Abstract
Entamoeba histolytica is an intestinal ameba that causes dysentery and liver abscesses. Cytotoxicity and phagocytosis of host cells characterize invasive E. histolytica infection. Prior to phagocytosis of host cells, E. histolytica induces apoptotic host cell death, using a mechanism that requires contact via an amebic galactose-specific lectin. However, lectin inhibition only partially blocks phagocytosis of already dead cells, implicating at least one additional receptor in phagocytosis. To identify receptors for engulfment of apoptotic cells, monoclonal antibodies against E. histolytica membrane antigens were screened for inhibition of phagocytosis. Of 43 antibodies screened, one blocked lectin-independent uptake of apoptotic cells, with >90% inhibition at a dose of 20 μg/ml (P < 0.0003 versus control). The same antibody also inhibited adherence to apoptotic lymphocytes and, to a lesser extent, adherence to and killing of viable lymphocytes. The antigen recognized by the inhibitory antibody was purified by affinity chromatography and identified by liquid chromatography-mass spectrometry as the serine-rich E. histolytica protein (SREHP). Consistent with this, the inhibitory antibody bound to recombinant SREHP present in bacterial lysates on immunoblots. The SREHP is an abundant immunogenic surface protein of unclear function. The results of this unbiased antibody screen strongly implicate the SREHP as a participant in E. histolytica phagocytosis and suggest that it may play an important role in adherence to apoptotic cells.
Entamoeba histolytica, the intestinal protozoan that causes amebic dysentery and liver abscess, is a global public health problem estimated to cause as many as 50 million symptomatic infections and 100,000 deaths annually (37). Phagocytosis of colonic bacteria is essential for E. histolytica nutrient acquisition and growth (17, 23, 28), and E. histolytica phagocytosis of host erythrocytes and immune cells is a prominent pathological feature of invasive amebiasis (12). In fact, light microscopic examination of clinical stool samples can distinguish E. histolytica infection from infection with the intestinal commensal Entamoeba dispar only if amebae that have ingested host erythrocytes are seen (11).
Despite the central role of phagocytosis in the biology of E. histolytica, the mechanism underlying E. histolytica phagocytosis is poorly defined. Several receptors have been suggested, including (i) an unusual 112-kDa adhesin that appears to be comprised of two proteins and also possesses proteinase activity (9), (ii) an as yet unidentified mannose-containing amebic surface molecule that interacts with bacterial mannose binding proteins (4), and (iii) a Gal/GalNAc-specific amebic surface lectin that is strongly implicated in amebic adherence to and killing of host cells (25, 30).
We previously demonstrated that E. histolytica induces caspase 3-dependent apoptosis of lymphocytes, using a mechanism that requires ameba-host cell contact via the Gal/GalNAc-specific adherence lectin (16). We also found that host cell caspase 3 activation precedes amebic phagocytosis and that E. histolytica preferentially phagocytoses apoptotic cells compared to healthy and necrotic cells (15). Subsequent studies demonstrated preferential uptake of Ca2+ ionophore-treated erythrocytes, which is not surprising given that Ca2+ ionophore treatment induces erythrocyte membrane changes reminiscent of apoptosis (2). Importantly, inclusion of d-galactose during phagocytosis assays (which inhibits the Gal/GalNAc-specific lectin) nearly completely blocks amebic adherence to and killing of host cells but inhibits phagocytosis of apoptotic cells poorly (15). Based on this observation, we hypothesize that at least one additional receptor participates in recognition and clearance of killed cells by E. histolytica. Here we report the results of a monoclonal antibody screen conducted to identify additional E. histolytica surface proteins that participate in phagocytosis. The screen identified the serine-rich E. histolytica protein (SREHP), a dominant surface antigen of unclear function, as an E. histolytica phagocytosis receptor with an apparent role in amebic adherence to apoptotic cells.
MATERIALS AND METHODS
Chemicals and reagents.
The fluorescent dye 5 (and 6)-carboxytetramethylrhodamine succinimidyl ester (TAMRA) and an anti-six-histidine-tag antibody were purchased from Invitrogen (Carlsbad, CA). d-Galactose was purchased from Fisher Scientific (Fair Lawn, NJ). A fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse polyclonal antibody, peroxidase-conjugated anti-immunoglobulin G (anti-IgG) antibody, and actinomycin D were purchased from Sigma (St. Louis, MO). Anti-SREHP ascites was provided by Samuel Stanley (Washington University, St. Louis, MO).
Cell lines and tissue culture.
Entamoeba histolytica trophozoites (strain HM-1:IMSS) were grown axenically in TYI-S-33 (Trypticase-yeast extract-iron-serum) medium supplemented with 100 U of penicillin/ml and 100 μg of streptomycin sulfate/ml at 37°C (8). Trophozoites were used during mid-log-phase growth for all experiments and were harvested by incubation on ice for 10 min, centrifugation at 200 × g and 4°C for 5 min, and suspension in medium 199 (Gibco BRL, Grand Island, NY) supplemented with 5.7 mM cysteine, 25 mM HEPES, and 0.5% bovine serum albumin at pH 6.8 (M199s medium).
The human leukemia T-cell line Jurkat (clone E6-1; American Type Culture Collection, Manassas, VA) was grown in RPMI 1640 medium (Gibco BRL) supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg/ml of streptomycin sulfate (36). Prior to use, cultures were enriched for viable cells by centrifugation at 800 × g for 10 min at room temperature through Ficoll-Paque Plus (Amersham Biosciences, Piscataway, NJ), as previously described (3). Where indicated, Jurkat cell apoptosis was induced either by treatment with actinomycin D (5 μg/ml for 14 h) or by placing culture flasks on a UV light box for 10 min, followed by a 3-h incubation at 37°C. Each of these treatments consistently resulted in >85% cell death, as determined by altered forward and side scatter characteristics upon flow cytometry (data not shown).
Preparation of the membrane fraction of E. histolytica.
Approximately 1 × 108 E. histolytica trophozoites were washed twice with ice-cold phosphate-buffered saline (PBS) and resuspended in 10 mM sodium phosphate buffer (pH 8.0) containing 2 mM phenylmethylsulfonyl fluoride, 5 mM EDTA, 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), and 2 mM polyhexamethylene biguanide. Cells were then incubated at 37°C for 5 min and sonicated on ice with three sequential bursts at the microtip maximum level. Lysis was confirmed microscopically, and resultant cell lysates were centrifuged at 50,000 × g (1 h, 4°C). The pellet was then resuspended in 10 mM sodium phosphate buffer and centrifuged at 100,000 × g (1 h, 4°C) to pellet the membrane fraction.
Monoclonal antibodies against E. histolytica cell membrane antigens.
Hybridoma cell lines producing mouse monoclonal antibodies directed against E. histolytica surface antigens were produced at the Lymphocyte Culture Center at the University of Virginia Medical School according to a standard protocol (6). Hybridoma culture supernatants were screened by enzyme-linked immunosorbent assay for production of antibodies specific for E. histolytica membrane-associated antigens, and antibody-positive hybridomas were cloned twice by limiting dilution. Thirteen hybridomas were produced in mice immunized with E. histolytica membrane preparations (see above). Supernatants from an additional 30 hybridomas generated at the Lymphocyte Culture Center by immunizing animals with whole fixed E. histolytica trophozoites were kindly provided by TechLab, Inc. (Blacksburg, VA). Antibodies present in hybridoma culture supernatants were screened for the ability to inhibit E. histolytica phagocytosis by flow cytometry. Bulk quantities of promising antibodies were subsequently produced from 500-ml hybridoma cultures and purified by affinity chromatography on recombinant protein G columns. Antibody purity was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining.
Phagocytosis assay.
E. histolytica phagocytosis was assayed by flow cytometry, using a previously described method (15). Apoptotic Jurkat lymphocytes were fluorescently labeled by incubation for 20 min at 37°C in PBS containing 47 μM TAMRA. Unbound dye was quenched by incubation with an equal volume of fetal bovine serum at 37°C for 20 min, and cells were washed twice with RPMI 1640 and then resuspended in complete RPMI 1640 medium until use in phagocytosis assays. To assay phagocytosis, fluorescently labeled Jurkat cells were mixed with amebic trophozoites in M199s medium in the presence of 110 mM d-galactose (which inhibits the Gal/GalNAc-specific amebic adherence lectin), and the mixture was centrifuged (200 × g, 5 min, 4°C) and incubated at 37°C for 10 min. Following incubation, ameba-host cell rosettes were disrupted by being washed with a solution of 110 mM d-galactose in ice-cold PBS. Cells were then fixed with 3% paraformaldehyde and analyzed using a FACScan flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Amebae and Jurkat cells were distinguished on the basis of differences in forward and side scatter characteristics. E. histolytica ingestion of fluorescent Jurkat cells was measured as increased amebic fluorescence and quantitated as the percentage of amebae with fluorescence above background levels. Data were analyzed and histogram overlays created for publication using the program WinMDI (version 2.8).
Confocal microscopy.
Samples analyzed using the flow cytometry phagocytosis assay described above were examined using confocal microscopy to verify the fluorescence-activated cell sorter assay results. For this, 15-μl aliquots of samples analyzed by flow cytometry were pipetted onto glass slides and covered with glass coverslips, and coverslips were sealed with nail polish. Slides were examined using a Zeiss LSM 410 confocal microscope.
To localize the antigens recognized by the 10D11 and 5E12 antibodies within amebic trophozoites, E. histolytica trophozoites (5 × 105) were allowed to adhere to acetone-washed glass coverslips in 24-well tissue culture plates by incubation in TYI-S-33 medium at 37°C for 20 to 30 min, washed twice with warm PBS, and fixed with 3% paraformaldehyde. Cells were permeabilized or not, as indicated, by treatment with 0.1% Triton X-100 in PBS (60 s, 25°C), washed with 50 mM NH4Cl in PBS, and blocked with 20% goat serum-5% bovine serum albumin in PBS (1 h, 37°C). Trophozoites were then stained with either the 10D11 or 5E12 monoclonal antibody (50 μg/ml, 16 h, 4°C) diluted in blocking buffer and with a FITC-conjugated goat anti-mouse polyclonal antibody diluted 1:64 in blocking buffer (1 h, 37°C). After staining of the slides, the coverslips were washed twice with PBS, inverted onto Gel Mount (Biomeda, Foster City, CA) on glass slides, and examined using a Zeiss Pascal confocal microscope.
Adherence assay.
E. histolytica adherence was measured using a modified rosette formation assay (27). Assays for adherence to healthy cells were conducted with or without 110 mM d-galactose. Assays for adherence to apoptotic cells were conducted with or without 11 mM d-galactose, a dose previously shown to partially inhibit E. histolytica adherence to apoptotic lymphocytes (15). To assay adherence, E. histolytica trophozoites (105) were suspended in M199s medium and preincubated (20 min, 4°C) with or without antibodies and/or d-galactose, as indicated. Healthy or apoptotic Jurkat cells (5 × 105) in M199s medium were added, and the cell mixture was centrifuged (200 × g, 5 min, 4°C) and then incubated for 30 min on ice. After incubation, the supernatant was poured off, the pellet was resuspended using a Pasteur pipette, 1 drop was placed on a hemacytometer, and results were scored by a blinded observer. Adherent amebae were defined as trophozoites with two or more bound Jurkat cells, and data were normalized to those for control samples (trophozoites pretreated with PBS).
Cytotoxicity assay.
A 51Cr release assay was used to measure in vitro cell killing by E. histolytica trophozoites (16). Healthy Jurkat T cells were labeled with 51Cr (0.2 mCi/6 × 106 cells in 400 μl) for 2 h at 37°C in RPMI 1640 medium with 10% fetal bovine serum, washed twice, and resuspended in M199s medium. E. histolytica trophozoites were suspended in M199s medium and pretreated for 20 min at 4°C with 20 μg/ml of the 10D11 and 5E12 antibody or PBS, as indicated. Amebae (2.5 × 104) were mixed with labeled Jurkat cells (2.5 × 105), centrifuged (200 × g, 5 min, 4°C), and incubated at 37°C for the indicated times. After incubation, the cells were recentrifuged, the supernatant was transferred to a fresh tube, and released 51Cr was quantitated with a gamma counter (Micromedics, Horsham, PA). Percent specific 51Cr release was calculated as follows: (countssample − countsspontaneous lysis)/(counts100% lysis − countsspontaneous lysis) × 100. Data were normalized to % lysis at 60 min in the PBS control.
Immunoblotting.
Samples were resuspended in 30 μl of Laemmli sample buffer (19) with protease inhibitors (800 μM AEBSF, 50 μM bestatin, 0.4 μM leupeptin, 0.15 μM aprotinin, 0.6 μM E-64, and 400 μM EDTA) and then boiled for 5 min. The samples were microcentrifuged for 1 min. The proteins were separated by SDS-PAGE on 8 or 10% polyacrylamide gels, as indicated, and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore Corp., Bedford, MA) at 100 mA for 1 h at 25°C. The membranes were blocked overnight at 4°C in Tris-buffered saline-1% Tween 20 (pH 7.4) (TBS-Tween) containing 5% nonfat dry milk, probed with 10D11Ab at 0.5 μg/ml or anti-SREHP ascites at a 1:500 dilution in TBS-Tween with 5% dry milk for 1 h at 25°C, washed five times with TBS-Tween, and probed with peroxidase-conjugated secondary antibody for 1 h at 25°C. Antigen-antibody complexes were detected using enhanced chemiluminescence (Amersham Pharmacia, Piscataway, NJ).
Affinity purification and antigen identification by liquid chromatography-mass spectrometry (LC-MS).
An affinity column for purification of the antigen bound by the 10D11 antibody was made by coupling the antibody to Affi-Gel-10 medium (Bio-Rad, Hercules, CA). The 10D11 antibody (5.4 mg) in 0.1 M sodium bicarbonate (pH 8.5) was bound to 7 ml (solid) of activated Affi-Gel-10 affinity matrix for 48 h at 4°C according to the manufacturer's instructions. The starting material for antigen purification was approximately 108 E. histolytica trophozoites grown in culture to near confluence. Trophozoites were washed two times with 50 ml of 75 mM Tris base, 65 mM NaCl (pH 7.2) buffer and resuspended in 10 ml of lysis buffer (150 mM NaCl, 50 mM Tris base [pH 8.3], 0.5% NP-40 supplemented with 2 mM phenylmethylsulfonyl fluoride, 5 mM EDTA, 2 mM polyhexamethylene biguanide, and 1 mM AEBSF). This was mixed by vortexing, lysis was confirmed microscopically, and insoluble components were removed by centrifugation (10,000 × g, 25 min, 4°C). The resultant supernatant was applied to the affinity column three times, and the column was then washed with 500 ml of ice-cold wash buffer (0.487 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4·7H2O, and 1.2 mM KH2PO4 [pH 7.4]). Bound antigen was eluted with 0.1 M glycine in 0.5 M NaCl (pH 2.5), and 3-ml fractions were collected and neutralized with 300 μl of 1 M Tris-HCl (pH 8.8). Fractions were analyzed for the presence of 10D11 antigen by dot immunoblotting. Fractions 2 to 5, which contained the eluted 10D11 antigen, were pooled, dialyzed, lyophilized, resuspended in 500 μl PBS, and analyzed by SDS-PAGE followed by silver staining and immunoblotting. Silver staining was done using a protocol that is compatible with protein identification by MS (26).
Trypsin digestion and LC-MS to identify the purified proteins were performed by the W. M. Keck Biomedical Mass Spectrometry Laboratory at the University of Virginia. Gel pieces were transferred to siliconized tubes, washed and destained in 200 μl 50% methanol overnight, dehydrated in acetonitrile, rehydrated in 30 μl of 10 mM dithiothreitol in 0.1 M ammonium bicarbonate, and reduced at room temperature for 0.5 h. The dithiothreitol solution was removed, and the sample was alkylated in 30 μl 50 mM iodoacetamide in 0.1 M ammonium bicarbonate at room temperature for 0.5 h. The reagent was removed, and the gel pieces were dehydrated in 100 μl acetonitrile. The acetonitrile was removed, and the gel pieces were rehydrated in 100 μl 0.1 M ammonium bicarbonate. The pieces were dehydrated in 100 μl acetonitrile, the acetonitrile was removed, and the pieces were completely dried by vacuum centrifugation. The pieces were then rehydrated in 20 ng/μl trypsin in 50 mM ammonium bicarbonate on ice for 10 min. Excess trypsin solution was removed, and 20 μl of 50 mM ammonium bicarbonate was added. The samples were digested overnight at 37°C, and the peptides formed were extracted from the polyacrylamide in two 30-μl aliquots of 50% acetonitrile-5% formic acid. These extracts were combined and evaporated to 25 μl for MS analysis.
The LC-MS system consisted of a Finnigan LCQ ion-trap mass spectrometer system with a Protana nanospray ion source interfaced to a self-packed 8-cm by 75-μm (internal diameter) Phenomenex Jupiter 10-μm C18 reversed-phase capillary column. Volumes (0.5 to 5 μl) of the extract were injected, and the peptides were eluted from the column by an acetonitrile-0.1 M acetic acid gradient at a flow rate of 0.25 μl/min. The nanospray ion source was operated at 2.8 kV. The digest was analyzed using the double-play capability of the instrument, acquiring full-scan mass spectra to determine peptide molecular weights and product ion spectra to determine amino acid sequences in sequential scans. This mode of analysis produces approximately 400 collisionally activated dissociation spectra of ions, ranging in abundance over several orders of magnitude. The data were analyzed by searching the nonredundant GenBank database using the Sequest search algorithm (20), and data are presented according to guidelines set by the Human Proteome Organization Proteomics Standards Initiative (24).
Cloning and expression of recombinant SREHP.
DNA carrying the full-length SREHP gene, excluding the first 39 nucleotides, which encode the predicted N-terminal signal peptide, was amplified by PCR using E. histolytica genomic DNA as the template and the following primers: For-SREHP (5′-ACTAATATCATTCTTGATTTGGA-3′) and Rev-BamHI-SREHP (5′-CGGGATCCCGCTTCATTTAGAAGATGATAGCT-3′). The amplified DNA fragment was then cloned into the vector pCRT7/NT-TOPO, which incorporates an N-terminal six-histidine tag, using a TOPO TA cloning kit and the manufacturer's protocol (Invitrogen, Carlsbad, CA). Miniprep DNA was screened for correct gene orientation by BamHI restriction digestion.
For recombinant SREHP expression, competent Escherichia coli strain BL21(DE3)/pLysS was transformed and grown overnight in 10 ml Luria broth supplemented with 100 μg/ml ampicillin and 34 μg/ml chloramphenicol. Fresh 10-ml cultures were then inoculated with 500 μl of overnight culture and grown to an optical density at 600 nm of 0.6, and recombinant protein expression was induced for 2.5 h by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells transformed with the vector pCRT7/NT-E3 (Invitrogen), an expression control vector, were used as a control to ensure that the antibodies did not detect the six-histidine tag.
Statistics.
All quantitative data were expressed as mean percentages of the control and standard deviations of the means. Significance was determined using unpaired Student's t test.
RESULTS
Identification of a monoclonal antibody that inhibits amebic phagocytosis.
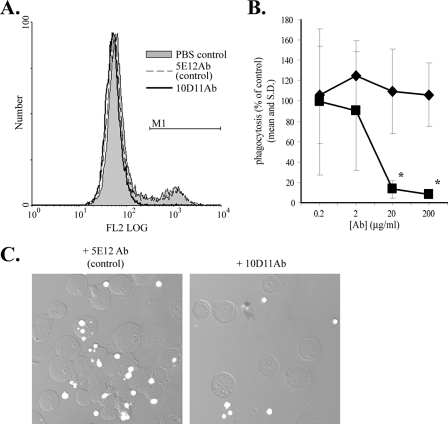
Galactose, which nearly completely blocks E. histolytica adherence to and killing of host cells, only partially inhibits phagocytosis of apoptotic cells, which suggests that receptors other than the Gal/GalNAc-specific lectin participate in phagocytosis (15). We screened a panel of mouse monoclonal antibodies for ones that inhibit amebic phagocytosis as one approach to identifying these receptors. The following two types of antibodies were used: 13 monoclonal antibodies produced by immunizing mice with the E. histolytica membrane fraction and 30 monoclonal antibodies produced by immunizing mice with fixed, intact E. histolytica trophozoites. An initial screen, performed in the presence of 110 mM d-galactose with hybridoma cell culture supernatants and a rapid flow cytometry-based phagocytosis assay, was used to identify hybridomas producing monoclonal antibodies that warranted purification and further study. Of three purified antibodies, one, 10D11Ab (IgG1 isotype), dramatically inhibited Gal/GalNAc lectin-independent E. histolytica phagocytosis of apoptotic Jurkat lymphocytes (Fig. 1). Entamoeba histolytica has a very active cell surface that responds vigorously to incubation with some antibodies by capping and reorganization of the antibody target and of unrelated proteins (5). Therefore, we purified a second IgG1 antibody, 5E12Ab, raised against the E. histolytica membrane preparation, to control for the possibility that antibody binding to any E. histolytica surface antigen might cause a nonspecific effect. As shown in Fig. 1, only 10D11Ab inhibited phagocytosis. Inhibition was dose dependent, with >90% inhibition at an antibody concentration of 20 μg/ml (Fig. 1B), and the results obtained by flow cytometry were verified by confocal microscopy (Fig. 1C). We concluded that 10D11Ab is a potent and specific inhibitor of E. histolytica phagocytosis.
FIG. 1.
Inhibitory effect of monoclonal antibody 10D11 on E. histolytica phagocytosis of apoptotic lymphocytes. Antibody-treated E. histolytica trophozoites (4°C, 20 min) were incubated with fluorescent apoptotic Jurkat T cells (E. histolytica-to-Jurkat cell ratio of 1:2, 37°C, 10 min) in the presence of 110 mM d-galactose to inhibit the Gal/GalNAc lectin. Phagocytosis was assayed using flow cytometry. (A) Representative fluorescence-activated cell sorter histograms showing amebic fluorescence (20-μg/ml antibody dose). The M1 gate indicates phagocytic amebae. (B) Dose effect of 10D11Ab on E. histolytica phagocytosis (mean percentage of PBS effect with standard deviation [SD]) (n = 6). Squares, 10D11Ab; diamonds, 5E12Ab (anti-E. histolytica membrane IgG1 control). Asterisks indicate P values of <0.0003 compared to the control. (C) Confocal microscopy verifying reduced amebic phagocytosis of apoptotic lymphocytes following treatment with 10D11Ab. Original magnification, ×200.
Initial characterization of the antigen recognized by the inhibitory antibody 10D11Ab.
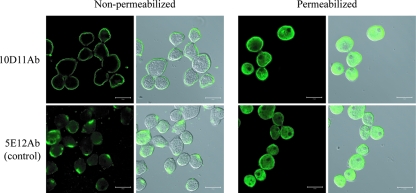
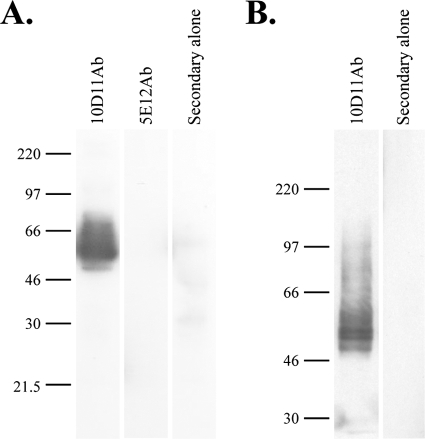
It was important to confirm the cell surface localization of the antigens bound by 10D11Ab and the 5E12Ab control. Immunofluorescence confocal microscopy was performed on both nonpermeabilized E. histolytica trophozoites and cells permeabilized with Triton X-100 (Fig. 2). Both antibodies recognized antigens exposed on the amebic cell membrane in nonpermeabilized cells, although the staining patterns were distinct (Fig. 2, left panels). The antigen bound by 10D11Ab was distributed diffusely on the cell membrane, and the antigen bound by the 5E12Ab control was most often found in a single focal patch. In addition to staining localized to the cell membrane, there was substantial intracellular staining of permeabilized cells, which appeared to be within small vesicles, with both antibodies (Fig. 2, right panels). No staining was seen with the secondary antibody alone (data not shown). 10D11Ab bound to two specific bands with approximate molecular masses of 50 kDa and to several less intense higher-molecular-mass bands on immunoblots of whole amebic lysates (Fig. 3A). Consistent with localization to the cell membrane, the antigen bound by 10D11Ab was present in the amebic membrane fraction (Fig. 3B). Unfortunately, 5E12Ab detected no protein by Western blotting performed with whole amebic lysates under denaturing (Fig. 3A) or nondenaturing (data not shown) conditions. Collectively, these data indicate that the antigens recognized by the 10D11Ab and 5E12Ab antibodies are both present on the amebic surface but are distinct from one another and, based on its apparent molecular weight, that 10D11Ab is not directed against any component of the known Gal/GalNAc lectin receptor.
FIG. 2.
Confocal immunofluorescence microscopy showing localization of the antigens recognized by the 10D11 and 5E12 monoclonal anti-E. histolytica antibodies. Amebic trophozoites were fixed, either permeabilized or not, as indicated, and stained with the 10D11 and 5E12 antibodies followed by a FITC-conjugated secondary antibody. Cells were examined by confocal microscopy. Examination of nonpermeabilized cells (left) demonstrated the antigen recognized by the 10D11 antibody diffusely on the amebic surface and the antigen bound by the 5E12 antibody present in focal patches on the amebic surface. No staining was seen with the secondary antibodies used alone (data not shown). Original magnification, ×800. Bars, 20 μm.
FIG. 3.
Immunoblots using the 10D11 antibody show two specific bands of approximately 50 kDa in whole amebic lysates and in the amebic membrane fraction. (A) Whole amebic lysates were separated by 8% SDS-PAGE, transferred to a PVDF membrane, and probed with 10D11Ab, 5E12Ab, or secondary antibody alone, as indicated. (B) The amebic membrane fraction was probed as described above, using the 10D11Ab or secondary antibody alone.
Effect of 10D11Ab on E. histolytica adherence and cytotoxic ability.
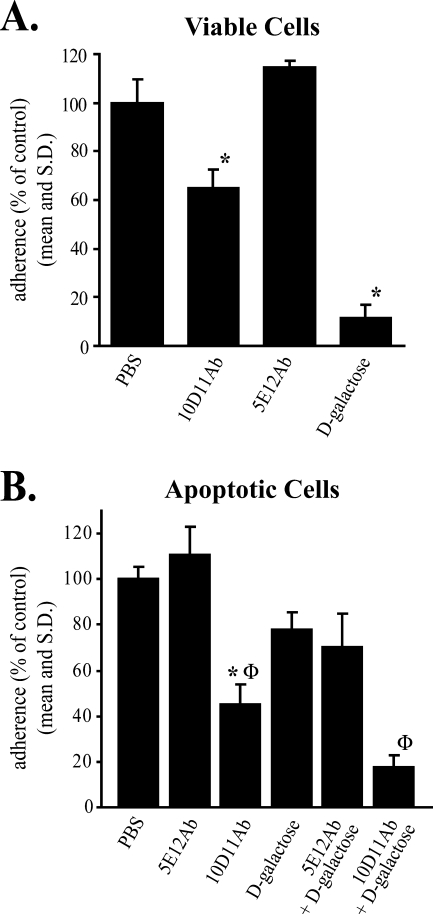
Entamoeba histolytica adherence, cell killing, and phagocytosis are sequential events, suggesting that the antigen recognized by the phagocytosis inhibitory 10D11 antibody may participate in all three of these processes (15). Consistent with a role in adherence, it appeared that fewer apoptotic host cells were bound to the surfaces of E. histolytica trophozoites pretreated with 10D11Ab than to the surfaces of amebae treated with the 5E12Ab control (Fig. 1C). We therefore conducted rosetting assays to quantify the effect of 10D11Ab on amebic adherence to viable and apoptotic cells (Fig. 4). 10D11Ab (20 μg/ml) reduced amebic adherence to viable lymphocytes to 65% of control levels (P = 0.007 versus PBS control), while the 5E12Ab control had no effect (Fig. 4A). d-Galactose was used as a positive control and inhibited adherence to viable cells nearly 90%, as previously shown (27). Interestingly, 10D11Ab was more efficient at reducing E. histolytica adherence to apoptotic lymphocytes, reducing this to 45% of the PBS control level (P = 0.0007) (Fig. 4B). 10D11Ab in combination with d-galactose at a dose that partially inhibits adherence to apoptotic lymphocytes reduced adherence to 17% of the control level.
FIG. 4.
Effect of 10D11 antibody on E. histolytica adherence to healthy (A) and apoptotic (B) lymphocytes. Adherence was measured following preincubation of amebic trophozoites with 10D11Ab, 5E12Ab (control), or PBS (4°C, 15-min preincubation, antibody concentration of 20 μg/ml) in the presence or absence of d-galactose. Data are expressed as percentages of amebae adherent to Jurkat cells in the absence of d-galactose and antibodies (PBS control). An adherent ameba was defined as one having at least two adherent lymphocytes (data are means and SD; n = 3). Asterisks indicate P values of <0.001 compared to the PBS control; Φ indicates a P value of <0.008 compared to treatment with d-galactose alone.
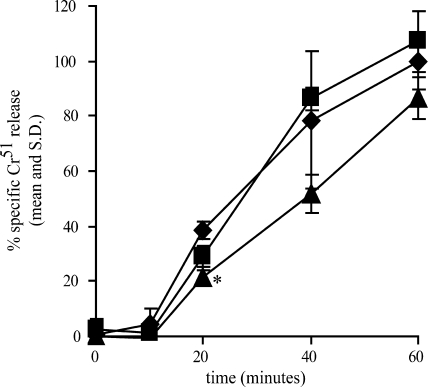
We also tested the effect of 10D11Ab on in vitro killing of Jurkat lymphocytes by E. histolytica trophozoites by measuring 51Cr released from Jurkat cells at various times following exposure to antibody-treated amebae. 10D11Ab significantly reduced cell killing after 20 min of incubation with trophozoites (P = 0.002 versus PBS control) (Fig. 5). Although it was statistically significant, the effect on cell killing was small. This was consistent with the modest effect that 10D11Ab had on adherence to viable cells.
FIG. 5.
Effect of 10D11 antibody on host cell killing by E. histolytica. Entamoeba histolytica trophozoites were preincubated with 10D11Ab, 5E12Ab, or PBS (4°C, 20 min, antibody concentration of 20 μg/ml) prior to incubation with 51Cr-loaded Jurkat lymphocytes for the indicated times (E. histolytica-to-Jurkat cell ratio of 1:10, 37°C). Data are expressed as percentages of specific 51Cr released in the absence of antibodies (PBS control) at 60 min (means and SD; n = 3). Triangles, 10D11Ab; squares, 5E12Ab (control); diamonds, PBS control. An asterisk indicates a P value of 0.002 versus the PBS control at the same time point.
The phagocytosis inhibitory antibody 10D11Ab recognizes SREHP.
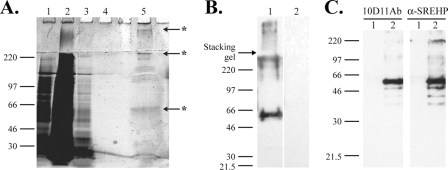
The antigen recognized by 10D11Ab was purified by affinity chromatography, using approximately 108 E. histolytica trophozoites as starting material. SDS-PAGE and silver staining of the eluted protein demonstrated a predominant band running at approximately 50 kDa, with a number of higher-molecular-weight proteins (Fig. 6A). This observation and immunoblots performed with the purified antigen (Fig. 6B) were consistent with the immunoblots of whole amebic lysates. Three predominant bands recognized by Western blotting (running at approximately 50 kDa, at 230 kDa, and within the stacking gel [indicated with an asterisk in each case]) were excised, digested with trypsin, and analyzed by LC-MS for protein identification. Unique peptides from the SREHP were identified from each band (Table 1). Altogether, five unique peptides spanning 47 of 233 amino acids (20.2% coverage) within the amino terminus of the SREHP were identified with high confidence.
FIG. 6.
Identification of antigen recognized by 10D11Ab. (A and B) Partial purification of the 10D11-reactive antigen by immunoaffinity chromatography. (A) Silver-stained 8% SDS-PAGE gel of proteins applied to and eluted from a 10D11 antibody column. The contents of the numbered lanes are as follows: 1, cleared lysate applied to column; 2, column flowthrough; 3, proteins eluted with first wash; 4, proteins eluted with final wash; 5, eluted 10D11-reactive antigen. Asterisks indicate bands excised and identified by MS. (B) 10D11 antibody immunoblot performed on purified antigen separated by 8% SDS-PAGE. A predominant band of approximately 50 kDa is bound. Lane 1, purified antigen probed with 10D11 antibody; lane 2, purified antigen probed with secondary antibody alone. (C) Specific recognition of the SREHP by 10D11 antibody. Six-His-tagged SREHP or human E3 kinase (negative control) was expressed in E. coli, and bacterial proteins were separated by 10% SDS-PAGE, transferred to a PVDF membrane, and blotted with 10D11 antibody (left) or anti-SREHP ascites (right). Lane 1, His-E3 kinase; lane 2, His-SREHP.
TABLE 1.
Peptides present in the SREHP,a identified by MS of excised gel bands
| Peptide sequence | No. of times identified (Xcorr) | Position of gel band |
|---|---|---|
| NEASPEKLEEAEEKEK | 2 (3.35, 2.85) | 50 kDa |
| LEEAEEKEK | 1 (2.02) | 50 kDa |
| NEASPEKLEEAEEKEK | 1 (2.87) | 230 kDa |
| SSSAKPESSSNEDNEDDEDEK | 1 (2.22) | 230 kDa |
| NEASPEKLEEAEEKEK | 1 (4.67) | Stacking gel |
| NEASPEKLEEAEEK | 1 (2.47) | Stacking gel |
| DTNIYGVFLK | 1 (2.11) | Stacking gel |
GenBank NR database accession number gi 2350960 dbj BAA22007.1, for serine-rich 25-kDa antigen protein of Entamoeba histolytica.
To verify independently that 10D11Ab is an anti-SREHP antibody, we cloned the SREHP gene by PCR (excluding the predicted amino-terminal signal peptide), expressed the recombinant protein in E. coli, and performed immunoblots on whole E. coli lysates. An irrelevant six-histidine-tagged protein (human E3 kinase) was expressed to control for the possibility that 10D11Ab might recognize a six-histidine tag incorporated at the amino terminus of the recombinant SREHP. As shown by the immunoblots in Fig. 6C, 10D11Ab bound specifically to recombinant SREHP present in the E. coli lysates. A band of the same apparent molecular weight was bound by another anti-SREHP antibody. Furthermore, immunoblots performed using a monoclonal antibody specific for the six-histidine tag confirmed expression of both the SREHP and the control protein (data not shown). We concluded that the phagocytosis inhibitory antibody 10D11Ab is an anti-SREHP antibody.
DISCUSSION
The most important conclusion of this work is that the SREHP participates in E. histolytica phagocytosis of apoptotic host cells. Using an unbiased screen of monoclonal antibodies, we found one antibody (10D11Ab) that dramatically inhibits E. histolytica phagocytosis and identified the SREHP as the antigen to which it binds. 10D11Ab appears to block phagocytosis by inhibiting E. histolytica's ability to adhere to apoptotic cells. Interestingly, the antibody had more modest inhibitory effects on adherence to viable lymphocytes and on cell killing by E. histolytica. The Gal/GalNAc-specific lectin plays a dominant role in adherence to viable cells and in cell killing (27, 30), and the modest inhibitory effect of the anti-SREHP antibody on these processes suggests that the SREHP may become engaged as a secondary receptor following activation of E. histolytica by ligation of the lectin or following exposure of novel ligands on dying cells. This would be consistent with our previous work demonstrating that adherence, cell killing, and phagocytosis of host lymphocytes by E. histolytica are sequential processes (15) and may be analogous to models of rolling adhesion by neutrophils in which initial tethering by selectins activates integrin binding (31).
Several lines of evidence indicated that the phagocytosis inhibitory antibody 10D11Ab is an anti-SREHP antibody. First, the diffuse cell membrane staining we observed by immunofluorescence confocal microscopy and the apparent molecular weight of the antigen, as determined by immunoblotting, are both consistent with published studies on the SREHP (32, 34). Second, peptides from the SREHP were identified by LC-MS analysis of all three bands examined following affinity purification of the antigen bound by 10D11Ab. Finally, as evidenced by immunoblotting, 10D11Ab specifically bound to recombinant SREHP protein present in E. coli lysates. Collectively, these data provide strong evidence that the 10D11 antibody is directed against the SREHP.
Direct participation of the SREHP in E. histolytica phagocytosis is indicated by the specific nature of 10D11Ab's inhibitory effect. Of 43 antibodies screened, only 1 inhibited phagocytosis. The effect, furthermore, was dose dependent and potent. It was also not due simply to reorganization of the amebic cell membrane in response to antibody binding, since the control antibody, 5E12Ab, bound another antigen present on the cell surface but did not affect phagocytosis, adherence, or cell killing. Consistent with the specificity of 10D11Ab's effect, the anti-Gal/GalNAc lectin antibody 8C12, which inhibits both adherence and cell killing (22), had no effect on phagocytosis of apoptotic lymphocytes (data not shown). Interestingly, the anti-SREHP ascites used for the experiment shown in Fig. 6C had no specific effect on E. histolytica phagocytosis (data not shown), suggesting that the effect of 10D11Ab is epitope dependent. This is not uncommon. For example, various inhibitory, stimulatory, or indifferent effects on adherence and cell killing have been described for a number of monoclonal antibodies specific for the Gal/GalNAc-specific lectin (22). We are currently mapping the SREHP epitope that is bound by 10D11Ab.
The serine-rich and asparagine-rich E. histolytica proteins (SREHP and Ariel) are members of a family of immunogenic amebic surface proteins (18, 21, 32). Immunization with the SREHP is protective in animal models of amebiasis, and the SREHP is a leading vaccine candidate (29, 33, 35, 39, 40). Both the SREHP and Ariel proteins contain central regions of tandem repeat units, a run of charged amino acids upstream of these, hydrophobic amino acids at the carboxy terminus, and predicted amino-terminal signal peptides (18, 21, 32). The amino acid sequences of the HM-1-IMSS strain SREHP and Ariel1 proteins are 53% identical and 89% similar. Their high degree of sequence similarity suggests that they may have common functions; however, posttranslational modifications may be important. The SREHP is modified by predicted phosphorylation, acetylation, and O-linked terminal N-acetylglucosamine residues, but Ariel proteins lack the predicted phosphorylation sites and contain multiple possible N-linked glycosylation sites (34, 38).
The functions of the SREHP and Ariel proteins remain unclear. Entamoeba histolytica trophozoites migrate toward recombinant SREHP in transwell migration assays (34), and inhibition of adherence in vitro by polyclonal anti-SREHP serum suggests a possible role in amebic adhesion to host cells (32). Our data provide further evidence that the SREHP may function in adherence and the first evidence that the SREHP plays a critical role in phagocytosis. Consistent with a potential role in phagocytosis, Ariel1 localizes to phagosomes containing ingested E. coli, but no functional studies of the Ariel proteins have been published (10).
Additional work is needed to directly test the function of the SREHP as an adhesin. The adherence studies presented here suggest the possibility that the SREHP is an adhesin primarily for apoptotic cells. However, given the methodology used in this study, only limited comparisons of adherence to living versus apoptotic cells are possible. We are currently working to express the SREHP on the surfaces of mammalian cell monolayers to enable a more detailed analysis of its role in adherence.
As noted earlier, the SREHP is highly polymorphic, with remarkable variation among E. histolytica clinical isolates in the number of tandem repeat units in the protein's central region (1, 7, 13, 14). It is therefore tempting to speculate that serine-rich proteins with variable numbers of tandem repeats may mediate adherence with variable affinity. This would seem likely if the tandem repeats are actually binding domains and, if so, may have important implications regarding the phagocytic and/or pathogenic ability of different E. histolytica strains. In addition to clarifying the functions of the SREHP in adherence to apoptotic versus living cells, the heterotopic expression system described above may also enable direct comparison of polymorphic serine-rich proteins from different E. histolytica isolates.
Finally, although the SREHP may function in phagocytosis primarily as an adhesin, it may participate primarily in cell signaling required for particle uptake or may participate both in adherence and in cell signaling. We have hypothesized that E. histolytica likely uses multiple receptors for phagocytosis, and we therefore believe that the nearly complete inhibition of phagocytosis induced by treatment with 10D11Ab may result from an ability to interfere with a common signaling pathway. Further studies to define the effect (if any) of 10D11Ab on cell signaling will help to address this possibility.
Acknowledgments
This work was supported by National Institutes of Health grants K08 AI053678 and P20 RR021905 to C.D.H. and by a young investigator's award from the American Society for Clinical Investigation to C.D.H.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 17 December 2007.
REFERENCES
- 1.Ayeh-Kumi, P. F., I. M. Ali, L. A. Lockhart, C. A. Gilchrist, W. A. Petri, and R. Haque. 2001. Entamoeba histolytica: genetic diversity of clinical isolates from Bangladesh as demonstrated by polymorphisms in the serine-rich gene. Exp. Parasitol. 9980-88. [DOI] [PubMed] [Google Scholar]
- 2.Boettner, D. R., C. D. Huston, J. A. Sullivan, and W. A. Petri. 2005. Entamoeba histolytica and Entamoeba dispar utilize externalized phosphatidylserine for recognition and phagocytosis of erythrocytes. Infect. Immun. 733422-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyum, A. 1968. Isolation of mononuclear cells and granulocytes from human blood. Scand. J. Clin. Investig. 21(Suppl. 97)107-111. [PubMed] [Google Scholar]
- 4.Bracha, R., D. Kobiler, and D. Mirelman. 1982. Attachment and ingestion of bacteria by trophozoites of Entamoeba histolytica. Infect. Immun. 36396-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderon, J., M. De Lourdes-Munoz, and H. M. Acosta. 1980. Surface redistribution and release of antibody-induced caps in Entamoebae. J. Exp. Med. 151184-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, J.-H., W. M. Sutherland, and S. J. Parsons. 1995. Monoclonal antibodies to oncoproteins. Methods Enzymol. 254430-445. [DOI] [PubMed] [Google Scholar]
- 7.Clark, C. G., and L. S. Diamond. 1993. Entamoeba histolytica: a method for isolate identification. Exp. Parasitol. 77450-455. [DOI] [PubMed] [Google Scholar]
- 8.Diamond, L. S., D. R. Harlow, and C. Cunnick. 1978. A new medium for axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72431-432. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Rivera, G., M. A. Rodriguez, R. Ocadiz, M. C. Martinez-Lopez, R. Arroyo, A. Gonzalez-Robles, and E. Orozco. 1999. Entamoeba histolytica: a novel cysteine protease and an adhesin form the 112 kDa surface protein. Mol. Microbiol. 33556-568. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh, S. K., J. Field, M. Frisardi, B. Rosenthal, Z. Mai, R. Rogers, and J. Samuelson. 1999. Chitinase secretion by encysting Entamoeba invadens and transfected Entamoeba histolytica trophozoites: localization of secretory vesicles, endoplasmic reticulum, and Golgi apparatus. Infect. Immun. 673073-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Ruiz, A., R. Haque, A. Aguirre, G. Castanon, A. Hall, F. Guhl, G. Ruiz-Palacios, M. A. Miles, and D. C. Warhurst. 1994. Value of microscopy in the diagnosis of dysentery associated with invasive Entamoeba histolytica. J. Clin. Pathol. 47236-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin, J. L. 1972. Human amebic dysentery: electron microscopy of Entamoeba histolytica contacting, ingesting, and digesting inflammatory cells. Am. J. Trop. Med. Hyg. 21895-906. [PubMed] [Google Scholar]
- 13.Haghighi, A., S. Kobayashi, T. Takeuchi, G. Masuda, and T. Nozaki. 2002. Remarkable genetic polymorphism among Entamoeba histolytica isolates from a limited geographic area. J. Clin. Microbiol. 404081-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haghighi, A., S. Kobayashi, T. Takeuchi, N. Thammapalerd, and T. Nozaki. 2003. Geographic diversity among genotypes of Entamoeba histolytica field isolates. J. Clin. Microbiol. 413748-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huston, C. D., D. R. Boettner, V. Miller-Sims, and W. A. Petri. 2003. Apoptotic killing and phagocytosis of host cells by the parasite Entamoeba histolytica. Infect. Immun. 71964-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huston, C. D., E. R. Houpt, B. J. Mann, C. S. Hahn, and W. A. Petri. 2000. Caspase 3-dependent killing of host cells by the parasite Entamoeba histolytica. Cell. Microbiol. 2617-625. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs, L. 1947. The elimination of viable bacteria from cultures of Endamoeba histolytica and the subsequent maintenance of such cultures. Am. J. Hyg. 46172-176. [DOI] [PubMed] [Google Scholar]
- 18.Kohler, S., and E. Tannich. 1993. A family of transcripts (K2) of Entamoeba histolytica contains polymorphic repetitive regions with highly conserved elements. Mol. Biochem. Parasitol. 5949-58. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 20.Link, A. J., J. Eng, D. M. Schieltz, E. Carmarck, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17676-682. [DOI] [PubMed] [Google Scholar]
- 21.Mai, Z., and J. Samuelson. 1998. A new gene family (ariel) encodes asparagine-rich Entamoeba histolytica antigens, which resemble the amebic vaccine candidate serine-rich E. histolytica protein. Infect. Immun. 66353-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mann, B. J., C. Y. Chung, J. M. Dodson, L. S. Ashley, L. L. Braga, and T. L. Snodgrass. 1993. Neutralizing monoclonal antibody epitopes of the Entamoeba histolytica galactose adhesin map to the cysteine-rich extracellular domain of the 170-kilodalton subunit. Infect. Immun. 611772-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura, M. 1953. Nutrition and physiology of Endamoeba histolytica. Bacteriol. Rev. 17189-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orchard, S., H. Hermjakob, C. Taylor, P. A. Binz, C. Hoogland, R. Julian, J. S. Garavelli, R. Aebersold, and R. Apweiler. 2006. Autumn 2005 workshop of the Human Proteome Organisation Proteomics Standards Initiative (HUPO-PSI) Geneva, September, 4-6, 2005. Proteomics 6738-741. [DOI] [PubMed] [Google Scholar]
- 25.Petri, W. A., Jr., R. D. Smith, P. H. Schlesinger, C. F. Murphy, and J. I. Ravdin. 1987. Isolation of the galactose binding lectin of Entamoeba histolytica. J. Clin. Investig. 801238-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabilloud, T. 1992. A comparison between low background silver diammine and silver nitrate protein stains. Electrophoresis 13429-439. [DOI] [PubMed] [Google Scholar]
- 27.Ravdin, J. I., and R. L. Guerrant. 1981. Role of adherence in cytopathogenic mechanisms of Entamoeba histolytica. Study with mammalian tissue culture cells and human erythrocytes. J. Clin. Investig. 681305-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rees, C. W., L. V. Reardon, and L. Jacobs. 1941. The cultivation of the parasitic protozoa without bacteria. Am. J. Trop. Med. 21695-716. [Google Scholar]
- 29.Ryan, E. T., J. R. Butterton, T. Zhang, M. A. Baker, S. L. Stanley, Jr., and S. B. Calderwood. 1997. Oral immunization with attenuated vaccine strains of Vibrio cholerae expressing a dodecapeptide repeat of the serine-rich Entamoeba histolytica protein fused to the cholera toxin B subunit induces systemic and mucosal antiamebic and anti-V. cholerae antibody responses in mice. Infect. Immun. 653118-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saffer, L. D., and W. A. Petri, Jr. 1991. Role of the galactose lectin of Entamoeba histolytica in adherence-dependent killing of mammalian cells. Infect. Immun. 594681-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, C. W. 2000. Possible steps involved in the transition to stationary adhesion of rolling neutrophils: a brief review. Microcirculation 7385-394. [PubMed] [Google Scholar]
- 32.Stanley, S. L., Jr., A. Becker, C. Kunz-Jenkins, L. Foster, and E. Li. 1990. Cloning and expression of a membrane antigen of Entamoeba histolytica possessing multiple tandem repeats. Proc. Natl. Acad. Sci. USA 874976-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanley, S. L., Jr., J. L. Blanchard, N. Johnson, L. Foster, C. Kunz-Jenkins, T. Zhang, K. Tian, and F. B. Cogswell. 1995. Immunogenicity of the recombinant serine rich Entamoeba histolytica protein (SREHP) amebiasis vaccine in the African green monkey. Vaccine 13947-951. [DOI] [PubMed] [Google Scholar]
- 34.Stanley, S. L., Jr., K. Tian, J. P. Koester, and E. Li. 1995. The serine-rich Entamoeba histolytica protein is a phosphorylated membrane protein containing O-linked terminal N-acetylglucosamine residues. J. Biol. Chem. 2704121-4126. [DOI] [PubMed] [Google Scholar]
- 35.Sultan, F., L. L. Jin, M. G. Jobling, R. K. Holmes, and S. L. Stanley, Jr. 1998. Mucosal immunogenicity of a holotoxin-like molecule containing the serine-rich Entamoeba histolytica protein (SREHP) fused to the A2 domain of cholera toxin. Infect. Immun. 66462-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss, A., R. L. Wiskocil, and J. D. Stobo. 1984. The role of T3 surface molecules in the activation of human T cells: a two-stimulus requirement for IL 2 productions reflects events occurring at a pre-translational level. J. Immunol. 133123-128. [PubMed] [Google Scholar]
- 37.WHO. 1997. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis, Mexico City, Mexico, 28-29 January, 1997. WHO Epidemiol. Bull. 1813-14. [PubMed] [Google Scholar]
- 38.Willhoeft, U., H. Buss, and E. Tannich. 1999. DNA sequences corresponding to the ariel gene family of Entamoeba histolytica are not present in E. dispar. Parasitol. Res. 85787-789. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, T., P. R. Cieslak, and S. L. Stanley. 1994. Protection of gerbils from amebic liver abscess by immunization with a recombinant Entamoeba histolytica antigen. Infect. Immun. 621166-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, T., and S. L. Stanley, Jr. 1996. Oral immunization with an attenuated vaccine strain of Salmonella typhimurium expressing the serine-rich Entamoeba histolytica protein induces an antiamebic immune response and protects gerbils from amebic liver abscess. Infect. Immun. 641526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]