Abstract
Campylobacter jejuni NCTC 11168 was capable of growth to levels comparable with FeSO4 in defined iron-limited medium (minimal essential medium alpha [MEMα]) containing ferrilactoferrin, ferritransferrin, or ferri-ovotransferrin. Iron was internalized in a contact-dependent manner, with 94% of cell-associated radioactivity from either 55Fe-loaded transferrin or lactoferrin associated with the soluble cell fraction. Partitioning the iron source away from bacteria significantly decreased cellular growth. Excess cold transferrin or lactoferrin in cultures containing 55Fe-loaded transferrin or lactoferrin resulted in reduced levels of 55Fe uptake. Growth of C. jejuni in the presence of ferri- and an excess of apoprotein reduced overall levels of growth. Following incubation of cells in the presence of ferrilactoferrin, lactoferrin became associated with the cell surface; binding levels were higher after growth under iron limitation. A strain carrying a mutation in the cj0178 gene from the iron uptake system Cj0173c-Cj0178 demonstrated significantly reduced growth promotion in the presence of ferrilactoferrin in MEMα compared to wild type but was not affected in the presence of heme. Moreover, this mutant acquired less 55Fe than wild type when incubated with 55Fe-loaded protein and bound less lactoferrin. Complementation restored the wild-type phenotype when cells were grown with ferrilactoferrin. A mutant in the ABC transporter system permease gene (cj0174c) showed a small but significant growth reduction. The cj0176c-cj0177 intergenic region contains two separate Fur-regulated iron-repressible promoters. This is the first demonstration that C. jejuni is capable of acquiring iron from members of the transferrin protein family, and our data indicate a role for Cj0178 in this process.
Campylobacter jejuni is the leading cause of bacterial food-borne enteritis in the developed world (55). It is a commensal organism in the intestine of warm-blooded animals and birds (31). Infection in humans is often derived from contaminated poultry meat (51) and is frequently self-limiting (55) but can be followed by autoimmune complications such as Guillain-Barré syndrome (37). Understanding of Campylobacter pathogenicity is incomplete, but factors including motility and chemotaxis (26, 32) and adhesion and invasion (34) are thought to play a key role. In addition, iron homeostasis is proposed to be vital (56), with a large number of genes in the relatively small genome of C. jejuni NCTC 11168 involved in iron uptake, metabolism, and storage (41).
Iron is essential for bacteria and the human host, participating in a variety of basic metabolic pathways (5). However, excess iron in combination with oxygen leads to the production of damaging hydroxyl radicals by the Haber-Weiss reactions, and so iron uptake and storage must be strictly regulated (9, 45). In bacteria, such regulation is achieved principally by the global iron-dependent transcription regulator Fur (17). Under high-iron conditions, Fe(II) binds as a corepressor to the Fur dimer, and the complex binds to specific operator sequences (Fur boxes) within the promoter regions of iron-responsive genes, blocking transcription (16).
Under oxidizing conditions and at physiological pH, iron exists principally in the ferric form [Fe(III)], the majority of which is biologically unavailable due to its very low solubility (45). To combat this, Fe(III) is complexed in the host by iron-binding and transport proteins such as hemoglobin, transferrin, and lactoferrin (5). This results in an iron-restricted environment within the host (a concentration of free iron of ∼10−18 M) that acts as a nonspecific defense mechanism, limiting microbial growth (5). To overcome iron limitation, bacteria have evolved a number of mechanisms to scavenge iron from the host environment, including uptake systems specific for iron associated with host iron-binding proteins (5, 14, 30, 47). The C. jejuni NCTC 11168 genome contains four ferric iron uptake systems, chuABCD for iron bound to heme and hemoglobin (47), cfrA and ceuBCDE for ferri-enterochelin (39), and two uncharacterized systems, cj1658-cj1663 and cj0173c-cj0178. All these systems are composed of an outer membrane receptor protein component, periplasmic binding protein, and ABC transport system, with the exception of Cj1658-Cj1663 for which there is no obvious receptor. In addition, a pseudogene, cj0444, with similarity to genes for outer membrane receptors, is present in the NCTC 11168 genome (41).
Members of the transferrin iron-binding protein family are glycosylated, bilobed, monomeric proteins of around 80 kDa (24). Each protein reversibly binds two atoms of Fe(III) in conjunction with two bicarbonate anions (1, 3) and under normal conditions is only partially iron saturated in the host (30%). Transferrin is primarily found in serum (24), and lactoferrin is present in exocrine secretions, principally milk (3, 50). The avian iron transport protein ovotransferrin, which is present in blood serum and egg white, shares 51% and 49% homology with human transferrin and lactoferrin, respectively (21). The use of iron bound to lactoferrin or transferrin has been described for pathogenic Neisseria (24), Pasteurella spp. (24), Haemophilus influenzae (27, 44), Pseudomonas aeruginosa (52), Bordetella spp. (23, 35, 46), Helicobacter pylori (13, 29), Staphylococcus aureus (40), and Candida albicans (33).
C. jejuni was previously considered incapable of utilizing iron bound to members of the transferrin protein family (43). Here, we demonstrate for the first time that C. jejuni can grow in liquid culture supplemented with iron bound to lactoferrin, transferrin, or ovotransferrin. The process of iron uptake is dependent upon proximity and demonstrates receptor specificity. Mutants defective in the expression of potential receptor proteins for lactoferrin/transferrin were constructed and characterized, and a role for Cj0178 is proposed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. All media were from Oxoid, and all chemicals were obtained from Sigma-Aldrich unless otherwise indicated. Escherichia coli DH5αe was cultured aerobically in either Luria-Bertani broth or on Luria-Bertani agar plates at 37°C (48). Ampicillin (100 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (20 μg/ml) were added as required. C. jejuni strains were grown microaerobically (85% N2, 10% CO2, and 5% O2) at 37°C in a variable atmosphere incubator (Don Whitley Scientific Ltd., United Kingdom) in Mueller-Hinton broth (MHB), on Mueller-Hinton agar (MHA), on Brucella agar (BA; Becton, Dickinson and Company), in SAPI medium (2.77 mM glucose, 6.25 mM NH4NO3, 1.84 mM KH2PO4, 3.35 mM KCl, 1.01 mM MgSO4) containing 50 mM Tris-HCl, pH 7.5 (19), or in minimal essential medium alpha (MEMα; Invitrogen). All media were supplemented with vancomycin (10 μg/ml) and trimethoprim (5 μg/ml). When necessary, further antibiotic selection was provided by adding kanamycin (50 μg/ml) or chloramphenicol (20 μg/ml). Addition of FeSO4 to a final concentration of 10 μM ensured iron-replete conditions. To induce iron-restricted conditions in MHB, MHA, and BA, deferoxamine mesylate salt (Desferal) was added to a final concentration of 20 μM (59). Growth in unsupplemented MEMα induced iron limitation, as described previously (59).
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Characteristics | Reference and/or source |
|---|---|---|
| Strains | ||
| E. coli DH5αe | Cloning host strain: F− φ80lacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17 (rK− mK+) gal phoA supE44 λ−thi-1 gyrA96 relA1 | 25; Invitrogen |
| C. jejuni strains | ||
| NCTC 11168 (11168-GS) | Wild-type genome sequenced strain | 41; NCTC |
| 480 (NCTC 12744) | Clinical isolate used as host strain for maintaining plasmids/reporter assays | 32; NCTC |
| KAR2 | NCTC 11168 cj0178::Kanr | This study |
| KAR3 | NCTC 11168 p19::Kanr | This study |
| JDR5 | NCTC 11168 ΔchuA::cat | 47 |
| JDR21 | NCTC 11168 Δcj0178::cat | This study |
| CEM3 | NCTC 11168 Δcj0174c::Kanr | This study |
| CEM5 | NCTC 11168 ΔcfrA::Kanr | This study |
| CEM8 | Complementation of cj0178::Kanr with a wild-type copy of cj0178, plus cat, into cj0752 | This study |
| Plasmids | ||
| pUC19 | Cloning/suicide vector; Ampr | New England Biolabs |
| pMW10 | E. coli/C. jejuni shuttle LacZ reporter vector; Kmr | 60 |
| p23E5 | Reporter construct; C. jejuni NCTC 11168 metK promoter region inserted into BamHI site of pMW10 | 59, 60 |
| pJDR13 | Reporter construct; C. jejuni NCTC 11168 chuA promoter region inserted into BamHI site of pMW10 | 47 |
| pAV35 | pBluescript vector containing a promotered Cmr (cat) cassette | 59 |
| pJMK30 | pUC19 containing C. coli (aph-3) Kmr gene flanked by multiple cloning regions | 59 |
| pcam193b8a | pUC19 containing C. jejuni NCTC 11168 genomic fragment (bases 174132 to 175858) at SmaI site | 41; from tiled genome library |
| pKAR2 | pcam193b8::Kanr; insertion of kan at PsiI site (base position 1128 of Cj0178) | This study |
| pGEMCWH01b | cj0752 flanks with internal multiple cloning region in pGEM T-vector backbone | 15 |
| pCEM3 | pUC19 containing C. jejuni NCTC 11168 cj0174c genomic fragment (bases 171562 to 169946) at KpnI and PstI sites | This study |
| pCEM5 | pUC19 containing C. jejuni NCTC 11168 cfrA genomic fragment (bases 705450 to 707540) at KpnI and PstI sites | This study |
| pCEM9 | pCEM3, cj0174c::Kanr; deletion of 1,477 bp of cj0174c ORF (bases 171520 to 170042) and insertion of Kanr (in forward orientation with respect to deleted gene) into BglII site created at point of deletion | This study |
| pCEM11 | pCEM5, ΔcfrA::Kanr; deletion of 1,966 bp of cfrA ORF (bases 705484 to 707451) and insertion of Kanr (in forward orientation with respect to deleted gene) into BglII site created at point of deletion | This study |
| pCEM14 | Reporter construct; C. jejuni NCTC 11168 cj0176c-cj0177 promoter region (bases 172915 to 172695) inserted into BamHI site of pMW10 (cj0176c orientation) | This study |
| pCEM15 | Reporter construct; C. jejuni NCTC 11168 cj0176c-cj0177 promoter region (bases 172695 to 172915) inserted into BamHI site of pMW10 (cj0177 orientation) | This study |
| pCEM16 | Reporter construct; C. jejuni NCTC 11168 cj0178 promoter region (bases 173477 to 173763) inserted into BamHI site of pMW10 | This study |
| pCEM17 | Reporter construct; C. jejuni NCTC 11168 exbB1 promoter region (bases 175801 to 176042) inserted into BamHI site of pMW10 | This study |
| pCEM18 | pUC19 containing C. jejuni NCTC 11168 genomic fragment (bases 172686 to 176113; promoter region, cj0177, and cj0178) at KpnI and PstI sites | This study |
| pCEM19 | pCEM18, Δcj0177; deletion of 764 bp of cj0177 ORF (bases 172943 to 173706) | This study |
| pCEM20 | pCEM19 plus cat; insertion of cat at PstI site downstream of cj0178 (in forward orientation with respect to wild type-copy of gene) | This study |
pcam193b8 was kindly provided by J. Parkhill, PSU, Sanger Centre, Cambridge, United Kingdom.
A gift from C. Penn, University of Birmingham, Birmingham, United Kingdom.
Agar plate growth assays.
C. jejuni NCTC 11168 was tested for the ability to use human ferrilactoferrin, human ferritransferrin, or ferri-ovotransferrin as a sole source of iron for growth as described by Pickett et al. (43), using BA or MHA plates containing 20 μM Desferal.
Liquid medium growth assays.
C. jejuni strains were grown in MEMα in the presence of human lactoferrin, human transferrin, or ovotransferrin in both ferri- and apo- forms (0.27 μM or 1.11 μM) or in the presence of porcine heme (25 μM). Apolactoferrin was produced by sequential dialysis of ferrilactoferrin against 0.2 M citric acid (pH 2.3), distilled water, and 100 mM Tris-HCl (pH 7.5) (20). Final protein concentration was determined by the Bradford assay (Sigma-Aldrich). Receptor specificity was tested by competing an excess of apoproteins or bovine serum albumin (BSA; 0.81 μM) against ferriproteins (0.27 μM). Proximity dependency was tested by separating the ferriproteins from the C. jejuni cells in narrow-bore dialysis tubing (6-kDa cutoff). Strains were harvested with phosphate-buffered saline (PBS; 2 ml/plate) following 24 h of growth on MHA plates. Cells were suspended in MEMα and used to inoculate 5 ml of MEMα cultures to an initial optical density at 600 nm (OD600) of 0.1. Cultures were incubated microaerobically for 12 to 16 h with agitation to achieve iron depletion. Following this, cells were harvested and used to inoculate fresh MEMα 10-ml cultures to an initial OD600 of 0.025. Cultures were supplemented with the appropriate iron sources and grown as described previously (47), and OD readings were taken at intervals. Growth differences were analyzed for significance by an unpaired Student's t test (P = <0.05; n = 6).
Molecular genetic methods.
Restriction enzymes and T4 DNA ligase were purchased from New England Biolabs; shrimp alkaline phosphatase was purchased from Roche. All procedures were as previously described (48), unless stated otherwise. PCR amplification of DNA was achieved using the primers listed in Table 2 and Taq polymerase (Abgene) or the Triple Master Mix System (Eppendorf), with reactions cycled in an Eppendorf Mastercycler. PCR products were purified using a Qiagen QIAquick or Minelute PCR purification kit according to the manufacturer's instructions. DNA sequencing was performed using a BigDye Terminator (version 3.1) Cycle Sequencing Kit and processed on an ABI 377 DNA sequencer. C. jejuni chromosomal DNA was prepared following a standard protocol (6). Small-scale and large-scale plasmid DNA preparations were performed using Qiagen QIAprep Spin Mini- and Midi-prep kits according to the manufacturer's instructions. Constructs were transformed into C. jejuni by electroporation (48, 58).
TABLE 2.
Primers
| Primer function and name | Base sequence (5′-3′)a | Product or annealing site |
|---|---|---|
| Cloning | ||
| 0172cF | GGGGTACCGAGCTATGGCAAGAGGATGTAAG | pCEM1 insert |
| 0172cR | AAAACTGCAGGAGCCTTATGGCATTGATATAG | |
| 0173cF | GGGGTACCGCTTTAATCACTGCTTTAATTACC | pCEM2 insert |
| 0173cR | AAAACTGCAGCATAGCCATGATCACCTGCATTG | |
| 0174cF | GGGGTACCGATAGAGATCAAGCCAGACAAG | pCEM3 insert |
| 0174cR | AAAACTGCAGGTTACCATAATAGCCGTAATGCC | |
| 0177F1 | GGGGTACCGAGAATGTTCAGAGCAAGCAAAAG | pCEM4 insert 1 |
| 0177R1 | TCCCCCGGGCATTAGTAAGTTCATTGGCTTTAATGG | |
| 0177F2 | TCCCCCGGGGTTGTGGTGAATTTAAGCTATGG | pCEM4 insert 2 |
| 0177R2 | AAAACTGCAGCCCACATATCCATTATTTTCAAGTTC | |
| cfrAF | GGGGTACCCATTCGAAGGCTCTTGTATAG | pCEM5 insert |
| cfrAR | AAAACTGCAGCTTCTACCTCCATAAACAAGAC | |
| Inverse PCR | ||
| 0172cInvF | GAAGATCTCCTTGGAAGATTATCGAAATG | pCEM1 |
| 0172cInvR | GAAGATCTCATTGCGCATTTTACAGTAG | |
| 0173cInvF | GAAGATCTCTTAAAATAGGAAATCGCTTTGAG | pCEM2 |
| 0173cInvR | GAAGATCTCTCCTTCTTTGACATGAAGATTG | |
| 0174cInvF | GAAGATCTGATGCAGCTTTGCCTTCCTTG | pCEM3 |
| 0174cInvR | GAAGATCTGTAAAATTGCCCCGAGTTTATAG | |
| cfrAInvF | GAAGATCTTCGCAAGTGGGAGTGGTAG | pCEM5 |
| cfrAInvR | GAAGATCTTCCCAATGGCGCAAACTG | |
| KAR2 complementation | ||
| 0752upF | GGAATAATCAAGCCTACAAAATC | pGEMCWH01 |
| 0752upR | GATTTTGTAGGCTTGATTATTCC | pGEMCWH01 |
| 0752downF | GTCAATCCACAATATACAAGCAAAAC | pGEMCWH01 |
| 0752downR | GTTTTGCTTGTATATTGTGGATTGAC | pGEMCWH01 |
| 0178compF | GGGGTACCCATAGACATTAATTCCTTTTAGATATTTTG | pCEM18 insert |
| 0178compR | AACTGCAGGTATCTGTTGCATTTGCTTCTG | |
| 0177comp InvF | GAAGATCTGCTATGGAGAAATAGATTTAAAAG | pCEM18 |
| 0177comp InvR | GAAGATCTGAATGCTTAAAATAGTATAAAATTTC | |
| Promoter cloning and gel shifts | ||
| 0176c-77 promF | CGGGATCCTAATTCCTTTTAGATATTTTG | pCEM14/15 insert |
| 0176c-77 promR | CGGGATCCTATTATTGTTACTTTCTTAG | |
| 0178promF | CGGGATCCGACATTATATCCTTAAGCCATAAGC | pCEM16 insert |
| 0178promR | CGGGATCCTCCTCCCCCTTATAAATCAAAAC | |
| exbB1promF | CGGGATCCCAAAAATTCCTATTATAGTAGATTTATATTG | pCEM17 insert |
| exbB1promR | CGGGATCCTTGTATATTCCTTAATATTTATATTCAAAAC | |
| 0176c-p-split | TTATTATAATAAAATGAAAATATTTATC | Promoter probes |
| 0177-p-split | CATTTTATTATAATAAATTTTGATAATA |
Restriction enzyme sites are shown in bold.
Construction of C. jejuni mutants.
Genes cj0174c and cfrA were inactivated by inverse PCR mutagenesis (61) to produce the strains detailed in Table 1. The genes were amplified from the C. jejuni NCTC 11168 genome using primers designed to incorporate 500 bp of DNA flanking the deletion (Table 2). Fragments were cloned directionally into pUC19 to produce constructs pCEM3 and pCEM5 (Table 1). Inverse PCR was followed by insertion of a kanamycin resistance marker to generate plasmids pCEM9 and pCEM11 (Table 1). Insertional inactivation of cj0178 was achieved by cloning a kanamycin resistance marker into the distal PsiI site within the cj0178 open reading frame (ORF) in construct pcam193b8 to produce pKAR2 (Table 1). The cj0178 mutant JDR21 was constructed by inverse PCR mutagenesis (61) with insertion of a chloramphenicol resistance marker derived from pAV35 (Table 1). Each final construct was confirmed by sequencing and transformed into C. jejuni NCTC 11168.
Radioactive iron uptake assays.
Assays involving 55Fe-loaded transferrin and lactoferrin were performed as described previously (20). SAPI medium (5 ml) was supplemented with 8.33 nmol 55Fe/ml of filter-sterilized 55Fe-transferrin or -lactoferrin, either directly into the medium (mixed) or enclosed within a 1-cm diameter dialysis membrane (partitioned). Reaction mixtures were incubated at 37°C for a minimum of 5 h. Unincorporated 55Fe was removed using a Bio-Rad Bio-Spin 6 chromatography column. The radioactivity in each sample was measured by scintillation counting (Minaxi Tri-Carb 400 series; Canberra-Packard). Approximately 5 ml of a C. jejuni overnight culture (2 × 108 to 3 × 108 CFU/ml) was washed once in PBS and three times in SAPI and then added to either the mixed or partitioned experiments and incubated for 4 h at 37°C microaerobically. After incubation, three aliquots (1 ml) of bacteria were removed from both the mixed and partitioned experiments, pelleted, washed once in PBS, and resuspended in 50 μl of PBS. The bacterial cells were then measured for 55Fe incorporation in the tritium counting channel, using 2 ml of Emulsifier Safe scintillation fluid per aliquot of culture or 100 μl of supernatant.
Demonstration of lactoferrin binding by C. jejuni.
C. jejuni cells were harvested from six MHA plates with confluent bacterial growth. The bacterial cells were washed twice in MEMα and used to inoculate iron-restricted and iron-replete 5-ml MEMα cultures to an initial OD600 of 0.1. Cultures were incubated microaerobically for 12 to 16 h with agitation. Following this, cells were harvested and used to inoculate fresh 5-ml MEMα cultures to an initial OD600 of 0.3 prior to incubation with ferrilactoferrin for 60 min. Cells were again pelleted, washed three times in MEMα, and suspended in 50 μl of 100 mM Tris-HCl (pH 6.8) containing 10% (vol/vol) glycerol and 2% (wt/vol) sodium dodecyl sulfate (SDS). This suspension was heated to 100°C for 15 min, pelleted, electrophoresed on 10% SDS-polyacrylamide gels (47, 48), and electroblotted onto polyvinylidene difluoride membranes at 4°C in 25 mM Tris, 192 mM glycine, 0.037% (wt/vol) SDS, and 10% (vol/vol) methanol. Blots were probed with anti-lactoferrin polyclonal antiserum, and binding was determined using enhanced chemiluminescence (20).
Complementation of cj0178::Kanr with a functional copy of cj0178.
Complementation of cj0178::Kanr was performed by inserting a wild-type copy of cj0178 into the putative insertion element transposase pseudogene, cj0752, of NCTC 11168 (15). Genes cj0178 and cj0177 and the associated promoter region were amplified from the C. jejuni NCTC 11168 chromosome using primers detailed in Table 2 and cloned into pUC19 to form pCEM18 (Table 1). Inverse PCR (61) was used to delete the cj0177 ORF, resulting in fusion of the promoter region to cj0178, producing construct pCEM19 (Table 1). A chloramphenicol resistance (cat) cassette (Table 1, pAV35) was inserted at the PstI site downstream of cj0178 in pCEM19, generating pCEM20 (Table 1). The entire region was excised using KpnI and ligated to pGEMCWH01 between flanking cj0752 sequences (Table 1, pCEM21); pCEM21 was introduced into KAR2 by electroporation.
Determination of promoter activity levels.
The region between the cj0176c and cj0177 genes and possible promoter regions immediately upstream of genes cj0178 and exbB1 were amplified using the primers detailed in Table 2 and cloned into pMW10 (Table 1), producing a transcriptional fusion between each putative promoter and the promoterless lacZ gene present in the vector (57, 60) (Table 1). The resulting constructs pCEM14 to pCEM17 (Table 1) were transformed into C. jejuni strain 480. Each construct was sequenced to confirm that the promoter regions contained no errors. Promoter activity levels were determined by measuring β-galactosidase activity in cultures containing reporter constructs grown in iron-restricted or iron-replete MHB over a 5-h period at 37°C, as previously described (36).
EMSAs.
C. jejuni Fur was expressed and purified as described previously (28). Electrophoretic mobility shift assays (EMSAs) were used to show the binding of Fur to Fur-regulated promoter regions between cj0176c and cj0177. Each half of the intergenic spacer region was amplified using PCR with the primers detailed in Table 2. EMSAs were completed following a procedure described previously (28).
RESULTS
Acquisition of iron for growth from members of the transferrin protein family.
C. jejuni NCTC 11168 was previously thought to be incapable of acquiring iron for growth from human ferrilactoferrin or human ferritransferrin (43). In order to test this, iron-restricted MHA or BA plates inoculated with C. jejuni (5 × 107 CFU) were spotted (10 μl) with 10 μM FeSO4 (positive control), double-distilled H2O (negative control), and 30 μM human ferrilactoferrin, human ferritransferrin, or ferri-ovotransferrin. Following microaerobic incubation, a halo of growth was visible surrounding the FeSO4 (diameter, 21 mm) (data not shown), but no halo of growth was visible around the negative control or the other iron sources, although limited growth appeared to be present directly beneath each ferriprotein spot (diameter, ∼5 mm) (data not shown). These results, which suggest that iron bound to proteins of the transferrin family is essentially unavailable to C. jejuni in a plate assay format, are in agreement with those of Pickett et al. (43).
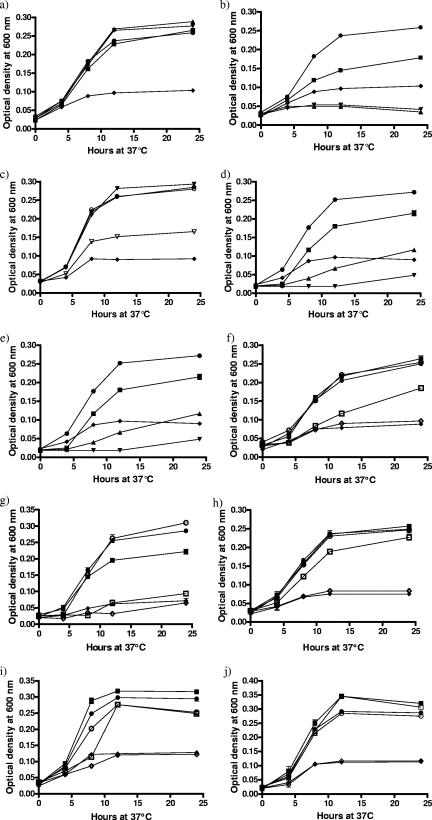
The situation is different, however, in liquid culture (Fig. 1). C. jejuni NCTC 11168 grew poorly in a defined iron-limited medium, MEMα, as reported previously (59), but supplementation with human ferrilactoferrin or human ferritransferrin or ferri-ovotransferrin (0.27 μM) resulted in growth to the same levels as with 10 μM FeSO4 (Fig. 1a). By contrast, bacterial growth in the presence of equivalent concentrations of human apotransferrin or apo-ovotransferrin was poorer than in unsupplemented medium alone (Fig. 1b). There was limited growth in the presence of the human apolactoferrin preparation (Fig. 1b), although significantly less than with the ferriprotein. This is probably due to incomplete iron removal during the preparation of apolactoferrin.
FIG. 1.
Growth assays of wild-type and iron utilization gene mutants with iron supplied bound to lactoferrin, transferrin, and ovotransferrin or as heme. All assays were conducted in MEMα with cultures incubated microaerobically with agitation. All conditions were tested in triplicate in two independent assays. Each data point is the mean of six replicates with sample error plotted. (a) C. jejuni NCTC 11168 with ferriproteins. •, 10 μM FeSO4 (iron-replete conditions); ⧫, unsupplemented medium (iron-limited conditions); ▪, 0.27 μM human ferrilactoferrin; ▾, 0.27 μM human ferritransferrin; ▴, 0.27 μM ferri-ovotransferrin. (b) C. jejuni NCTC 11168 with apoproteins. •, 10 μM FeSO4 (iron-replete conditions); ⧫, unsupplemented medium (iron-limited conditions); ▪, 0.27 μM human apolactoferrin; ▾, 0.27 μM human apotransferrin; ▴, 0.27 μM apo-ovotransferrin. (c) C. jejuni NCTC 11168 mixed with the iron source or partitioned from the iron source within dialysis tubing. •, 10 μM FeSO4 (iron-replete conditions) mixed; ○, 10 μM FeSO4 (iron-replete conditions) partitioned; ⧫, unsupplemented medium (iron-limited conditions); ▾, 0.27 μM human ferritransferrin mixed; ▿, 0.27 μM human ferritransferrin separated. (d) C. jejuni NCTC 11168 with competing apo- and ferriproteins. •, 10 μM FeSO4 (iron-replete conditions); ⧫ unsupplemented medium (iron-limited conditions); ▪, 0.27 μM human ferrilactoferrin and 0.81 μM human apolactoferrin; ▾, 0.27 μM human ferritransferrin and 0.81 μM human apotransferrin; ▴, 0.27 μM ferri-ovotransferrin and 0.81 μM apo-ovotransferrin. (e) C. jejuni NCTC 11168 with competition of BSA against ferriprotein. •, 10 μM FeSO4 (iron-replete conditions); ⧫, unsupplemented medium (iron-limited conditions); ▪, 0.27 μM human ferrilactoferrin and 0.81 μM BSA; ▾, 0.27 μM human ferritransferrin and 0.81 μM BSA; ▴, 0.27 μM ferri-ovotransferrin and 0.81 μM BSA. (f) C. jejuni NCTC 11168 and CEM5 (ΔcfrA::kanr) with human ferrilactoferrin. •, 11168 and 10 μM FeSO4 (iron-replete conditions); ○, CEM5 and 10 μM FeSO4 (iron-replete conditions); ⧫, 11168 with unsupplemented medium (iron-limited conditions); ⋄, CEM5 with unsupplemented medium (iron-limited conditions); ▪, 11168 with 0.27 μM human ferrilactoferrin; □, CEM5 with 0.27 μM human ferrilactoferrin. (g) C. jejuni NCTC 11168 and KAR2 (cj0178::Kanr) with human ferrilactoferrin. •, 11168 with 10 μM FeSO4 (iron-replete conditions); ○, KAR2 with 10 μM FeSO4 (iron-replete conditions); ⧫, 11168 with unsupplemented medium (iron-limited conditions); ⋄, KAR2 with unsupplemented medium (iron-limited conditions); ▪, 11168 with 0.27 μM human ferrilactoferrin; □, KAR2 with 0.27 μM human ferrilactoferrin. (h) C. jejuni NCTC 11168 and CEM3 (Δcj0174c::Kanr) with human ferrilactoferrin. •, 11168 with 10 μM FeSO4 (iron-replete conditions); ○, CEM3 with 10 μM FeSO4 (iron-replete conditions); ⧫, 11168 with unsupplemented medium (iron-limited conditions); ⋄, CEM3 with unsupplemented medium (iron-limited conditions); ▪, 11168 with 0.27 μM human ferrilactoferrin; □, CEM3 with 0.27 μM human ferrilactoferrin. (i) C. jejuni NCTC 11168 and CEM8 (complemented KAR2) with human ferrilactoferrin. •, 11168 with 10 μM FeSO4 (iron-replete conditions); ○, CEM8 with 10 μM FeSO4 (iron-replete conditions); ⧫, 11168 with unsupplemented medium (iron-limited conditions); ⋄, CEM8 with unsupplemented medium (iron-limited conditions); ▪, 11168 with 0.27 μM human ferrilactoferrin; □, CEM8 with 0.27 μM human ferrilactoferrin. (j) C. jejuni NCTC 11168 and KAR2 (cj0178::Kanr) with porcine heme. •, 11168 with 10 μM FeSO4 (iron-replete conditions); ○, KAR2 with 10 μM FeSO4 (iron-replete conditions); ⧫, 11168 with unsupplemented medium (iron-limited conditions); ⋄, KAR2 with unsupplemented medium (iron-limited conditions); ▪, 11168 with 25 μM porcine heme; □, KAR2 with 25 μM porcine heme.
Characterization of iron uptake from the transferrin protein family.
In order to demonstrate that C. jejuni can take up transferrin- or lactoferrin-bound iron, the ability to acquire radioactive iron from 55Fe-loaded transferrin (55Fe-transferrin) or 55Fe-loaded lactoferrin (55Fe-lactoferrin) was determined. Bacteria were either in direct contact (mixed) with 55Fe-transferrin or 55Fe-lactoferrin or were separated by a dialysis membrane (partitioned). Cells in contact with 55Fe-transferrin accumulated approximately sixfold more radioactive iron than those partitioned from the 55Fe-loaded protein (Fig. 2a), suggesting that acquisition of iron from transferrin requires close proximity or direct contact with the bacterial cells. Similar data were obtained with 55Fe-lactoferrin (data not shown). Fractionation of cells that had acquired iron from transferrin indicated that the majority of cell-associated radioactivity was present in the soluble fraction, which represents the periplasm and cytoplasm, and only 6% was associated with the insoluble membrane fraction. Again, similar data were obtained with 55Fe-lactoferrin (data not shown). To confirm the necessity for proximity between ferriprotein and the bacterial cell surface for successful iron acquisition, strain NCTC 11168 was grown in MEMα liquid culture with iron supplied in the form of each ferriprotein (0.27 μM) either enclosed within dialysis membrane or in direct contact with the cells (Fig. 1c). When in contact with the iron source, C. jejuni grew to levels comparable with the iron-replete controls, but when the ferriproteins were partitioned from the C. jejuni cells, growth was significantly reduced (Fig. 1c; only transferrin is shown). As expected, there was no significant difference between levels of C. jejuni growth when FeSO4 was mixed with or separated from the cells. These results support the observation that proximity is necessary for successful acquisition of iron from members of the transferrin protein family and implies that iron uptake from these sources does not require the ability to utilize siderophores.
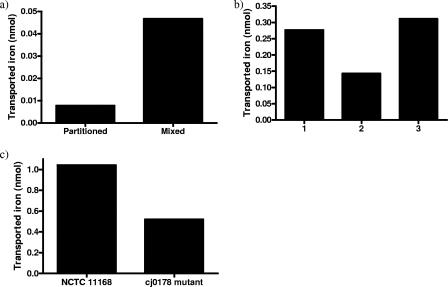
FIG. 2.
Radioactive 55Fe uptake from members of the transferrin protein family by C. jejuni. All experiments were completed using 1 ml of culture at 2 × 108 to 3 × 108 CFU/ml. Radioactivity is represented as nmol of 55Fe transported into the cell. (a) Radioactivity associated with the C. jejuni cell pellet from three dialysis partition experiments using 55Fe-transferrin as an iron source. 55Fe-transferrin was partitioned away from the bacteria using a dialysis membrane (partitioned) or in contact with the bacteria (mixed), and associated radioactivity was measured. (b) C. jejuni incorporation of 55Fe when incubated in direct contact with 55Fe-transferrin either alone (1), with an equivalent concentration of nonradioactive iron saturated transferrin (2), or with an equivalent concentration of BSA (3). Each column displays the mean values of three determinations from one experimental run. (c) Cell-associated radioactivity of C. jejuni NCTC 11168 and the cj0178 (JDR21) mutant strain measured from cells incubated in SAPI medium containing 55Fe-lactoferrin as an iron source. The uptake associated with the mutant strain is around half (49.93%) that of the wild-type strain.
To investigate the association of transferrin or lactoferrin with C. jejuni, 55Fe uptake assays were set up with C. jejuni in direct contact with 55Fe-transferrin either alone or with an equal concentration of nonradioactive ferritransferrin or with an equivalent concentration of BSA (Fig. 2b). The presence of cold ferritransferrin reduced the accumulation of radioactivity by half compared with cells incubated with 55Fe-transferrin alone, indicating competition for C. jejuni binding sites between transferrin molecules loaded with 55Fe and those bound to nonradioactive iron. On the other hand, cells incubated with 55Fe-transferrin and BSA showed an equivalent level of uptake to cells incubated with 55Fe-transferrin alone. An essentially identical pattern of 55Fe accumulation was observed with 55Fe-lactoferrin (data not shown).
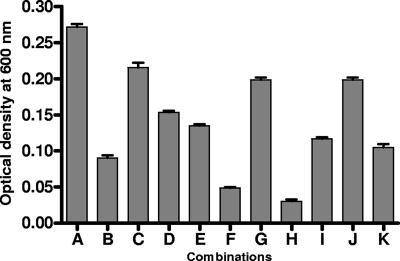
Similar effects were seen in the growth response to ferriproteins. Wild-type cells were grown in MEMα liquid culture with iron supplied solely as each ferriprotein (0.27 μM) with competing concentrations of either the respective apoprotein or BSA (0.81 μM) (Fig. 1d and e). Competition by excess apoprotein limited growth to levels almost comparable with apoprotein alone, again suggesting the presence of specific receptor sites on the C. jejuni cell surface for which apo- and ferri- forms of the protein compete. Overall growth levels were higher with human apolactoferrin, probably due, as mentioned earlier, to the fact that the preparation used may not have been completely devoid of iron. Competition of each ferriprotein with BSA resulted in levels of growth comparable to ferriprotein alone, indicating that BSA did not interfere with the process of iron uptake (Fig. 1e). All combinations of ferri- and apoproteins (0.27 μM and 0.81 μM, respectively) were tested; growth limitation comparable to that illustrated in Fig. 1d was observed with all protein combinations (Fig. 3).
FIG. 3.
Summary of C. jejuni growth in the presence of competing concentrations of different combinations of holo- and apoproteins. Growth assays were conducted in MEMα with cultures of C. jejuni NCTC 11168 incubated microaerobically with agitation. Combinations of holo- and apoproteins were tested in triplicate in two independent assays. Each bar is the mean of the six replicates at 24 h with sample error shown. Combinations are as follows: A, 10 μM FeSO4 (iron-replete conditions); B, unsupplemented medium (iron-limited conditions); C, 0.27 μM human ferrilactoferrin and 0.81 μM human apolactoferrin (reference); D, 0.27 μM human ferrilactoferrin and 0.81 μM human apotransferrin; E, 0.27 μM human ferrilactoferrin and 0.81 μM apo-ovotransferrin; F, 0.27 μM human ferritransferrin and 0.81 μM human apotransferrin (reference); G, 0.27 μM human ferritransferrin and 0.81 μM human apolactoferrin; H, 0.27 μM human ferritransferrin and 0.81 μM apo-ovotransferrin; I, 0.27 μM ferri-ovotransferrin and 0.81 μM apo-ovotransferrin (reference); J, 0.27 μM ferri-ovotransferrin and 0.81 μM human apolactoferrin; K, 0.27 μM ferri-ovotransferrin and 0.81 μM human apotransferrin.
Binding of lactoferrin to the C. jejuni cell surface.
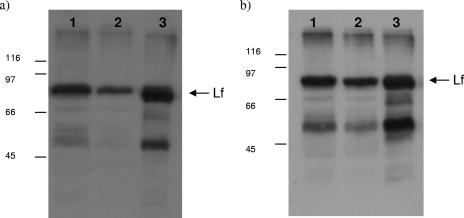
C. jejuni NCTC 11168 cells were incubated in the presence of ferrilactoferrin under iron-restricted and iron-replete conditions, pelleted, washed, and lysed, and the proteins were separated by SDS-polyacrylamide gel electrophoresis. Proteins were blotted to polyvinylidene difluoride membranes and probed with polyclonal anti-lactoferrin antiserum (Fig. 4a). The presence of an 80-kDa band in lanes 1 and 2 running to the same position as the mature pure lactoferrin protein (lane 3) indicates that lactoferrin associates sufficiently strongly to the cell surface of C. jejuni to avoid being removed by the washing step, and iron regulation of this binding is shown by the decreased intensity of the band in lane 2 (42% compared to lane 1). Transferrin also bound to cells (data not shown), indicating that C. jejuni is capable of binding either glycoprotein.
FIG. 4.
Lactoferrin binding to C. jejuni. Western blot analysis of solubilized C. jejuni proteins (approximately 25 μg of protein separated by SDS-polyacrylamide gel electrophoresis) prepared from iron-restricted NCTC 11168 cells (a, lane 1) and iron-replete NCTC 11168 cells (a, lane 2) incubated in the presence of lactoferrin and from iron-restricted NCTC 11168 cells (b, lane 1) and iron-restricted cj0178 (KAR2) mutant strain cells (b, lane 2) incubated in the presence of lactoferrin. Lanes 3 contained pure lactoferrin protein (positive control), and blots were probed with anti-lactoferrin antibodies. The arrow indicates the position of the mature lactoferrin (Lf) protein. Protein marker sizes are indicated in kilodaltons to the left of each blot.
Role of known C. jejuni iron uptake systems in acquisition of iron from lactoferrin.
To determine if one or more of the C. jejuni outer membrane receptor proteins is involved in iron acquisition from the transferrin family, the following mutant strains were tested for growth in liquid medium when iron was supplied solely in the form of human ferrilactoferrin: CEM5 (ΔcfrA::Kanr), KAR2 (cj0178::Kanr), KAR3 (p19::Kanr, periplasmic binding protein mutant), and JDR5 (ΔchuA::cat) (Table 1). The growth of all mutant strains under iron-replete conditions in MHB showed no significant difference from wild-type growth levels (data not shown). In MEMα all strains grew similarly, with comparable growth promotion under iron-replete conditions (10 μM FeSO4) and growth restriction under iron-limited conditions (unsupplemented MEMα) (Fig. 1). In the presence of human ferrilactoferrin (0.27 μM), strains KAR3 (p19::Kanr), JDR5 (ΔchuA::cat), and wild-type C. jejuni NCTC 11168 showed no significant difference in growth (data not shown). Strain CEM5 (ΔcfrA::Kanr) showed significantly less growth than wild type in the presence of 0.27 μM human ferrilactoferrin (Fig. 1f). CfrA has been previously characterized as the ferric enterochelin receptor (39), so the role that this protein plays in the uptake of iron from members of the transferrin protein family requires further investigation. KAR2 (cj0178::Kanr) showed the most significantly decreased growth compared to wild type with 0.27 μM human ferrilactoferrin as the sole iron source (Fig. 1g). Furthermore, the cj0178 mutant exhibited a significantly reduced ability to acquire iron from 55Fe-lactoferrin (Fig. 2c). In the presence of equivalent concentrations of human ferritransferrin and ferri-ovotransferrin, KAR2 had the same phenotype as with human ferrilactoferrin (data not shown). KAR2 mutant cells were tested for the ability to bind lactoferrin in comparison to NCTC 11168 cells (Fig. 4b). Less lactoferrin was associated with cj0178 mutant cells (lane 2, 73.2%) than NCTC 11168 cells, supporting a direct role for Cj0178 in lactoferrin binding. These results indicate a primary role for Cj0178 in iron utilization from members of the transferrin protein family for growth, although the possibility that other systems are involved cannot be excluded. Under the same conditions, a strain with a deletion in the gene encoding the cognate ABC transporter system permease (CEM3, cj0174c::Kanr) showed a small, but significant decrease in growth (Fig. 1 h). A BLAST comparison of Cj0178 showed 99% amino acid identity with a TonB-dependent colicin receptor protein of C. jejuni strain RM1221, but transferrin-associated iron uptake proteins of H. influenzae Rd KW20 (31%) and Neisseria meningitidis Z2491 (24%) were also highlighted (http://www.ncbi.nlm.nih.gov/BLAST/).
Complementation of cj0178::Kanr with a functional copy of cj0178.
Strain CEM8 grew to the same level as wild type in MEMα when iron was supplied solely as human ferrilactoferrin (Fig. 1i), human ferritransferrin, or ferri-ovotransferrin (data not shown).
Growth of the cj0178 mutant strain KAR2 is not affected when iron is supplied solely in the form of heme.
In addition to transferrin-binding proteins, Cj0178 also demonstrates sequence similarity to a heme-utilization protein of H. influenzae (30% identity), and along with Cj0177 has been proposed to have a role in the uptake of heme in C. jejuni NCTC 11168 (10). Although no direct role was demonstrated for Cj0178 in the uptake of heme, Cj0177 was shown to bind heme in vitro (10). In order to ascertain whether Cj0178 is necessary for the acquisition of heme for growth, the mutant strain KAR2 (cj0178::Kanr) was grown with iron supplied solely in the form of heme (25 μM) in liquid medium growth assays (Fig. 1j). There was no significant difference between KAR2 (cj0178::Kanr) and wild-type C. jejuni growth levels in the presence of heme (Fig. 1j).
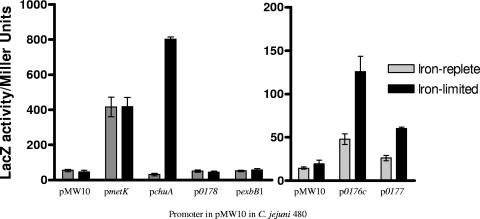
The operons cj0176c-cj0173c and cj0177-tonB1 are Fur regulated and expressed from two separate iron-responsive promoters within the cj0176c-cj0177 intergenic region.
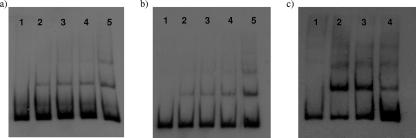
Previous expression level profiling of the C. jejuni iron regulon indicated that the cj0173c-tonB1 region of the C. jejuni genome is expressed at high levels under iron restriction and at low levels when iron is in excess (28). In a fur mutant strain, derepression of the genes was observed under iron-replete conditions, indicating the involvement of Fur (28, 39). The intergenic region between cj0176c and cj0177 contains Fur box consensus sequences (39, 56). To investigate and confirm Fur- and iron-responsive regulation of this region, the intergenic spacer between genes cj0176c and cj0177 was cloned into the E. coli/C. jejuni shuttle vector pMW10 (60). The region was incorporated into pMW10 immediately upstream of the lacZ gene in both orientations, producing constructs pCEM14 carrying pcj0176c::lacZ and pCEM15 carrying pcj0177::lacZ (Table 1). In addition, to determine whether promoter regions are present adjacent to cj0178 or exbB1, 200-bp regions immediately upstream of each gene were cloned into pMW10 using the same strategy, producing constructs pCEM16 carrying pcj0178::lacZ and pCEM17 carrying pexbB1::lacZ (Table 1). A constitutively active promoter region from the housekeeping gene metK (57) (p23E5 carrying pmetK::lacZ) and the iron-responsive promoter region of chuA (47) (pJDR13 carrying pchuA::lacZ) cloned into pMW10 and pMW10 with no promoter region were also tested. Promoter activity levels were determined from β-galactosidase measurements of iron-restricted or iron-replete C. jejuni 480 cultures containing reporter constructs grown in MHB over a 5-h period at 37°C (36, 60). Strain 480 showed identical growth patterns to NCTC 11168 in MHB, MEMα, and MEMα with iron (data not shown). The promoter regions of both cj0176c and cj0177 demonstrated significantly increased promoter activity under iron limitation (Fig. 5). The regions immediately upstream of cj0178 and exbB1 demonstrated no promoter activity (Fig. 5). These data confirm the prediction by Holmes et al. (28) that cj0177-tonB1 are cotranscribed from the promoter region located upstream of cj0177. To demonstrate Fur binding to the cj0176c-cj0177 intergenic region and confirm the presence of two separate Fur boxes, the cj0176c-cj0177 intergenic region was divided (using primers described in Table 2) to produce two fragments containing either the putative cj0176c Fur box (114-bp DNA fragment) or the putative cj0177 Fur box (140-bp DNA fragment). EMSAs were performed on the two regions using recombinant purified C. jejuni Fur protein (28). As the amount of Fur protein was increased, a concentration-dependent shift in the mobility of each DNA fragment was observed (Fig. 6a and b), demonstrating the presence of two separate functional Fur boxes in this intergenic region. To confirm specificity of binding, competitive EMSAs were completed as described previously (28) in which unlabeled competitor DNA (to a 1,500- or 2,000-fold excess) was added to the reaction mixture just before the Fur and labeled DNA. A decrease in the DNA shift was visible with an increase of unlabeled self-competitors (Fig. 6c).
FIG. 5.
β-Galactosidase assay to determine activity levels of promoter regions across the cj0173c-tonB1 region of the C. jejuni NCTC 11168 genome. Reporter gene assay of pMW10 with no promoter region (negative control) and the promoter regions upstream of housekeeping gene metK (positive control) (59), chuA (iron-responsive control) (47), and cj0176c, cj0177, cj0178, and exbB1. Promoter activity levels were determined from β-galactosidase activity of iron-limited (20 μM Desferal) or iron-replete (40 μM FeSO4) C. jejuni 480 cultures containing reporter constructs (Table 1) and grown in MHB for 5 h at 37°C. All conditions were tested in triplicate in two independent assays. Each data point is the mean of six replicates, with the standard error of the mean shown.
FIG. 6.
EMSAs of the cj0176c-cj0177 intergenic region using Fur. Digoxigenin-labeled DNA was present at 0.0775 nM. Lane 1, no protein, lanes 2 to 5, labeled DNA fragments incubated with 30, 60, 90, and 210 ng/μl C. jejuni Fur, respectively. (a) Labeled probe of the cj0176c side of intergenic region split by PCR to contain cj0176c Fur box (113 bp). (b) Labeled probe of the cj0177 side of intergenic region split by PCR to contain cj0177 Fur box (140 bp). (c) Competitive EMSA. Digoxigenin-labeled intergenic spacer region DNA was present at 0.0775 nM. Lane 1, no protein; lane 2, labeled DNA fragment incubated with 210 ng/μl C. jejuni Fur; lane 3, labeled DNA fragment incubated with Fur (210 ng/μl) with a 1,500-fold increase of competitors (unlabeled intergenic spacer fragment); lane 4, labeled DNA fragment incubated with Fur (210 ng/μl) with a 2,000-fold increase of competitors (unlabeled intergenic spacer fragment).
DISCUSSION
We have demonstrated for the first time that C. jejuni NCTC 11168 is capable of utilizing iron bound to members of the transferrin protein family. The process is receptor specific with a requirement for proximity between the iron source and the cell surface. There appears to be a primary role for the outer membrane receptor protein Cj0178 and a potential role for the associated ABC transport system Cj0173c-Cj0175c in this process. Promoter analysis demonstrated that the cj0173c-cj0178 region of the C. jejuni genome is iron responsive and classically Fur regulated and has two separate promoter regions between cj0176c and cj0177, with the genes cj0177-tonB1 coexpressed. We propose the redesignation of Cj0178 as the Campylobacter transferrin-bound iron utilization receptor (CtuA).
Repetition of previous work (43) has confirmed that lactoferrin-, transferrin-, and also ovotransferrin-bound iron is largely unavailable to C. jejuni for growth in a plate assay. However, liquid culture assays demonstrated that iron bound to human ferrilactoferrin, human ferritransferrin, and ferri-ovotransferrin is biologically available to C. jejuni for growth. Analysis of fractionated C. jejuni cells incubated in the presence of either 55Fe-transferrin or 55Fe-lactoferrin showed that iron obtained from these host proteins accumulated primarily within the cell. It is probable that iron is liberated from the proteins at the cell surface, transported across the outer membrane, and then transported across the inner membrane by one of the ABC transporter systems. Another possibility is that the ferri-glycoprotein is transported directly into the cell, but this is unlikely, based on the known mechanisms of iron acquisition from transferrin/lactoferrin in other gram-negative bacteria (49). The use of transferrin/lactoferrin as a bacterial iron source has been described extensively for pathogenic Neisseria spp., in which specific cell surface receptors that bind transferrin or lactoferrin consist of two proteins: transferrin or lactoferrin binding proteins A (TbpA/LbpA) and B (TbpB/LbpB). Protein A demonstrates homology to siderophore-receptor outer membrane proteins (7, 12), and protein B is a bilobed lipoprotein (4, 42). An interaction between transferrin, for example, and TbpAB induces conformational change, allowing liberation of iron at the outer membrane without internalization of the protein, and iron uptake occurs via the TbpA pore (8, 22). Once in the periplasm, iron is then bound by FbpA (11) and delivered to the inner membrane ABC transport system FbpBC, which transports it into the cytoplasm (2). P. aeruginosa employs a siderophore-mediated mechanism of iron acquisition from transferrin (52). Bordetella pertussis and Bordetella bronchiseptica are proposed to be capable of both siderophore-mediated and direct contact methods of iron uptake from transferrin (23, 35, 46). Further examples of pathogens capable of iron acquisition from transferrin or lactoferrin include H. pylori (13, 29), Pasteurella spp. (24), S. aureus (40), and the human fungal pathogen C. albicans (33).
Competition between 55Fe-transferrin/55Fe-lactoferrin and nonradioactive ferritransferrin/ferrilactoferrin decreased the amount of cell-associated radioactivity accumulated. Cellular growth was also reduced by competing excess apoproteins against ferriproteins. These results imply that apo- and ferri- forms of the proteins are competing for sites on the C. jejuni cell surface. As BSA causes no inhibition of radioactive iron uptake or growth in the presence of ferriprotein, the sites involved are likely to be specific receptors. As competing concentrations of all combinations of apo- and ferriproteins also reduced growth, C. jejuni is not likely to possess separate receptors or binding sites specific for individual proteins, as seen with Neisseria spp. LbpAB and TbpAB (49) but, rather, a single receptor capable of binding lactoferrin, transferrin, and ovotransferrin, indicating a novel system in C. jejuni. Moreover, iron uptake assays in which transferrin or lactoferrin was partitioned from the cells support the conclusion that C. jejuni must be in contact or close proximity with the protein in order to obtain iron. The demonstration that both lactoferrin and transferrin can bind to C. jejuni whole cells is evidence that there is direct contact between lactoferrin or transferrin and C. jejuni. The demonstration of iron-responsive binding, taken with the iron uptake and growth data, indicates the presence of an iron-regulated iron acquisition system which requires the interaction of ferritransferrins. There is no evidence for the presence of siderophore biosynthesis genes in C. jejuni NCTC 11168 (41) or other Campylobacter genomes sequenced to date (18; also http://xbase.bham.ac.uk/campydb/). Together, this implies that siderophores or other exogenous substrates are not essential for the uptake of iron from lactoferrin or transferrin by C. jejuni. C. jejuni appears to acquire iron in a receptor-specific, proximity-dependent manner. In the plate growth assay, contact between the ferriproteins and C. jejuni cell surface was limited to directly beneath the protein spot, explaining the limited growth observed only where the protein was in contact with the agar.
The putative ferric iron uptake outer membrane receptor protein Cj0178 demonstrates some similarity to several transferrin binding proteins. In addition, mutation of cj0178 results in reduced colonization ability in a chick cecum model and rabbit ileal loop model (39, 53). Comparison of growth patterns between wild-type and mutant C. jejuni strains supplied only with human lactoferrin-bound iron showed that the most significant decrease in growth is seen when cj0178 of the putative Cj0173c-Cj0178 iron uptake system is mutated. Moreover, 55Fe uptake experiments demonstrated that mutation of cj0178 reduced uptake of iron from ferrilactoferrin into the cell to approximately half that seen with the wild-type strain, and ferrilactoferrin was bound less by cj0178 mutant whole cells than NCTC 11168. The Cj0178 outer membrane receptor is unlikely to be a close ortholog of TbpA. However, a BLAST comparison of TbpA with the NCTC 11168 genome showed that Cj0178 and CfrA demonstrate 24% amino acid identity, whereas ChuA has 20% identity. In addition, the protein product of the functional copy of the NCTC 11168 pseudogene cj0444 from C. jejuni strain 81-176 shows 26% sequence identity to TbpA. Growth and uptake were not completely abolished in the cj0178 mutant strain, and disruption of the ferri-enterobactin receptor CfrA (39) also affected the utilization of iron from the transferrin protein family. Therefore, Cj0178 may not be the sole receptor, and the mechanism involved in uptake is likely to be different from that involving TbpA. In addition, decreased growth of a cj0178 mutant with human ferritransferrin and ferriovotransferrin suggests a multiprotein specificity for Cj0178, unlike Neisseria spp. (24). In the C. jejuni genome, there are no obvious accessory proteins equivalent to TbpB/LbpB, which are thought to assist TbpA/LbpA in distinguishing between iron-bound and iron-free transferrin or lactoferrin in Neisseria. This supports the proposed theory of a novel multiprotein-specific receptor in C. jejuni rather than separate receptors specific for transferrin or lactoferrin as seen in Neisseria. Complementation of the cj0178 mutation restored the wild-type phenotype, indicating that the growth changes observed are not due to polarity.
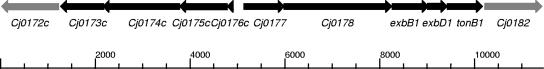
The small, but significant decrease in growth observed with CEM3 (Δcj0174c::Kanr) suggests a minor role for the associated ABC transport system Cj0173c-Cj0175c. Further ABC transporter systems in the NCTC 11168 genome (28, 39, 47) may complement Cj0173c-Cj0175c, resulting in the limited phenotype observed. Some redundancy of inner membrane ABC transporter systems has been observed for heme uptake in a range of bacteria (47), and ABC transporter system double mutants grew significantly less than single mutant strains when supplied with heme as the sole source of iron (data not shown). Alternatively, the limited phenotype of the cj0174c mutant may be due to an as yet uncharacterized mechanism for uptake. The Fur-regulated genes cj0173c-cj0178 are organized into two operons (cj0176c-cj0173c and cj0177-tonB1) (Fig. 7), with Cj0175c-Cj0173c demonstrating homology to the FbpABC inner-membrane transport system for lactoferrin- and transferrin-derived iron of Neisseria spp. and H. influenzae (28, 39, 54). Cj0175c is the proposed periplasmic binding protein, and Cj0174 and Cj0173c are the permease and ATPase constituents, respectively. Cj0176c and Cj0177 are putative lipoproteins of uncharacterized function. The operons are flanked by cj0172c (a putative gene of unknown function demonstrating homology to saccharopine dehydrogenase genes involved in the metabolism of lysine) (http://xbase.bham.ac.uk/campydb/) and cj0182 (encoding a putative transmembrane transport protein) (Fig. 7).
FIG. 7.
Genomic context of genes cj0173c-tonB1 in the C. jejuni NCTC 11168 genome. Distances are indicated in base pairs, with cj0173c-tonB1 in black and unrelated flanking genes in gray (41) (http://xbase.bham.ac.uk/campydb/).
A previous study demonstrated the ability of a Cj0177 homodimer to bind two cofacial heme groups (10). In addition, when a homolog of cj0177 in P. aeruginosa, phuW, was mutated and the strain was supplied solely with heme, growth was impaired (38). Comparison of wild-type C. jejuni and cj0178 mutant growth profiles when only porcine heme was supplied showed no significant difference. Reliance upon sequence similarity and an in vitro demonstration of heme binding may not be sufficient to conclusively demonstrate the role, if any, that Cj0178 plays in heme uptake. Moreover, chuABCD has been identified as the major heme uptake cluster in C. jejuni NCTC 11168 (47).
Previous work has suggested the presence of consensus Fur-box-like sequences upstream of both the cj0176c and cj0177 genes (39, 56). The genes cj0173c-tonB1 have been shown to be part of the Fur regulon and are induced under iron limitation (28, 39). Our reporter gene studies of the intergenic spacer region between cj0176c and cj0177, as well as the regions upstream of cj0178 and exbB1, confirmed the iron responsiveness of the cj0176c and cj0177 promoter regions. The regions immediately upstream of cj0178 and exbB1 demonstrated no promoter activity when iron was restricted, which is consistent with cotranscription of cj0177-tonB1 from the promoter region located upstream of cj0177 (28). EMSAs demonstrated direct Fur binding to both sides of the intergenic spacer region between cj0176c and cj0177. Holmes et al. (28) showed that expression of cj0177-tonB1 and cj0176c-cj0173c was derepressed in a fur mutant, suggesting Fur regulation of the region, but they suggested that a putative Fur box upstream of cj0177 (56) was involved in regulation of both promoter regions. We have shown for the first time that Fur binds to both promoters, indicating the presence of two Fur boxes within the intergenic spacer region and therefore separate classical Fur-dependent, iron-repressible regulation of the two operons.
Acknowledgments
This work was supported by studentships to C.E.M. and J.D.R. and project grants (91/D19661 and EGA16166) from the Biotechnology and Biological Sciences Research Council, United Kingdom.
We thank Ran Ren for carrying out the EMSA, Primrose Freestone for assistance with the preparation of apolactoferrin, Charles Penn for providing us with pGEMCWH01, Ashley Thorpe for making a pMW10-promoter reporter construct, and Emma Davé for production of the kanamycin resistance cassettes.
Footnotes
Published ahead of print on 18 January 2008.
REFERENCES
- 1.Abdallah, F. B., and J.-M. El Hage Chahine. 2000. Transferrins: iron release from lactoferrin. J. Mol. Biol. 303255-266. [DOI] [PubMed] [Google Scholar]
- 2.Adhikari, P., S. A. Berish, A. J. Nowalk, K. L. Veraldi, S. A. Morde, and T. A. Mietzer. 1996. The fbpABC locus of Neisseria gonorrhoeae functions in the periplasm-to-cytosol transport of iron. J. Bacteriol. 1782145-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, B. F., H. M. Baker, E. J. Dodson, G. E. Norris, S. V. Rumball, J. M. Waters, and E. N. Baker. 1987. Structure of human lactoferrin at 3.2-Å resolution. Proc. Natl. Acad. Sci. 841769-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, J. E., P. F. Sparling, and C. N. Cornelissen. 1994. Gonococcal transferrin-binding protein 2 facilitates but is not essential for transferrin utilization. J. Bacteriol. 1763162-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andrews, S. C., A. K. Robinson, and F. Rodríguez-Quiñones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27215-237. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley and Sons, New York, NY.
- 7.Biswas, G. D., and P. F. Sparling. 1995. Characterization of lbpA, the structural gene for a lactoferrin receptor in Neisseria gonorrhoeae. Infect. Immun. 632958-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas, G. D., J. E. Anderson, and P. F. Sparling. 1997. Cloning and functional characterization of Neisseria gonorrhoeae tonB, exbB, and exbD genes. Mol. Microbiol. 24169-179. [DOI] [PubMed] [Google Scholar]
- 9.Braun, V., and M. Braun. 2002. Iron transport and signaling in Escherichia coli. FEBS Lett. 52978-85. [DOI] [PubMed] [Google Scholar]
- 10.Chan, A. C. K., B. Lelj-Garolla, F. I. Rosell, K. A. Pedersen, A. G. Mauk, and M. E. P. Murphy. 2006. Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J. Mol. Biol. 3621108-1119. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C.-Y., S. A. Berish, S. A. Morse, and T. A. Mietzner. 1993. The ferric iron-binding protein of pathogenic Neisseria spp. functions as a periplasmic transport protein in iron acquisition from human transferrin. Mol. Microbiol. 10311-318. [DOI] [PubMed] [Google Scholar]
- 12.Cornelissen, C. N., G. D. Biswas, J. Tsai, D. K. Paruchuri, S. A. Thompson, and P. F. Sparling. 1992. Gonococcal transferrin-binding protein 1 is required for transferrin utilization and is homologous to TonB-dependent outer membrane receptors. J. Bacteriol. 1745788-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhaenens, L., L. Szczebara, and M. O. Husson. 1997. Identification, characterization and immunogenicity of the lactoferrin-binding protein from Helicobacter pylori. Infect. Immun. 65514-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekins, A., A. G. Khan, S. R. Shouldice, and A. B. Schryvers. 2004. Lactoferrin receptors in gram-negative bacteria: insights into the iron acquisition process. Biometals 17235-243. [DOI] [PubMed] [Google Scholar]
- 15.Elvers, K. T., S. M. Turner, L. M. Wainwright, G. Marsden, J. Hinds, J. A. Cole, R. K. Poole, C. W. Penn, and S. F. Park. 2005. NssR, a member of the Crp-Fnr superfamily from Campylobacter jejuni, regulates a nitrosative stress-responsive regulon that includes both a single-domain and a truncated haemoglobin. Mol. Microbiol. 57735-750. [DOI] [PubMed] [Google Scholar]
- 16.Escolar, L., J. Pérez-Martín, and V. de Lorenzo. 1998. Binding of the Fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283537-547. [DOI] [PubMed] [Google Scholar]
- 17.Escolar, L., J. Pérez-Martín, and V. de Lorenzo. 1999. Opening the iron box: Transcriptional metalloregulation by the Fur protein. J. Bacteriol. 1816223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freestone, P. P. E., R. D. Haigh, P. H. Williams, and M. Lyte. 2003. Involvement of enterobactin in norepinephrine-mediated iron supply from transferrin to enterohaemorrhagic Escherichia coli. FEMS Microbiol. Lett. 22239-43. [DOI] [PubMed] [Google Scholar]
- 20.Freestone, P. P. E., M. Lyte, C. P. Neal, A. F. Maggs, R. D. Haigh, and P. H. Williams. 2000. The mammalian neuroendocrine hormone norepinephrine supplies iron for bacterial growth in the presence of transferrin or lactoferrin. J. Bacteriol. 1826091-6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giansanti, F., P. Rossi, M. T. Massucci, D. Botti, G. Antonini, P. Valenti, and L. Seganti. 2002. Antiviral activity of ovotransferrin discloses an evolutionary strategy for the defensive activities of lactoferrin. Biochem. Cell Biol. 80125-130. [DOI] [PubMed] [Google Scholar]
- 22.Gómez, J. A., M. T. Criado, and C. M. Ferreirós. 1998. Cooperation between the components of the meningococcal transferrin receptor, TbpA and TbpB, in the uptake of transferrin iron by the 37-kDa ferric-binding protein (FbpA). Res. Microbiol. 149381-387. [DOI] [PubMed] [Google Scholar]
- 23.Gorringe, A. R., G. Woods, and A. Robinson. 1990. Growth and siderophore production by Bordetella pertussis under iron-restricted conditions. FEMS Microbiol. Lett. 54101-105. [DOI] [PubMed] [Google Scholar]
- 24.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 4185-191. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166557-580. [DOI] [PubMed] [Google Scholar]
- 26.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52471-484. [DOI] [PubMed] [Google Scholar]
- 27.Herrington, D. A., and P. F. Sparling. 1985. Haemophilus influenzae can use human transferrin as a sole source for required iron. Infect. Immun. 48248-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmes, K., F. Mulholland, B. M. Pearson, C. Pin, J. McNicholl-Kennedy, J. M. Ketley, and J. M. Wells. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151243-257. [DOI] [PubMed] [Google Scholar]
- 29.Husson, M. O., D. Legrand, G. Spik, and H. Leclerc. 1993. Iron acquisition by Helicobacter pylori: importance of human lactoferrin. Infect. Immun. 612694-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kammler, M., C. Schön, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 1756212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ketley, J. M. 1997. Pathogenesis of enteric infection by Campylobacter. Microbiology 1435-21. [DOI] [PubMed] [Google Scholar]
- 32.King, V., T. M. Wassenaar, B. A. M. van der Zeijst, and D. G. Newell. 1991. Variations in Campylobacter jejuni flagellin, and flagellin genes, during in vivo and in vitro passage. Microb. Ecol. Health Dis. 4135-140. [Google Scholar]
- 33.Knight, S. A. B., G. Vilaire, E. Lesuisse, and A. Dancis. 2005. Iron acquisition from transferrin by Candida albicans depends on the reductive pathway. Infect. Immun. 735482-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konkel, M. E., and L. A. Joens. 1989. Adhesion to and invasion of HEp-2 cells by Campylobacter spp. Infect. Immun. 572984-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Menozzi, F. D., C. Gantiez, and C. Locht. 1991. Identification and purification of transferrin- and lactoferrin-binding proteins of Bordetella pertussis and Bordetella bronchiseptica. Infect. Immun. 593982-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Nachamkin, I., B. M. Allos, and T. Ho. 1998. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 11555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochsner, U. A., Z. Johnson, and M. L. Vasil. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146185-198. [DOI] [PubMed] [Google Scholar]
- 39.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 1864714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, R. Y., H. Y. Sun, M. H. Choi, Y. H. Bai, and S. H. Shin. 2005. Staphylococcus aureus siderophore-mediated iron-acquisition system plays a dominant and essential role in the utilization of transferrin-bound iron. J. Microbiol. 43183-190. [PubMed] [Google Scholar]
- 41.Parkhill, J. W., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M-A. Rajandream, K. M. Rutherford, A. H. M. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403665-668. [DOI] [PubMed] [Google Scholar]
- 42.Pettersson, A., T. Prinz, A. Umar, J. van der Biezen, and J. Tommassen. 1998. Molecular characterization of LbpB, the second lactoferrin-binding protein of Neisseria meningitidis. Mol. Microbiol. 27599-610. [DOI] [PubMed] [Google Scholar]
- 43.Pickett, C. L., T. Auffenberg, E. C. Pesci, V. L. Sheen, and S. S. Jusuf. 1992. Iron acquisition and hemolysin production by Campylobacter jejuni. Infect. Immun. 603872-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pidcock, K. A., J. A. Wooten, B. A. Daley, and T. L. Stull. 1988. Iron acquisition by Haemophilus influenzae. Infect. Immun. 56721-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54881-941. [DOI] [PubMed] [Google Scholar]
- 46.Redhead, K., T. Hill, and H. Chart. 1987. Interaction of lactoferrin and transferrins with the outer membrane of Bordetella pertussis. J. Gen. Microbiol. 133891-898. [DOI] [PubMed] [Google Scholar]
- 47.Ridley, K. A., Y. Li, J. D. Rock, and J. M. Ketley. 2006. Heme utilization in Campylobacter jejuni. J. Bacteriol. 1887862-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 49.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 321117-1123. [DOI] [PubMed] [Google Scholar]
- 50.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417552-555. [DOI] [PubMed] [Google Scholar]
- 51.Slader, J., G. Domingue, F. Jørgensen, K. McAlpine, R. J. Owen, F. J. Bolton, and T. J. Humphrey. 2002. Impact of transport crate reuse and of catching and processing on Campylobacter and Salmonella contamination of broiler chickens. Appl. Environ. Microbiol. 68713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sriyosachati, S., and C. D. Cox. 1986. Siderophore-mediated iron acquisition from transferrin by Pseudomonas aeruginosa. Infect. Immun. 52885-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stintzi, A., D. Marlow, K. Palyada, H. Naikare, R. Panciera, L. Whitworth, and C. Clarke. 2005. Use of genome-wide expression profiling and mutagenesis to study the intestinal lifestyle of Campylobacter jejuni. Infect. Immun. 731797-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tom-Yew, S. A. L., D. T. Cui, E. G. Bekker, and M. E. P. Murphy. 2005. Anion-independent iron coordination by the Campylobacter jejuni ferric binding protein. J. Biol. Chem. 2809283-9290. [DOI] [PubMed] [Google Scholar]
- 55.van Vliet, A. H. M., and J. M. Ketley. 2001. Pathogenesis of enteric Campylobacter infection. J. Appl. Microbiol. 9045S-56S. [DOI] [PubMed] [Google Scholar]
- 56.van Vliet, A. H. M., J. M. Ketley, S. F. Park, and C. W. Penn. 2002. The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26173-186. [DOI] [PubMed] [Google Scholar]
- 57.van Vliet, A. H. M., J. D. Rock, L. N. Madeleine, and J. M. Ketley. 2000. The iron-responsive regulator Fur of Campylobacter jejuni is expressed from two separate promoters. FEMS Microbiol. Lett. 188115-118. [DOI] [PubMed] [Google Scholar]
- 58.van Vliet, A. H. M., A. C. Wood, J. Henderson, K. Wooldridge, and J. M. Ketley. 1998. Genetic manipulation of enteric Campylobacter species, p.407-419. In P. H. Williams, Ketley, J. M. and Salmond, G. (ed.), Bacterial pathogenesis. Methods in microbiology, vol 27. Academic Press, London, United Kingdom. [Google Scholar]
- 59.van Vliet, A. H. M., K. G. Wooldridge, and J. M. Ketley. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J. Bacteriol. 1805291-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wösten, M. M. S. M., M. Boeve, M. G. A. Koot, A. C. van Nuenen, and B. A. M. van der Zeijst. 1998. Identification of Campylobacter jejuni promoter sequences. J. Bacteriol. 180594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wren, B. W., J. Henderson, and J. M. Ketley. 1994. A PCR-based strategy for the rapid construction of defined bacterial deletion mutants. BioTechniques 16994-996. [PubMed] [Google Scholar]