Abstract
The transcriptional regulator GlnR of Streptococcus pneumoniae is involved in the regulation of glutamine and glutamate metabolism, controlling the expression of the glnRA and glnPQ-zwf operons, as well as the gdhA gene. To assess the contribution of the GlnR regulon to virulence, D39 wild-type and mutant strains lacking genes of this regulon were tested in an in vitro adherence assay and murine infection models. All of the mutants, except the ΔglnR mutant, were attenuated in adherence to human pharyngeal epithelial Detroit 562 cells, suggesting a contribution of these genes to adherence during the colonization of humans. During murine colonization, only the ΔglnA mutant and the glnP-glnA double mutant (ΔglnAP) were attenuated, in contrast to ΔglnP, indicating that the effect is caused by the lack of GlnA expression. In our pneumonia model, only ΔglnP and ΔglnAP showed a significantly reduced number of bacteria in the lungs and blood, indicating that GlnP is required for survival in the lungs and possibly for dissemination to the blood. In intravenously infected mice, glnP and glnA were individually dispensable for survival in the blood whereas the ΔglnAP mutant was avirulent. Finally, transcriptome analysis of the ΔglnAP mutant showed that many genes involved in amino acid metabolism were upregulated. This signifies the importance of glutamine/glutamate uptake and synthesis for full bacterial fitness and virulence. In conclusion, several genes of the GlnR regulon are required at different sites during pathogenesis, with glnA contributing to colonization and survival in the blood and glnP important for survival in the lungs and, possibly, efficient transition from the lungs to the blood.
Streptococcus pneumoniae is often carried asymptomatically in the human nasopharynx. However, this gram-positive bacterium can cause disease upon dissemination to other sites of the body, such as otitis media, pneumonia, septicemia, and meningitis (3). To survive at various sites in the host, S. pneumoniae needs to metabolize available nutrients. In line with this, several systems involved in nutrition and metabolism have been suggested to be important for virulence. For example, PsaA is part of an Mn(II) uptake system and contributes to colonization and invasive disease (2, 13) while the Ami-AliA/AliB oligopeptide uptake system contributes to colonization only (15).
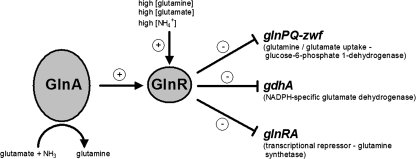
Nitrogen metabolism is of utmost importance for bacterial survival and is therefore strictly regulated. Glutamine and glutamate serve as main sources of nitrogen in the cell. We have recently shown that in the pneumococcus, expression of genes involved in glutamine metabolism is regulated by the transcriptional regulator GlnR, which has a similar function in Bacillus subtilis and Lactococcus lactis (5, 17, 19) (Fig. 1). In the pneumococcus, the GlnR regulon consists of two operons, glnRA and glnPQ-zwf, and the gdhA gene. The gene glnA encodes glutamine synthetase GlnA, which forms glutamine out of glutamate and ammonium while hydrolyzing ATP. The glnPQ genes encode the main glutamine/glutamate transporter, and zwf encodes glucose-6-phosphate 1-dehydrogenase, an enzyme involved in pentose metabolism (17). The gene gdhA encodes glutamate dehydrogenase, which converts 2-oxoglutarate and ammonium to glutamate, thereby hydrolyzing NAD(P)H. We have demonstrated that GlnR binds to a conserved operator sequence present in the promoter regions of its target genes (17). Regulation by GlnR is dependent on GlnA, as the GlnR targets are derepressed in a glnA mutant and GlnA stimulates binding of GlnR to its operator sequence (17). In addition, GlnR regulation is responsive to glutamine and ammonium (17, 19).
FIG. 1.
The pneumococcal GlnR regulon. GlnR regulates the expression of the glnRA and glnPQ-zwf operons and the gdhA gene. Repression of gene expression by GlnR is responsive to high concentrations glutamine, glutamate, and ammonium. Moreover, regulation by GlnR requires cellular GlnA for full repression of target genes.
Importantly, expression of gdhA has been shown to be under the control of a second regulatory protein, CodY (9, 17). This transcriptional repressor has been studied extensively in B. subtilis and L. lactis (6, 23, 28, 30) and recently in pneumococcus (9). Targets of CodY have been shown to consist mainly of genes involved in amino acid biosynthesis and uptake. It can be expected that GdhA plays an important role in central amino acid metabolism, as the expression of its gene is tightly regulated by GlnR and CodY. In B. subtilis, a GlnR ortholog is present, TnrA, which regulates various genes, including genes involved in nitrogen metabolism (30, 33). TnrA activates several genes during low nitrogen availability, and its DNA recognition site is similar to that of GlnR (5, 30, 33). In the pneumococcus, no TnrA homologue is present, indicating differences in the regulation of nitrogen metabolism from B. subtilis.
Signature-tagged mutagenesis screens have suggested that several pneumococcal genes involved in glutamine metabolism are required for full virulence (8, 20, 27). In group B streptococci, a mutant deficient in glnQ, the gene encoding a glutamate transporter, is less able to adhere to and invade A549 respiratory epithelial cells in vitro. Furthermore, this mutant showed a decreased virulence in a rat model of infection (29). In Mycobacterium tuberculosis, glutamine synthetase GlnA1 is essential for growth in macrophages (31). In another intracellular pathogen, Salmonella enterica serovar Typhimurium, glnA is regulated by the Ntr system (18). This gene shares an operon with genes encoding the two-component system NtrB/C. Mutation of glnA, ntrB, or ntrC resulted in a marked reduction of virulence and a reduced ability to survive within host cells (18).
In this study, we assessed the importance of glutamine and glutamate metabolism in S. pneumoniae by evaluating the contribution of the glutamine/glutamate regulator GlnR and its target genes, glnP, glnA, and gdhA, to pneumococcal virulence. To this end, we investigated the ability of mutants with changes in these genes to adhere to a human pharyngeal cell line in vitro. Furthermore, we tested the individual contributions of these genes in three murine infection models, representing the three major phases of pneumococcal disease: colonization, pneumonia, and bacteremia. In addition, we used microarray analysis to examine the global gene expression of the glnA-glnP mutant, which was severely affected in virulence.
MATERIALS AND METHODS
Bacterial strains and media.
All of the strains used in this study were constructed in an S. pneumoniae D39 genetic background (NCTC 7466, serotype 2) and are listed in Table 1. The glnAP mutant was constructed by transformation of chromosomal DNA from the glnP deletion mutant to the glnA mutant and selecting for erythromycin resistance (0.25 μg/ml) on GM17 agar plates supplemented with 1% sheep blood (Johnny Rottier, Kloosterzande, The Netherlands). Bacteria were checked for spectinomycin (150 μg/ml) resistance and for the glnA mutation by PCR. Stocks were frozen in 10% glycerol and stored at −80°C until further use.
TABLE 1.
Bacterial strains used in this study
| Strain | Mutation(s) | Function | D39/TIGR4 accession no. | Resistance cassette(s) | Reference |
|---|---|---|---|---|---|
| Δcps | cps | Type 2 capsule locus | SPD_0315-SPD_0328 | Kanamycin | 26 |
| ΔglnR | glnR | Glutamine synthetase repressor | SPD_0447/SP_0501 | Nonea | 17 |
| ΔglnA | glnA | Glutamine synthetase | SPD_0448/SP_0502 | Spectinomycin | 17 |
| ΔglnRA | glnR, glnA | Spectinomycin | 17 | ||
| ΔglnP | glnP | Glutamine/glutamate transporter | SPD_1098/SP_1241 | Erythromycin | 17 |
| ΔglnAP | glnA, glnP | Spectinomycin, erythromycin | This study | ||
| ΔgdhA | gdhA | Glutamate dehydrogenase | SPD_1158/SP_1306 | Erythromycin | 17 |
Premature stop codon introduced.
In vitro adherence.
Adherence of pneumococci to epithelial cells was studied essentially as described previously (4, 17). In short, the human pharyngeal cell line Detroit 562 (ATCC CCL-138) was cultured in RPMI 1640 medium without phenol red (Invitrogen) supplemented with 1 mM sodium pyruvate and 10% fetal calf serum (FCS). Aliquots of bacteria (grown to mid-log phase in GM17 broth) stored at −80°C were thawed rapidly, harvested by centrifugation, and resuspended in RPMI 1640 medium without phenol red supplemented with 1% FCS to 1 × 107 CFU/ml. Monolayers of Detroit 562 cells in 24-well tissue culture plates were washed twice with 1 ml phosphate-buffered saline (PBS), and 1 ml of bacterial suspension was allowed to adhere for 2 h at 37°C in a 5% CO2 atmosphere. Nonadherent bacteria were removed by three washes with 1 ml PBS, after which the Detroit 562 cells were detached by adding 200 μl of trypsin-EDTA and lysed by the addition of 800 μl of ice-cold 0.025% Triton X-100 in PBS. Serial 10-fold dilutions were plated on blood agar plates to count the adherent bacteria and corrected to account for small differences in the count in the initial inoculum. All experiments were performed in triplicate and repeated at least three times. Significant differences between the wild-type and mutant strains were determined with the Mann-Whitney U test (P < 0.05). The wild-type and mutant strains grew comparably in RPMI medium (without phenol red supplemented with 1% FCS) alone, without the addition of extra glutamine.
Experimental virulence in mice.
Infection models were essentially performed as described previously (9, 10, 15). Nine-week-old female outbred CD-1 mice (Harlan, Horst, Netherlands) were used for all infection models. Prior to the infection experiments, D39 wild-type and mutant strains were passaged in mice as described previously (15). Cultures of mouse-passaged S. pneumoniae strains were grown to an optical density at 600 nm of 0.3 and stored in aliquots at −80°C in 10% glycerol. Prior to infection, these aliquots were spun down and the bacteria were resuspended in sterile PBS to 106 CFU in volumes, depending on the infection model used. Upon intranasal infection, mice were anesthetized with 2.5% (vol/vol) isoflurane-O2. At predetermined time points after infection, depending on the infection model used, groups of mice were sacrificed by cervical dislocation and samples were taken from various sites to determine the bacterial load. During infection, signs of disease were closely monitored. If animals reached a moribund state, they were sacrificed by cervical dislocation and excluded from the experiment prematurely. All animal experiments were performed with approval from the Animal Experimentation Committee (DEC) of Erasmus Medical Centre, Rotterdam, The Netherlands.
Colonization model of infection.
In the colonization model, 10 μl of PBS containing 106 CFU of D39 wild-type or mutant bacteria was administered to the nostrils of groups of five mice as described previously (14). Due to this small volume, only the nose (nasopharynx) of the mouse becomes infected. Bacteria were recovered from the nasopharynx by flushing the nose with 2 ml sterile PBS (16), and lungs were removed from the body and homogenized in 2 ml of sterile PBS with a hand-held homogenizer (polytron PT 1200; Kinematica AG). Viable bacteria from the nasal lavage fluid, homogenized lungs, and blood samples were quantified by plating serial 10-fold dilutions on Columbia blood agar (Oxoid) supplemented with 5% (vol/vol) defibrinated sheep blood (Biotrading). Time points for sampling were 30 min, 24 h, 48 h, 96 h, and 192 h postinfection. The 30-min time point is considered to be the start of the infection and is therefore referred to as t = 0. Bacteriology results are expressed as the geometric mean ± the standard error of the mean. Comparison of bacterial loads in the time course experiment was performed with Student's t test with P < 0.05 considered statistically significant.
Pneumonia model of infection.
In the pneumonia model, five mice per group were infected with 50 μl of PBS containing 106 CFU of pneumococci. Bacteria were recovered from the different sites as described above, with the addition of a blood sample obtained by cardiac puncture. Viable bacteria isolated from the nasal wash, homogenized lungs, and blood were quantified as described above. Time points for sampling were 0, 12, 24, and 36 h postinfection. Bacteriology results are expressed as the geometric mean ± the standard error of the mean. Comparison of bacterial loads in the time course experiment was performed with Student's t test with P < 0.05 considered statistically significant.
Bacteremia model of infection.
In the bacteremia model, groups of six mice were infected by a tail vein with 106 CFU resuspended in 100 μl of sterile PBS. Bacteria were recovered from the blood by puncture of a lateral tail vein of the same mouse at three predetermined time points after infection (0, 12, and 24 h) and by cardiac puncture at the last time point, 36 h. In addition, mouse survival times were scored. Two separate experiments were carried out, the first being three groups of mice infected with the D39 wild-type, ΔglnA, and ΔglnR strains and the second being four groups of mice infected with, again, D39 wild-type, ΔglnP, ΔglnAP, and ΔgdhA bacteria. Data from the two experiments were combined, after which analysis of survival times was performed with the log-rank test with P < 0.05 considered statistically significant.
DNA microarray analysis.
Microarray analysis was performed as described previously (9, 10). In short, cultures of D39 wild-type and ΔglnAP bacteria were grown in GM17 in static flask cultures at 37°C. Cells were harvested when they reached an optical density at 600 nm of 0.3 (mid-exponential growth phase). Total RNA was isolated from both cultures as described previously and used to generate fluorescent DNA probes by indirect labeling by standard methods (9, 10). After overnight hybridization, dual-channel array images were acquired with a GeneTac LS IV confocal laser scanner (Genomics Solutions Inc.) and analyzed with ArrayPro 4.5 software (Media Cybernetics Inc.). Spots were screened visually to identify those of low quality. Slide data were processed with MicroPreP as previously described (9, 10, 32). Prior to analysis, automatically and manually flagged spots and spots with very low background subtracted signal intensity (5% of the weakest spots [sum of Cy3 and Cy5 net signals]) were filtered out of all data sets. Net signal intensities were calculated by grid-based background subtraction. A grid-based Lowess transformation was performed for slide normalization, negative and empty values were removed, and outliers were removed by the deviation test. Further analysis was performed with a Cyber-T Student t test for paired data (22). For identification of differentially expressed genes, only genes with a minimum of six reliable measurements, a Bayesian P value of <0.001, a false-discovery rate of <0.05, and a standard deviation < ratio were included. Since these criteria are purely statistical measurements of differential gene expression and reproducibility across replicates, an additional change cutoff of twofold was applied.
Microarray data accession number.
The microarray data have been deposited in the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under GEO Series accession number GSE9850.
RESULTS
Contribution of the GlnR regulon to in vitro adherence.
To assess the contribution of glutamine and glutamate metabolism to pneumococcal virulence, directed mutants with changes in glnR and genes regulated by GlnR were created. Deletions had no effect on in vitro growth, except for the glnA mutant and the glnA-glnP double mutant. These strains displayed slower growth in GM17 broth, but upon addition of 0.5 mg/ml glutamine, normal growth was restored. Similar observations were made during growth in THY broth (data not shown).
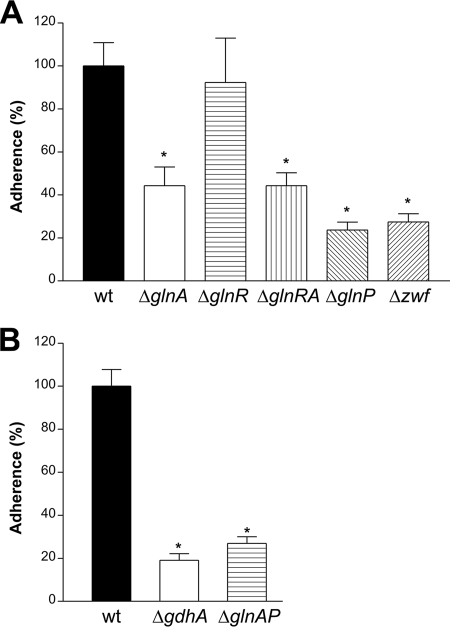
Pneumococcal colonization is mediated by adherence to respiratory epithelial cells. To assess the relevance of genes of the GlnR regulon in the process of adhesion, we performed in vitro adherence assays with the individual mutant strains. Since unencapsulated strains tend to display significantly higher levels of adherence than encapsulated strains (4), all mutants were constructed in a capsule-negative isogenic derivative of D39 (D39Δcps) (26). The adherence capacity of the ΔglnA, ΔglnR, ΔglnRA, and ΔglnP mutant strains has been described previously (17) and is included in this study for completeness of analysis. All of the strains, except ΔglnR, showed a severe reduction in adherence of over 50% (P < 0.05, Fig. 2A and B). For the glnA mutant and the glnR-glnA double mutant, adherence was approximately 44% of that of the wild type. This effect was most likely caused by the glnA mutation, as the glnR mutant adhered at wild-type levels. The number of adherent glnP and glnA-glnP double-mutant bacteria was approximately 25% of that of the adherent wild-type bacteria. In this case, this appeared to be mainly the result of the lack of glnP, as the glnA mutant adhered at significantly higher levels, i.e., 44% of wild-type adherence. Finally, deletion of zwf and gdhA also impaired adherence to Detroit cells (27% and 20% of the wild-type level, respectively).
FIG. 2.
In vitro adherence of pneumococcal mutants to the human pharyngeal epithelial cell line Detroit 562. All strains were constructed in a D39Δcps genetic background. The adherence of the mutants is given as a percentage of that of the wild type (wt). *, P < 0.05.
Contribution of the GlnR regulon to colonization.
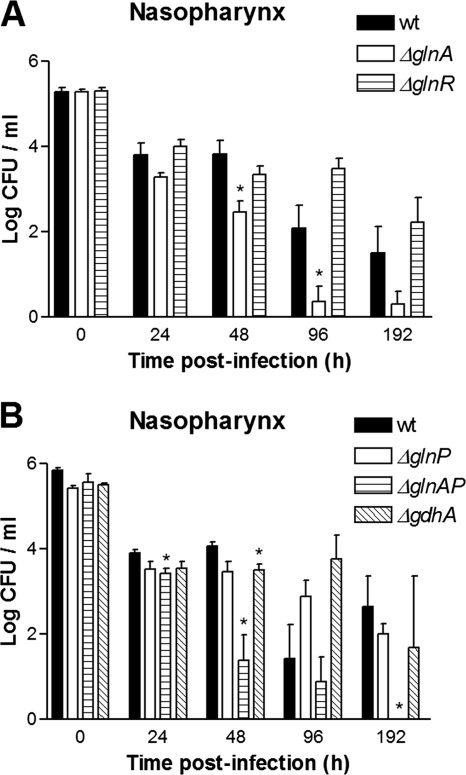
In the colonization model, extended colonization of the nasopharynx without development of invasive disease is achieved by a small-volume inoculum. Indeed, wild-type pneumococci were able to colonize the murine nasopharynx for a period of 192 h (Fig. 3). The level of colonization was fairly consistent for 48 h, i.e., 104 CFU/ml, after which the colonization level slowly started to decrease to 103 CFU/ml at 192 h.
FIG. 3.
Colonization model. Shown are the bacterial loads in the nasal lavage fluid of mice infected with D39 wild-type (wt), ΔglnA, or ΔglnR bacteria (A) and D39 wild-type, ΔglnP, ΔglnAP, and ΔgdhA (B) bacteria. *, P < 0.05.
The colonization kinetics of the glnR mutant did not differ significantly from those of the wild type during 192 h of colonization (Fig. 3A). In contrast, the glnA mutant displayed a clear attenuated phenotype. After 48 h throughout 96 h of infection, mice infected with the glnA mutant had significantly lower bacterial loads in the nasopharynx (Fig. 3A). At 192 h, the difference was still present but it was not statistically significant.
Neither the glnP mutant nor the gdhA mutant was significantly attenuated during 192 h of colonization, whereas the glnA-glnP double mutant was significantly attenuated after 24 h of colonization and severely attenuated from 48 h onward, to the point of being cleared at 192 h. Due to the relatively low level of colonization by the wild type at 96 h, no significant difference was reached at this time point (Fig. 3B). The more severe attenuation of the glnA-glnP double mutant in comparison to that of the glnA mutant is suggestive of an additive effect of the glnP mutation to the colonization phenotype of the ΔglnA mutant.
Contribution of the GlnR regulon to pneumonia.
In the pneumonia model, the infection is monitored at three distinct sites, i.e., the nasopharynx, the lungs, and the blood compartment. This model allows assessment of the two ends of the spectrum of pneumococcal infection, i.e., the progression from nasopharyngeal colonization to invasive disease.
In agreement with the colonization model, we observed that in the pneumonia model, mice infected with the glnA mutant or the glnA-glnP double mutant had lower numbers of bacteria in the nasal lavage fluid at 36 postinfection than wild-type-infected mice, while bacterial loads in mice infected with the ΔglnR, ΔglnP, or ΔgdhA mutant did not differ from those in wild-type-infected mice (data not shown).
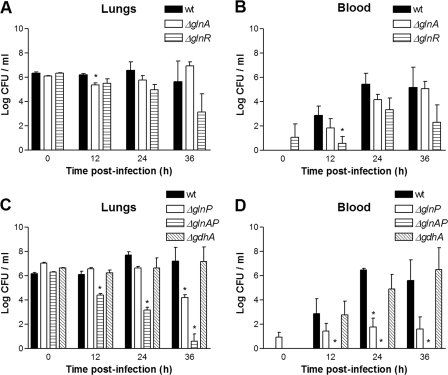
During the 36-h infection period, we observed no significant differences between bacterial loads in the lungs of mice infected with the glnR mutant and those infected with wild-type bacteria (Fig. 4A). Moreover, the glnR mutant was equally able to enter the blood component after 12 h and onward (Fig. 4B). Similarly, we did not observe significant differences between the glnA mutant and the wild type, although the bacterial loads in the lungs and blood tended to be higher than for the glnR mutant at later time points. However, this difference did not reach statistical significance (Fig. 4A). The glnP mutant was attenuated after 36 h of infection in the lungs, indicating that GlnP is required for full virulence in this model (Fig. 4C). In addition, in ΔglnP-infected mice fewer bacteria reached the systemic circulation, suggesting that GlnP plays a role in the dissemination of S. pneumoniae from the lungs to the bloodstream (Fig. 4D). Bacterial counts of the glnA-glnP double mutant decreased in time and were significantly lower from 12 h postinfection onward compared to wild-type counts (Fig. 4C). Moreover, no bacteria reached the bloodstream in mice infected with the glnA-glnP double mutant (Fig. 4D), again indicating a role for GlnP in the transition from the lungs to the blood, with an additive effect of GlnA. The gdhA mutant showed no difference in bacterial loads compared to the wild type in either the lungs or the systemic circulation (Fig. 4C and D), indicating that this gene is not required for full virulence in this model.
FIG. 4.
Pneumonia model. Shown are the bacterial loads in the lungs and blood of mice infected with D39 wild-type (wt), ΔglnA, ΔglnR, ΔglnP, ΔglnAP, or ΔgdhA bacteria. A and C, homogenized lungs; B and D, blood. *, P < 0.05.
Contribution of the GlnR regulon to bacteremia.
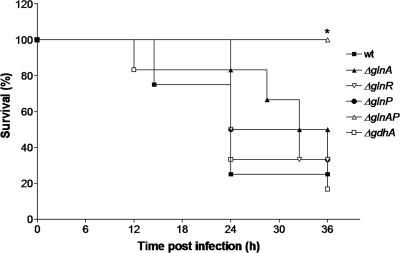
In the bacteremia model, infection of the bloodstream is followed for 36 h, allowing tracking of bacterial survival and growth within individual mice. Moreover, comparison of results obtained with the blood compartment in the pneumonia model enabled us to discriminate between attenuation in bacterial survival in blood or in the capability to disseminate from the lungs to the blood component. As a measurement of disease potential, the mean survival times of mice infected with the different strains were compared.
Mice infected with D39 wild-type bacteria had a calculated median survival time of 24 h, whereas ΔglnR and ΔglnA mutant-infected mice had calculated median survival times of 28.25 and 34.25 h, respectively (Fig. 5). These differences in median survival times were not statistically significantly different from that of wild-type-infected mice. In addition, survival times of mice infected with ΔglnP (i.e., 30 h) and ΔgdhA (i.e., 24 h) were not significantly different from those of mice infected with the wild type. However, all of the mice infected with the glnA-glnP double mutant survived until the end of the experiment (36 h), indicating reduced virulence of the double mutant during bacteremia (Fig. 5). Furthermore, all bacteria were cleared from the bloodstream after 24 h (data not shown).
FIG. 5.
Bacteremia model. Survival of mice infected with D39 wild-type (wt), ΔglnA, ΔglnR, ΔglnP, ΔglnAP, and ΔgdhA bacteria. *, P < 0.05.
Expression profile of the glnA-glnP double mutant.
To investigate the effect of the concurrent deletion of glnA and glnP on global gene expression, we performed microarray analysis of D39 wild-type and ΔglnAP bacteria.
Twelve genes were downregulated in the glnAP double mutant. In addition to GlnR-regulated glnA, glnP, and glnQ, these included genes encoding transcriptional regulators (e.g., SPD_0096, SPD_1524), ABC transporters (e.g., SPD_1526, SPD_1607), the starving-cell-induced protein Dpr, and the serine protease HtrA (Table 2).
TABLE 2.
Differentially expressed genes in D39 ΔglnAP
| Gene identifiera
|
Gene name | Annotation | Ratiob | |
|---|---|---|---|---|
| D39 | TIGR4/R6 | |||
| SPD_0051 | SP_0044 | purC | Phosphoribosylaminoimidazole-succinocarboxamide synthase | 1.1 |
| SPD_0111 | spr0103 | Argininosuccinate lyase | 1.1 | |
| SPD_0142 | SP_0139 | Hypothetical protein | 1.3 | |
| SPD_0143 | spr0139 | UDP-glucose 6-dehydrogenase | 1.9 | |
| SPD_0144 | SP_0141 | mutR | Transcriptional regulator | 2.0 |
| SPD_0145 | SP_0142 | Hypothetical protein | 3.8 | |
| SPD_0146 | SP_0143 | CAAX amino-terminal protease family protein | 3.9 | |
| SPD_0147 | SP_0144 | CAAX amino-terminal protease family protein | 4.0 | |
| SPD_0148 | SP_0145 | ABC transporter, substrate-binding protein | 3.9 | |
| SPD_0149 | SP_0146 | Hypothetical protein | 1.7 | |
| SPD_0161 | SP_0159 | Hypothetical protein | 3.9 | |
| SPD_0283 | SP_0310 | PTS system, IIC component | 1.5 | |
| SPD_0300 | SP_0327 | Hypothetical protein | 1.5 | |
| SPD_0334 | SP_0366 | aliA | Oligopeptide ABC transporter | 2.1 |
| SPD_0335 | SP_0368 | Cell wall surface anchor family protein | 1.4 | |
| SPD_0361 | SP_0395 | Transcriptional regulator, putative | 1.4 | |
| SPD_0364 | spr0361 | ABC-type polar amino acid transport system | 3.5 | |
| SPD_0404 | SP_0445 | ilvB | Acetolactate synthase, large subunit | 3.2 |
| SPD_0405 | SP_0446 | ilvN | Acetolactate synthase, small subunit | 3.4 |
| SPD_0406 | SP_0447 | ilvC | Ketol acid reductoisomerase | 3.1 |
| SPD_0407 | SP_0448 | Hypothetical protein | 2.5 | |
| SPD_0408 | SP_0449 | Hypothetical protein | 2.7 | |
| SPD_0409 | SP_0450 | ilvA | Threonine dehydratase | 2.7 |
| SPD_0424 | SP_0474 | PTS system, cellobiose-specific IIC component | 1.1 | |
| SPD_0541 | spr0470 | blpO | Bacteriocin | 2.4 |
| SPD_0652 | SP_0749 | livJ | Branched-chain amino acid ABC transporter | 1.0 |
| SPD_0653 | SP_0750 | livH | Branched-chain amino acid ABC transporter | 1.4 |
| SPD_0654 | SP_0751 | livM | Branched-chain amino acid ABC transporter, permease protein | 1.7 |
| SPD_0655 | SP_0752 | livG | Branched-chain amino acid ABC transporter | 1.7 |
| SPD_0656 | SP_0753 | livF | Branched-chain amino acid ABC transporter | 1.7 |
| SPD_0749 | SP_0856 | ilvE | Branched-chain amino acid aminotransferase | 1.8 |
| SPD_0751 | SP_0858 | Hypothetical protein | 1.8 | |
| SPD_0752 | SP_0859 | Hypothetical protein | 1.6 | |
| SPD_0753 | SP_0860 | pcp | Pyrrolidone-carboxylate peptidase | 1.7 |
| SPD_0778 | spr0786 | Hypothetical protein | 2.4 | |
| SPD_0778 | SP_0882 | Hypothetical protein | 2.3 | |
| SPD_0780 | SP_0884 | Hypothetical protein | 2.0 | |
| SPD_0780 | spr0788 | Hypothetical protein | 1.9 | |
| SPD_0781 | SP_0885 | Hypothetical protein | 2.3 | |
| SPD_0844 | SP_0955 | celB | Competence protein | 1.1 |
| SPD_0900 | SP_1013 | asd | Aspartate-semialdehyde dehydrogenase | 2.0 |
| SPD_0901 | SP_1014 | dapA | Dihydrodipicolinate synthase | 2.1 |
| SPD_1004 | SP_1119 | gapN | Glyceraldehyde-3-phosphate dehydrogenase, NADP dependent | 2.2 |
| SPD_1011 | SP_1126 | glxK | Glycerate kinase | 1.1 |
| SPD_1156 | spr1179 | Putative iron-dependent peroxidase | 1.0 | |
| SPD_1158 | SP_1306 | gdhA | NADP-specific glutamate dehydrogenase | 2.1 |
| SPD_1190 | SP_1356 | Atz/Trz family protein | 1.1 | |
| SPD_1191 | SP_1357 | ABC transporter, ATP-binding/permease protein | 1.1 | |
| SPD_1192 | SP_1358 | ABC transporter, ATP-binding/permease protein | 1.3 | |
| SPD_1276 | spr1302 | Hypothetical protein | 1.3 | |
| SPD_1464 | SP_1651 | psaD | Thiol peroxidase | 1.3 |
| SPD_1472 | SP_1659 | ileS | Isoleucyl-tRNA synthetase | 1.1 |
| SPD_1500 | SP_1688 | ABC transporter, permease protein | 1.1 | |
| SPD_1563 | spr1598 | Dicarboxylate/amino acid:cation (Na+ or H+) symporter | 1.3 | |
| SPD_1564 | SP_1754 | Hypothetical protein | 1.1 | |
| SPD_1585 | spr1620 | ABC transporter, sugar-binding protein | 1.7 | |
| SPD_1596 | SP_1811 | trpA | Tryptophan synthase, alpha subunit | 1.1 |
| SPD_1602 | SP_1817 | trpE | Anthranilate synthase component I | 1.1 |
| SPD_1649 | SP_1869 | fatD | Iron compound ABC transporter, permease protein | 1.5 |
| SPD_1650 | SP_1870 | fatC | ABC transporter membrane-spanning permease, ferric iron transport | 1.5 |
| SPD_1651 | SP_1871 | fecA | ABC transporter ATP-binding protein, ferric iron transport | 1.1 |
| SPD_1652 | SP_1872 | fatB | Iron compound ABC transporter, iron compound-binding protein | 1.1 |
| SPD_1667 | SP_1887 | amiF | ABC transporter, ATP-binding protein, oligopeptide transport | 1.1 |
| SPD_1668 | SP_1888 | amiE | ABC transporter, ATP-binding protein, oligopeptide transport | 1.3 |
| SPD_1669 | SP_1889 | amiD | ABC transporter, membrane-spanning permease, oligopeptide transport | 1.2 |
| SPD_1670 | SP_1890 | amiC | ABC transporter membrane-spanning permease, oligopeptide transport | 1.1 |
| SPD_1783 | SP_1986 | Hypothetical protein | 1.1 | |
| SPD_1840 | SP_2031 | Predicted Zn-dependent hydrolases of the β-lactamase fold | 1.3 | |
| SPD_1845 | SP_2036 | PTS system, ascorbate-specific IIA component, PTS-EII | 1.3 | |
| SPD_1954 | SP_2125 | Hypothetical protein | 2.2 | |
| SPD_1981 | spr1961 | Hypothetical protein | 1.1 | |
| SPD_1985 | SP_2157 | adh2 | Alcohol dehydrogenase, iron containing | 1.0 |
| SPD_1988 | SP_2160 | Hypothetical protein | 1.1 | |
| SPD_1994 | SP_2166 | fucA | l-Fuculose-phosphate aldolase | 1.2 |
| SPD_0096 | SP_0100 | Transcriptional regulator, PadR family protein | −1.0 | |
| SPD_0381 | SP_0418 | acp | Acyl carrier protein | −1.0 |
| SPD_0448 | SP_0502 | glnA | glnA, glutamine synthetase | −6.5 |
| SPD_0803 | SP_0910 | Hypothetical protein | −1.1 | |
| SPD_1098 | SP_1241 | glnP | Glutamine/glutamate transporter | −6.2 |
| SPD_1099 | SP_1242 | glnQ | Glutamine/glutamate transporter | −3.2 |
| SPD_1402 | SP_1572 | dpr | Starved-cell-like peroxide resistance protein | −1.1 |
| SPD_1524 | SP_1714 | Transcriptional regulator, GntR family protein | −1.3 | |
| SPD_1526 | spr1560 | Hypothetical protein (ABC-2 transporter) | −1.1 | |
| SPD_1526 | SP_1715 | Hypothetical protein, ABC-2 type transporter | −1.2 | |
| SPD_1607 | SP_1824 | ABC transporter, permease protein [Fe(III)] | −1.2 | |
| SPD_2068 | SP_2239 | htrA | Serine protease | −1.1 |
Gene identifier as deposited in the NCBI Gene Expression Omnibus (GEO) database under GEO Series accession number GSE9850 (TIGR4, SP; R6, SPR). Genes directly regulated by GlnR are in boldface.
The microarray ratio is a log2-transformed expression of ΔglnPA expression with respect to that of the wild-type strain.
Approximately 80 genes were upregulated in the glnA-glnP double mutant, most of them involved in amino acid metabolism (Table 2). Strikingly, many of these genes belong to the CodY regulon, such as the Ami operon, the Ilv operon, and the Liv operon (9). Also, gdhA, regulated by both GlnR and CodY, was strongly upregulated. Interestingly, two putative transcriptional regulators (SPD_0144 and SPD_0361) were upregulated, which suggests that other regulatory systems are active to complement the glnA and glnP mutations.
Of note, besides the genes within the GlnR regulon, no overlap in differentially expressed genes of the glnA mutant (described in reference 17) and the glnAP double mutant was observed.
DISCUSSION
The ability to adequately adapt to changes in the availability of nutrients is a prerequisite for bacterial survival. Several studies have suggested that nitrogen metabolism, and especially glutamine metabolism, is important for the virulence of pathogens (18, 29, 31). The transcriptional repressor GlnR regulates, together with glutamine synthetase GlnA, genes involved in glutamine/glutamate uptake and conversion in S. pneumoniae, L. lactis, and B. subtilis (7, 17, 19). DNA-binding assays have demonstrated that GlnR repression is dependent on GlnA, but the exact mechanism remains unknown (17). As a result, GlnR targets are derepressed in a glnA mutant. In this study, we investigated the contribution of the transcriptional regulator GlnR and its target genes to pneumococcal virulence, in particular, its contribution during colonization, pneumonia, and bacteremia in mice.
Absence of GlnR had no significant effect on bacterial virulence in any of the three infection models we used. It could well be that levels of available glutamine in the nasopharynx and in the lungs are quite low and, hence, no repression by GlnR is triggered. Alternatively, all genes of the GlnR regulon might be expressed in the ΔglnR strain, and redundant expression of these genes might not influence the ability to colonize or cause disease.
The conversion of glutamate to glutamine appears to be required during colonization, as the mutant lacking GlnA was found to be attenuated in colonization of the murine nasopharynx. Alternatively, the lack of GlnA might affect regulation by GlnR through derepression of its gene targets, although this appears less likely given the lack-of-colonization phenotype of the glnR mutant. Attenuation of the glnA mutant was not observed in the lungs and bloodstream in the pneumonia and bacteremia models of infection, indicating that GlnA is not required for bacterial survival in the lungs and the transition from the lungs to the blood in our infection models.
GlnP, which is part of the main glutamine/glutamate ABC transporter GlnPQ, is not required for colonization of the murine nasopharynx. Interestingly, GlnP was found to be involved in adherence to Detroit 562 cells, suggesting a role in colonization in humans. The possibility cannot be excluded that the difference between in vitro adherence and the murine colonization model reflects the differences between the in vitro and in vivo settings. For instance, expression of glnP might be required in RPMI medium, but not in the murine (or human) nasopharynx. In group B streptococci, it has been proposed that glnQ, also part of the glutamine/glutamate transporter, is involved in adherence to fibronectin and virulence in rats, possibly by modulating cytoplasmic glutamine levels (29). GlnP was also identified as a candidate for fibronectin binding by phage display library analysis (1). However, binding of fibronectin by GlnP of streptococci has never been confirmed, suggesting an indirect involvement (1, 29). In the pneumonia model, mice infected with the glnP mutant showed a lower bacterial load in the lungs at 36 h than wild-type-infected mice. Strikingly, while the number of wild-type bacteria increased with time, fewer glnP mutants reached the bloodstream from 24 h postinfection onward. This suggests a role for GlnP in the transition from the lungs to the bloodstream. Uptake of glutamine could be of importance at this stage of infection, i.e., entering the bloodstream. The impaired glutamine uptake in the glnP mutant might therefore create a growth disadvantage, leading to a smaller number of bacteria in the blood circulation. However, we cannot rule out the possibility that the ΔglnP phenotype, i.e., impaired transition from lungs to blood, is caused by multiple factors. Fewer bacteria cause less tissue damage in the lungs, and this will consequently lead to less spillover into the blood. One such example might be lower pneumolysin concentrations due to lower bacterial loads in the lungs (11).
The glnA-glnP double mutant displayed an attenuated phenotype similar to that of the glnA mutant in the colonization model, most likely caused by the glnA mutation only, given the lack-of-colonization phenotype of the glnP mutant. Moreover, since GlnP is part of the main glutamine and glutamate transporter (17), our data suggest that pneumococcus, while colonizing the murine nasopharynx, is able to acquire glutamine and glutamate through other processes such as peptide uptake and degradation. Different results were obtained in the lungs; here, the glnA mutation alone did not result in attenuation whereas mutation of both glnA and glnP did, suggesting that pneumococcus cannot easily use other sources for acquisition of glutamine at this particular site. The double mutant is not able to convert glutamate to glutamine (GlnA) and is not able to take up free glutamate or glutamine (GlnP). Consequently, the double mutant has to rely on another system for acquiring these amino acids. If this is so, such a system is apparently not able to complement the lack of GlnA and GlnP sufficiently when pneumococci reach the lungs or blood circulation, in contrast to the nasopharynx, where only glutamine synthesis (GlnA) contributes to survival. The glnAP mutant was able to grow in defibrinated blood in vitro, but this growth was slower and to a lower cell density than that of the wild type. Addition of glutamine rescued the phenotype of ΔglnAP to wild-type growth (data not shown). This suggests that, when added in high concentrations, glutamine can enter the cell, possibly by other uptake systems with very low affinity for glutamine or by passive diffusion.
The microarray data showed that predominantly genes involved in amino acid metabolism are upregulated in the glnAP mutant. These genes, most of them belonging to the CodY regulon (9), are probably upregulated because the double mutant is starved for amino acids, glutamine in particular. Although glutamine was present in the medium used for the expression study, the gene-regulatory network sensing and controlling general amino acid metabolism seemed perturbed. This is underscored by the differential expression of four transcriptional regulators; however, their function is unknown. This suggests that regulation of amino acid metabolism within the pneumococcal cell is quite complex and has severe effects on fitness and virulence. The impact on virulence could also be explained by the twofold downregulation of the expression of htrA in the glnAP mutant, as it was shown previously that this gene is required for virulence in D39 (12).
The gene gdhA, encoding glutamate dehydrogenase, does not appear to be required for virulence at any site in our infection models. No paralog of gdhA is present in the pneumococcal genome, which could complement its metabolic function. Regulation of gdhA by CodY has been described and might be the principal regulator in vivo by repressing gdhA expression (9, 17). Possibly, gdhA expression is coordinated in such a way that it is only activated under specific conditions (e.g., nitrogen limitation) and these conditions are not encountered by the pneumococcus in our mouse models.
We have described the behavior of mutants lacking genes of the GlnR regulon, the regulatory system of glutamine and glutamate metabolism, during adherence to human pharyngeal epithelial cells in vitro and experimental virulence in mice. We have identified two genes that play a role in virulence, namely, glnA and glnP. glnA encodes glutamine synthetase, which is required for colonization, and glnP encodes a membrane glutamine/glutamate permease which is required for survival in the lungs and possibly for the transition from the lungs to the blood circulation. This study provides novel insight into the nutritional requirements of S. pneumoniae within its host, more specifically, the glutamine and glutamate requirements. The different phenotypes of mutants during in vitro adherence to human pharyngeal cells and colonization of the murine nasopharynx suggest that different host species (i.e., mice and humans) may have different adhesion properties and nutritional supplies for the bacterium.
Many studies have shown that expression of pneumococcal proteins during pathogenesis is required at different stages of infection (15, 21, 25). One study, performed by Oggioni and coworkers, presents evidence that the “transcriptional state” of the pneumococcal cells differs at particular niches within the host (24). The bacterium has to deal with the supply of nutrients that each site of the body offers. For this reason, coregulation of site-specific metabolic and virulence factors by nutritional regulators is likely to occur. In this study, we report on the role of glutamine/glutamate metabolism and its regulation in our experimental models of virulence in mice. The site-specific requirements for GlnA and especially GlnP reported in this study might offer novel strategies to explore these molecules as future drug targets that specifically target invasive disease while leaving colonization unaffected.
In conclusion, genes within the GlnR regulon are of definite importance for bacterial survival within our experimental models of infection, with different subsets of the genes involved in glutamine/glutamate metabolism contributing to pneumococcal survival during different stages of infection.
Acknowledgments
W.T.H. is supported by the Sophia Foundation for Medical Research (SSWO 356, Rotterdam, The Netherlands), and T.G.K. and H.J.B. are supported by IOP Genomics grant IGE03002 from the Dutch Ministry of Economic Affairs.
We thank P. Burghout for critical reading of the manuscript and Anne de Jong for depositing the microarray data in the NCBI GEO database.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 3 January 2008.
REFERENCES
- 1.Beckmann, C., J. D. Waggoner, T. O. Harris, G. S. Tamura, and C. E. Rubens. 2002. Identification of novel adhesins from group B streptococci by use of phage display reveals that C5a peptidase mediates fibronectin binding. Infect. Immun. 702869-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry, A. M., and J. C. Paton. 1996. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect. Immun. 645255-5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogaert, D., R. De Groot, and P. W. Hermans. 2004. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect. Dis. 4144-154. [DOI] [PubMed] [Google Scholar]
- 4.Bootsma, H. J., M. Egmont-Petersen, and P. W. Hermans. 2007. Analysis of the in vitro transcriptional response of human pharyngeal epithelial cells to adherent Streptococcus pneumoniae: evidence for a distinct response to encapsulated strains. Infect. Immun. 755489-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, S. W., and A. L. Sonenshein. 1996. Autogenous regulation of the Bacillus subtilis glnRA operon. J. Bacteriol. 1782450-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.den Hengst, C. D., S. A. van Hijum, J. M. Geurts, A. Nauta, J. Kok, and O. P. Kuipers. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 28034332-34342. [DOI] [PubMed] [Google Scholar]
- 7.Fisher, S. H. 1992. Glutamine synthesis in Streptomyces—a review. Gene 11513-17. [DOI] [PubMed] [Google Scholar]
- 8.Hava, D. L., and A. Camilli. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol. Microbiol. 451389-1406. [PMC free article] [PubMed] [Google Scholar]
- 9.Hendriksen, W. T., H. J. Bootsma, S. Estevao, T. Hoogenboezem, A. de Jong, R. de Groot, O. P. Kuipers, and P. W. Hermans. 2008. CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190590-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendriksen, W. T., N. Silva, H. J. Bootsma, C. E. Blue, G. K. Paterson, A. R. Kerr, A. de Jong, O. P. Kuipers, P. W. Hermans, and T. J. Mitchell. 2007. Regulation of gene expression in Streptococcus pneumoniae by response regulator 09 is strain dependent. J. Bacteriol. 1891382-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirst, R. A., A. Kadioglu, C. O'Callaghan, and P. W. Andrew. 2004. The role of pneumolysin in pneumococcal pneumonia and meningitis. Clin. Exp. Immunol. 138195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibrahim, Y. M., A. R. Kerr, J. McCluskey, and T. J. Mitchell. 2004. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 723584-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston, J. W., D. E. Briles, L. E. Myers, and S. K. Hollingshead. 2006. Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect. Immun. 741171-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadioglu, A., N. A. Gingles, K. Grattan, A. Kerr, T. J. Mitchell, and P. W. Andrew. 2000. Host cellular immune response to pneumococcal lung infection in mice. Infect. Immun. 68492-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kerr, A. R., P. V. Adrian, S. Estevao, R. de Groot, G. Alloing, J. P. Claverys, T. J. Mitchell, and P. W. Hermans. 2004. The Ami-AliA/AliB permease of Streptococcus pneumoniae is involved in nasopharyngeal colonization but not in invasive disease. Infect. Immun. 723902-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerr, A. R., J. J. Irvine, J. J. Search, N. A. Gingles, A. Kadioglu, P. W. Andrew, W. L. McPheat, C. G. Booth, and T. J. Mitchell. 2002. Role of inflammatory mediators in resistance and susceptibility to pneumococcal infection. Infect. Immun. 701547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kloosterman, T. G., W. T. Hendriksen, J. J. Bijlsma, H. J. Bootsma, S. A. van Hijum, J. Kok, P. W. Hermans, and O. P. Kuipers. 2006. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem. 28125097-25109. [DOI] [PubMed] [Google Scholar]
- 18.Klose, K. E., and J. J. Mekalanos. 1997. Simultaneous prevention of glutamine synthesis and high-affinity transport attenuates Salmonella typhimurium virulence. Infect. Immun. 65587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsen, R., T. G. Kloosterman, J. Kok, and O. P. Kuipers. 2006. GlnR-mediated regulation of nitrogen metabolism in Lactococcus lactis. J. Bacteriol. 1884978-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40555-571. [DOI] [PubMed] [Google Scholar]
- 21.LeMessurier, K. S., A. D. Ogunniyi, and J. C. Paton. 2006. Differential expression of key pneumococcal virulence genes in vivo. Microbiology 152305-311. [DOI] [PubMed] [Google Scholar]
- 22.Long, A. D., H. J. Mangalam, B. Y. Chan, L. Tolleri, G. W. Hatfield, and P. Baldi. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 27619937-19944. [DOI] [PubMed] [Google Scholar]
- 23.Molle, V., Y. Nakaura, R. P. Shivers, H. Yamaguchi, R. Losick, Y. Fujita, and A. L. Sonenshein. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 1851911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oggioni, M. R., C. Trappetti, A. Kadioglu, M. Cassone, F. Iannelli, S. Ricci, P. W. Andrew, and G. Pozzi. 2006. Switch from planktonic to sessile life: a major event in pneumococcal pathogenesis. Mol. Microbiol. 611196-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orihuela, C. J., G. Gao, K. P. Francis, J. Yu, and E. I. Tuomanen. 2004. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J. Infect. Dis. 1901661-1669. [DOI] [PubMed] [Google Scholar]
- 26.Pearce, B. J., F. Iannelli, and G. Pozzi. 2002. Construction of new unencapsulated (rough) strains of Streptococcus pneumoniae. Res. Microbiol. 153243-247. [DOI] [PubMed] [Google Scholar]
- 27.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 665620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonenshein, A. L. 2005. CodY, a global regulator of stationary phase and virulence in gram-positive bacteria. Curr. Opin. Microbiol. 8203-207. [DOI] [PubMed] [Google Scholar]
- 29.Tamura, G. S., A. Nittayajarn, and D. L. Schoentag. 2002. A glutamine transport gene, glnQ, is required for fibronectin adherence and virulence of group B streptococci. Infect. Immun. 702877-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tojo, S., T. Satomura, K. Morisaki, J. Deutscher, K. Hirooka, and Y. Fujita. 2005. Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA. Mol. Microbiol. 561560-1573. [DOI] [PubMed] [Google Scholar]
- 31.Tullius, M. V., G. Harth, and M. A. Horwitz. 2003. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 713927-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Hijum, S. A., J. Garcia de la Nava, O. Trelles, J. Kok, and O. P. Kuipers. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl. Bioinformatics 2241-244. [PubMed] [Google Scholar]
- 33.Zalieckas, J. M., L. V. Wray, Jr., and S. H. Fisher. 2006. Cross regulation of the Bacillus subtilis glnRA and tnrA genes provides evidence for DNA binding site discrimination by GlnR and TnrA. J. Bacteriol. 1882578-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]