reading frame and a portion of the EBNA-LP open reading frame are deleted from the EBV genomes in these cells) (21). By contrast, EBNA-1 mRNAs indicative of Qp usage could only be amplified from KemI and MutuI cells (Fig. 7B), as expected. Although the relative amount of unprocessed transcript from Qp differed by a factor of 10 between the lowest (LHF/latency III)- and highest (MutuI/latency I)-expressing lines, levels within a given cell line remained relatively constant, and three lines that do not use Qp for EBNA-1 expression (TN11/10, Oku, and Sal) contained at least as much unprocessed transcript as KemI, which uses Qp exclusively for EBNA-1 expression. We conclude, therefore, that autorepression by EBNA-1 is indeed likely to occur in the context of latent infection, at least within the latency III program in which Qp derived EBNA-1 mRNAs are undetectable.
DISCUSSION
Here we show that EBNA-1 posttranscriptionally regulates expression from its promoter Qp by inhibiting pre-mRNA processing. Further, by detection of unspliced transcripts originating from Qp during latency III, as well as the Wp-restricted program in variant BLs, we provide evidence that this previously unknown mode of EBNA-1 action is active in latently infected B cells, likely contributing to Qp “inactivity” during latency III, in which EBNA transcription originates from Cp or Wp. While the existence of Qp-specific EBNA-1 mRNAs in BL cells maintaining latency I prevents us from concluding with certainty that negative autoregulation occurs during restricted programs of latency, EBNA-1 occupancy of Qp in these BL cells (Fig. 6) suggests that autoregulation is active. Also, earlier studies demonstrated that deletion or mutation of the EBNA-1 binding sites results in a relative increase in Qp-driven reporter activity upon transfection of BL cells maintaining either latency I or III, suggesting that autoregulation is not limited to a particular latency program (42, 45). Further, the greater amount of EBNA-1 in KemIII compared to KemI cells (insert, Fig. 6) suggests that a higher Cp-driven EBNA-1 expression may be the primary basis for the lack of mRNAs derived from Qp in these cells. However, as others have also noted (25, 45), the difference in EBNA-1 levels between latency I and latency III are often modest or not apparent by immunoblotting techniques. We have found that Qp-driven reporter expression is exquisitely sensitive to EBNA-1 and, consistent with a previous report (45), requires only a single EBNA-1 binding site to observe repression. Thus, even a very modest increase in the EBNA-1 level, and one that is perhaps not easily discernible by immunoblotting, could significantly affect Qp usage.
The presence of incompletely processed transcripts originating from Qp during latency III (Fig. 7) also argues against transcriptional repression as the ultimate determinant of Qp usage in these cells, as suggested by reporter assays demonstrating moderate repression (relative to EBNA-1) of Qp by IRF-2 and IRF-7, in conjunction with higher levels of these IRFs in cell lines that maintain latency III (63, 64). Although initially identified as a transcriptional repressor, IRF-2 is a major transactivator of Qp (31, 43) and several cellular promoters (18, 34, 57). We have found that the context of its single binding site within Qp dictates that IRF-2 functions as a transcriptional activator and that upon IRF-2 overexpression in reporter assays, apparent repression of Qp does not require IRF-2 DNA-binding activity; i.e., observed repression is the likely consequence of coactivator squelching (unpublished observations). Though we are occasionally able to detect (by ChIP) IRF-7 bound to Qp in cells that maintain latency III, this contrasts to readily detectable occupancy by IRF-2 (unpublished observation), which in our experience has a much higher affinity for Qp than IRF-7 (31). We predict, therefore, that inactivity of Qp for EBNA-1 expression during latency III is likely to be due principally to posttranscriptional regulation by EBNA-1 rather than direct transcriptional repression of Qp by IRFs.
The ability of EBNA-1 to inhibit splicing and possibly other pre-mRNA processing appears to be strictly dependent on the context of the EBNA-1 binding sites within the gene template. Notably, reporter constructs in which EBNA-1 binding sites are placed as little as an additional 40 bp downstream from the transcription start site, albeit in a heterologous promoter, are unresponsive to EBNA-1 (45), and in the context of a Qp-driven hGH reporter construct, relocation of the EBNA-1 binding sites to +580, within the second hGH intron, results in a loss of EBNA-1 responsiveness (our unpublished observation). Further, our earlier work indicated that the presence of a functional Qp 5′ splice donor site is not essential for EBNA-1 repression (42). We propose, therefore, that EBNA-1 may directly target not splicing but rather a requisite event for pre-mRNA processing in general that occurs soon after initiation of transcription. Given the position of its binding sites within Qp (+10), bound EBNA-1 seems to be optimally positioned to interfere with 5′ capping of the nascent transcript, which occurs by the time the nascent transcript is 22 to 40 nucleotides in length, and/or generation of the cap binding complex, which are critical for pre-mRNA processing (65). Precisely how EBNA-1 might do this is unclear. Our initial prediction, ruled out here, was that this might be a consequence of EBNA-1 binding to the nascent transcript via its N terminus, specifically, its RGG RNA-binding motifs (28, 51). Our finding that more than the N-terminal half of EBNA-1 is dispensable for autorepression also argues against a role for EBNA-1-associated cellular proteins that require the N terminus for interaction, notably, the potential splicing regulator p32/TAP (7, 59). In general, the identities of EBNA-1-associated proteins do not provide obvious clues to the precise mechanism of action. Exceptions may be protein arginine methyltransferases (PRMTs) 1 and 5 (49). PRMTs have been implicated in the regulation of a number of cellular processes, including pre-mRNA processing (3). Although arginine residues within EBNA-1 are methylated as an apparent result of its interactions with PRMTs (49), the purpose of this modification and the possible influence EBNA-1 has on PRMT function are unknown.
Perhaps the most intriguing aspect of this autoregulatory function of EBNA-1 is why there is a need to so carefully control EBNA-1 levels. Whereas posttranscriptional regulation of an active Qp would offer an efficient means to ensure ample EBNA-1 to occupy its higher-affinity binding sites within oriP to promote genome maintenance, the exceptionally long half-life of EBNA-1 (>36 h) (9) suggests that, instead of a need to rapidly increase EBNA-1 levels, prevention of its overexpression is more critical. A possibility for this that warrants strong consideration is that negative autoregulation contributes to immune evasion. While earlier studies suggested that the host does not mount an MHC class I-mediated T-cell response to EBNA-1, this clearly is not the case (25, 54). Though difficulty detecting T-cell responses to EBNA-1 was initially directly attributed to the inhibitory effect of the GAr domain on proteasomal degradation of EBNA-1 (26, 27), it is now evident that EBNA-1 peptides displayed in association with class I antigen primarily arise not from the degradation of mature EBNA-1 but from defective ribosomal products produced during translation of the EBNA-1 mRNA (54, 55, 58), which, interestingly, is also inhibited in cis by the GAr domain (62). Therefore, while the slow turnover of EBNA-1 as a consequence of its GAr domain would reduce the need to continually synthesize new protein, the autoregulatory function of EBNA-1 described here would ultimately ensure that EBNA-1 is produced at a level just sufficient to perform its essential latency-associated functions. This would be particularly important upon the establishment of a persistent infection within memory B cells, the major reservoir of latent EBV and in which EBNA-1 may be one of the few, if not the only, viral proteins ever expressed (16). In summary, we propose that the autoregulatory function of EBNA-1 contributes an important piece to the puzzle of EBV immune evasion, serving to prevent excessive new synthesis of EBNA-1, which in turn limits MHC class I display of EBNA-1 peptides below an immunogenic threshold, above which latently infected cells would be at risk for elimination by the anti-EBNA-1 T-cell response.
Acknowledgments
We thank Bill Sugden for the NΔ450-641 EBNA-1 construct; Janet Hearing for EBNA-1 antiserum and advice on EBNA-1 ChIP; Alan Rickinson, Lindsey Hutt-Fletcher, and John Sixbey for cell lines; Carol Dickerson and Daniel Henson for excellent technical assistance; and Peter Murray, Janet Partridge, Christopher Norbury, and Clare Sample for helpful advice and discussions.
This work was supported by U.S. Public Health Service grants CA056639 and CA073544 and Cancer Center support grant CA21765, the American Lebanese Syrian Associated Charities (ALSAC), and the Penn State Cancer Institute.
Footnotes
Published ahead of print on 12 December 2007.
REFERENCES
- 1.Altmann, M., D. Pich, R. Ruiss, J. Wang, B. Sugden, and W. Hammerschmidt. 2006. Transcriptional activation by EBV nuclear antigen 1 is essential for the expression of EBV's transforming genes. Proc. Natl. Acad. Sci. USA 10314188-14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambinder, R. F., W. A. Shah, D. R. Rawlins, G. S. Hayward, and S. D. Hayward. 1990. Definition of the sequence requirements for binding of the EBNA-1 protein to its palindromic target sites in Epstein-Barr virus DNA. J. Virol. 642369-2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedford, M. T., and S. Richard. 2005. Arginine methylation: an emerging regulator of protein function. Mol. Cell 18263-272. [DOI] [PubMed] [Google Scholar]
- 4.Boffa, L. C., E. M. Carpaneto, M. R. Mariani, M. Louissaint, and V. G. Allfrey. 1997. Contrasting effects of PNA invasion of the chimeric DMMYC gene on transcription of its myc and PVT domains. Oncol. Res. 941-51. [PubMed] [Google Scholar]
- 5.Chau, C. M., and P. M. Lieberman. 2004. Dynamic chromatin boundaries delineate a latency control region of Epstein-Barr virus. J. Virol. 7812308-12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 9810085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, M. R., J. F. Yang, C. W. Wu, J. M. Middeldorp, and J. Y. Chen. 1998. Physical association between the EBV protein EBNA-1 and P32/TAP/hyaluronectin. J. Biomed. Sci. 5173-179. [DOI] [PubMed] [Google Scholar]
- 8.Cutrona, G., E. M. Carpaneto, A. Ponzanelli, M. Ulivi, E. Millo, S. Scarfi, S. Roncella, U. Benatti, L. C. Boffa, and M. Ferrarini. 2003. Inhibition of the translocated c-myc in Burkitt's lymphoma by a PNA complementary to the Eμ enhancer. Cancer Res. 636144-6148. [PubMed] [Google Scholar]
- 9.Davenport, M. G., and J. S. Pagano. 1999. Expression of EBNA-1 mRNA is regulated by cell cycle during Epstein-Barr virus type I latency. J. Virol. 733154-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day, L., C. M. Chau, M. Nebozhyn, A. J. Rennekamp, M. Showe, and P. M. Lieberman. 2007. Chromatin profiling of Epstein-Barr virus latency control region. J. Virol. 816389-6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng, Z., L. Lezina, C. J. Chen, S. Shtivelband, W. So, and P. M. Lieberman. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9493-503. [DOI] [PubMed] [Google Scholar]
- 12.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106287-296. [DOI] [PubMed] [Google Scholar]
- 13.Gahn, T. A., and B. Sugden. 1995. An EBNA-1-dependent enhancer acts from a distance of 10 kilobase pairs to increase expression of the Epstein-Barr virus LMP gene. J. Virol. 692633-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenburg, M. E., and T. P. Bender. 1995. Identification of newly transcribed RNA, p. 4.10.1-4.10.11. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., Indianapolis, IN. [DOI] [PubMed]
- 15.Gregory, C. D., M. Rowe, and A. B. Rickinson. 1990. Different Epstein-Barr virus-B cell interactions in phenotypically distinct clones of a Burkitt's lymphoma cell line. J. Gen. Virol. 711481-1495. [DOI] [PubMed] [Google Scholar]
- 16.Hochberg, D., J. M. Middeldorp, M. Catalina, J. L. Sullivan, K. Luzuriaga, and D. A. Thorley-Lawson. 2004. Demonstration of the Burkitt's lymphoma Epstein-Barr virus phenotype in dividing latently infected memory cells in vivo. Proc. Natl. Acad. Sci. USA 101239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hutchings, I. A., R. J. Tierney, G. L. Kelly, J. Stylianou, A. B. Rickinson, and A. I. Bell. 2006. Methylation status of the Epstein-Barr virus (EBV) BamHI W latent cycle promoter and promoter activity: analysis using novel EBV-positive Burkitt and lymphoblastoid cell lines. J. Virol. 8010700-10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jesse, T. L., R. LaChance, M. F. Iademarco, and D. C. Dean. 1998. Interferon regulatory factor-2 is a transcriptional activator in muscle where it regulates expression of vascular cell adhesion molecule-1. J. Cell Biol. 1401265-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, C. H., S. D. Hayward, and D. R. Rawlins. 1989. Interaction of the lymphocyte-derived Epstein-Barr virus nuclear antigen EBNA-1 with its DNA-binding sites. J. Virol. 63101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapoor, P., B. D. Lavoie, and L. Frappier. 2005. EBP2 plays a key role in Epstein-Barr virus mitotic segregation and is regulated by aurora family kinases. Mol. Cell. Biol. 254934-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly, G., A. Bell, and A. Rickinson. 2002. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA2. Nat. Med. 81098-1104. [DOI] [PubMed] [Google Scholar]
- 22.Kirchmaier, A. L., and B. Sugden. 1997. Dominant-negative inhibitors of EBNA-1 of Epstein-Barr virus. J. Virol. 711766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lear, A. L., M. Rowe, M. G. Kurilla, S. Lee, S. Henderson, E. Kieff, and A. B. Rickinson. 1992. The Epstein-Barr virus (EBV) nuclear antigen 1 BamHI F promoter is activated on entry of EBV-transformed B cells into the lytic cycle. J. Virol. 667461-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, M. A., M. E. Diamond, and J. L. Yates. 1999. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J. Virol. 732974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, S. P., J. M. Brooks, H. Al Jarrah, W. A. Thomas, T. A. Haigh, G. S. Taylor, S. Humme, A. Schepers, W. Hammerschmidt, J. L. Yates, A. B. Rickinson, and N. W. Blake. 2004. CD8 T cell recognition of endogenously expressed Epstein-Barr virus nuclear antigen 1. J. Exp. Med. 1991409-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levitskaya, J., M. Coram, V. Levitsky, S. Imreh, P. M. Steigerwald-Mullen, G. Klein, M. G. Kurilla, and M. G. Masucci. 1995. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature 375685-688. [DOI] [PubMed] [Google Scholar]
- 27.Levitskaya, J., A. Sharipo, A. Leonchiks, A. Ciechanover, and M. G. Masucci. 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. USA 9412616-12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu, C. C., C. W. Wu, S. C. Chang, T. Y. Chen, C. R. Hu, M. Y. Yeh, J. Y. Chen, and M. R. Chen. 2004. Epstein-Barr virus nuclear antigen 1 is a DNA-binding protein with strong RNA-binding activity. J. Gen. Virol. 852755-2765. [DOI] [PubMed] [Google Scholar]
- 29.Lupton, S., and A. J. Levine. 1985. Mapping genetic elements of Epstein-Barr virus that facilitate extrachromosomal persistence of Epstein-Barr virus-derived plasmids in human cells. Mol. Cell. Biol. 52533-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nonkwelo, C., E. B. Henson, and J. Sample. 1995. Characterization of the Epstein-Barr virus Fp promoter. Virology 206183-195. [DOI] [PubMed] [Google Scholar]
- 31.Nonkwelo, C., I. K. Ruf, and J. Sample. 1997. Interferon-independent and -induced regulation of Epstein-Barr virus EBNA-1 gene transcription in Burkitt lymphoma. J. Virol. 716887-6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nonkwelo, C., I. K. Ruf, and J. Sample. 1997. The Epstein-Barr virus EBNA-1 promoter Qp requires an initiator-like element. J. Virol. 71354-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nonkwelo, C., J. Skinner, A. Bell, A. Rickinson, and J. Sample. 1996. Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J. Virol. 70623-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oshima, S., T. Nakamura, S. Namiki, E. Okada, K. Tsuchiya, R. Okamoto, M. Yamazaki, T. Yokota, M. Aida, Y. Yamaguchi, T. Kanai, H. Handa, and M. Watanabe. 2004. Interferon regulatory factor 1 (IRF-1) and IRF-2 distinctively up-regulate gene expression and production of interleukin-7 in human intestinal epithelial cells. Mol. Cell. Biol. 246298-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulson, E. J., and S. H. Speck. 1999. Differential methylation of Epstein-Barr virus latency promoters facilitates viral persistence in healthy seropositive individuals. J. Virol. 739959-9968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rawlins, D. R., G. Milman, S. D. Hayward, and G. S. Hayward. 1985. Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA-1) to clustered sites in the plasmid maintenance region. Cell 42859-868. [DOI] [PubMed] [Google Scholar]
- 37.Reisman, D., and B. Sugden. 1986. trans activation of an Epstein-Barr viral transcriptional enhancer by the Epstein-Barr viral nuclear antigen 1. Mol. Cell. Biol. 63838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reisman, D., J. Yates, and B. Sugden. 1985. A putative origin of replication of plasmids derived from Epstein-Barr virus is composed of two cis-acting components. Mol. Cell. Biol. 51822-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rickinson, A. B., and E. Kieff. 2006. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 40.Robertson, K. D., and R. F. Ambinder. 1997. Methylation of the Epstein-Barr virus genome in normal lymphocytes. Blood 904480-4484. [PubMed] [Google Scholar]
- 41.Robertson, K. D., A. Manns, L. J. Swinnen, J. C. Zong, M. L. Gulley, and R. F. Ambinder. 1996. CpG methylation of the major Epstein-Barr virus latency promoter in Burkitt's lymphoma and Hodgkin's disease. Blood 883129-3136. [PubMed] [Google Scholar]
- 42.Sample, J., E. B. Henson, and C. Sample. 1992. The Epstein-Barr virus nuclear protein 1 promoter active in type I latency is autoregulated. J. Virol. 664654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaefer, B. C., E. Paulson, J. L. Strominger, and S. H. Speck. 1997. Constitutive activation of Epstein-Barr virus (EBV) nuclear antigen 1 gene transcription by IRF1 and IRF2 during restricted EBV latency. Mol. Cell. Biol. 17873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaefer, B. C., J. L. Strominger, and S. H. Speck. 1995. The Epstein-Barr virus BamHI F promoter is an early lytic promoter: lack of correlation with EBNA 1 gene transcription in group 1 Burkitt's lymphoma cell lines. J. Virol. 695039-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaefer, B. C., J. L. Strominger, and S. H. Speck. 1997. Host-cell-determined methylation of specific Epstein-Barr virus promoters regulates the choice between distinct viral latency programs. Mol. Cell. Biol. 17364-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 204588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sears, J., M. Ujihara, S. Wong, C. Ott, J. Middeldorp, and A. Aiyar. 2004. The amino terminus of Epstein-Barr Virus (EBV) nuclear antigen 1 contains AT hooks that facilitate the replication and partitioning of latent EBV genomes by tethering them to cellular chromosomes. J. Virol. 7811487-11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shire, K., D. F. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 732587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shire, K., P. Kapoor, K. Jiang, M. N. Hing, N. Sivachandran, T. Nguyen, and L. Frappier. 2006. Regulation of the EBNA1 Epstein-Barr virus protein by serine phosphorylation and arginine methylation. J. Virol. 805261-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith, P. R., and B. E. Griffin. 1992. Transcription of the Epstein-Barr virus gene EBNA-1 from different promoters in nasopharyngeal carcinoma and B-lymphoblastoid cells. J. Virol. 66706-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Snudden, D. K., J. Hearing, P. R. Smith, F. A. Grässer, and B. E. Griffin. 1994. EBNA-1, the major nuclear antigen of Epstein-Barr virus, resembles ‘RGG’ RNA binding proteins. EMBO J. 134840-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugden, B., and N. Warren. 1989. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J. Virol. 632644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sung, N. S., J. Wilson, M. Davenport, N. D. Sista, and J. S. Pagano. 1994. Reciprocal regulation of the Epstein-Barr virus BamHI-F promoter by EBNA-1 and an E2F transcription factor. Mol. Cell. Biol. 147144-7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tellam, J., G. Connolly, K. J. Green, J. J. Miles, D. J. Moss, S. R. Burrows, and R. Khanna. 2004. Endogenous presentation of CD8+ T cell epitopes from Epstein-Barr virus-encoded nuclear antigen 1. J. Exp. Med. 1991421-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tellam, J., M. H. Fogg, M. Rist, G. Connolly, D. Tscharke, N. Webb, L. Heslop, F. Wang, and R. Khanna. 2007. Influence of translation efficiency of homologous viral proteins on the endogenous presentation of CD8+ T cell epitopes. J. Exp. Med. 204525-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tierney, R. J., H. E. Kirby, J. K. Nagra, J. Desmond, A. I. Bell, and A. B. Rickinson. 2000. Methylation of transcription factor binding sites in the Epstein-Barr virus latent cycle promoter Wp coincides with promoter down-regulation during virus-induced B-cell transformation. J. Virol. 7410468-10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vaughan, P. S., F. Aziz, A. J. van Wijnen, S. Wu, H. Harada, T. Taniguchi, K. J. Soprano, J. L. Stein, and G. S. Stein. 1995. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature 377362-365. [DOI] [PubMed] [Google Scholar]
- 58.Voo, K. S., T. Fu, H. Y. Wang, J. Tellam, H. E. Heslop, M. K. Brenner, C. M. Rooney, and R. F. Wang. 2004. Evidence for the presentation of major histocompatibility complex class I-restricted Epstein-Barr virus nuclear antigen 1 peptides to CD8+ T lymphocytes. J. Exp. Med. 199459-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, Y., J. E. Finan, J. M. Middeldorp, and S. D. Hayward. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 23618-29. [DOI] [PubMed] [Google Scholar]
- 60.Wysokenski, D. A., and J. L. Yates. 1989. Multiple EBNA1-binding sites are required to form an EBNA1-dependent enhancer and to activate a minimal replicative origin within oriP of Epstein-Barr virus. J. Virol. 632657-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313812-815. [DOI] [PubMed] [Google Scholar]
- 62.Yin, Y., B. Manoury, and R. Fahraeus. 2003. Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science 3011371-1374. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, L., and J. S. Pagano. 1997. IRF-7, a new interferon regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 175748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, L., and J. S. Pagano. 1999. Interferon regulatory factor 2 represses the Epstein-Barr virus BamHI Q latency promoter in type III latency. Mol. Cell. Biol. 193216-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zorio, D. A., and D. L. Bentley. 2004. The link between mRNA processing and transcription: communication works both ways. Exp. Cell Res. 29691-97. [DOI] [PubMed] [Google Scholar]