Abstract
Integrated retroviral DNA is subject to epigenetic gene silencing, resulting in loss of expression of viral genes as well as reporter or therapeutic genes transduced by retroviral vectors. Possible mediators of such silencing include the histone deacetylase (HDAC) family of cellular proteins. We previously isolated HeLa cell populations that harbored silent avian sarcoma virus-based green fluorescent protein (GFP) vectors that could be reactivated by treatment with HDAC inhibitors. Here, we developed a small interfering RNA (siRNA)-based approach to identify specific host factors that participate in the maintenance of silencing. Knockdown of HDAC1, the transcriptional repressor Daxx (a binding partner of HDAC1), or heterochromatin protein 1 gamma resulted in robust and specific GFP reporter gene reactivation. Analyses of cell clones and diverse GFP vector constructs revealed that the roles of HDAC1 and Daxx in retroviral silencing are largely independent of the integration site or the promoter controlling the silent GFP reporter gene. Previous findings from our laboratory and those of others have suggested that Daxx and HDAC proteins may act broadly as part of an antiviral response to repress viral gene transcription. Expression of presumptive viral “countermeasure” proteins that are known to inhibit Daxx or HDACs (pp71, IE2, and Gam1) resulted in the reactivation of GFP reporter gene expression. This study has identified individual host factors that maintain retroviral silencing and supports the proposal that these factors participate in an antiviral response. Furthermore, our results indicate that siRNAs can be used as specific reagents to interrupt the maintenance of epigenetic silencing.
Epigenetic silencing of integrated retroviral DNA results in a transcriptionally dormant viral state that can be heritable through many rounds of cell division. This process may represent an active cellular response to invading foreign DNA (41, 59). However, epigenetic silencing may also provide an advantage to retroviruses by allowing some infected cells to escape detection by the host, as they show no evidence of viral gene expression (22, 33, 41, 47, 54). Silencing is observed for many retroviral systems and, most notably, can contribute to postintegration latency during human immunodeficiency virus (HIV) infection.
Epigenetic silencing is commonly observed after the transduction of therapeutic or reporter genes using retrovirus-based vectors, and this event can occur at various frequencies (16, 17, 51, 52, 57). Several parameters may influence the incidence or severity of retroviral silencing, including the differentiation state of the host cell (16, 17, 52), the location of the retroviral integration site in the host cell chromosomes (32), the composition of the retroviral reporter gene (12), host factor expression (20, 40), and repressive viral sequences (16, 17, 52, 56). Biological properties of the native retroviruses from which the retrovirus-based vectors are derived, as well as vector design, may also affect the frequency of silencing.
The epigenetic processes that lead to retroviral silencing overlap extensively with those that regulate gene expression during embryonic development and cell differentiation (26). In fact, it has been proposed that an epigenetic silencing system initially evolved as a protective mechanism to silence transposable elements (59). Epigenetic regulation is mediated mainly by two interrelated chromatin-marking systems, DNA methylation and histone modification (26). Combinations of histone modifications provide a “histone code” that imparts heritable instructions for either sustained gene expression or gene silencing (2, 3, 21, 30). These posttranslational modifications include acetylation, methylation, phosphorylation, sumoylation, and ubiquitination, primarily targeting the N-terminal histone tails. The families of chromatin-modifying enzymes (and DNA methyltransferases) are the ultimate mediators of epigenetic processes, and it is of significant interest to determine how these enzymatic activities are directed to specific regions of chromatin. Once this marking occurs, the histone modifications are “read,” leading to the recruitment of complexes that either promote or repress gene expression; the modifications also may affect chromatin structure and thereby regulate accessibility by the transcriptional apparatus.
The enzymes that add histone modifications have been designated “writers,” while those that remove modifications are termed “erasers” (48). The best characterized writer-eraser pairs are members of the histone acetyltransferase (HAT) and histone deacetylase (HDAC) families. HATs catalyze histone tail acetylation, a posttranslational modification that is characteristic of transcriptionally active regions of chromatin, while HDACs remove these modifications and are generally viewed as transcriptional repressors. HATs and HDACs are typically found within large protein coactivator and corepressor complexes, respectively. Another well-characterized histone code modification is histone 3 lysine 9 methylation (H3K9me). This modification is written and erased by enzymes of the histone methyltransferase and histone demethylase families, respectively (50). Closed heterochromatin is generally characterized by histone hypoacetylation and the H3K9me modification. The H3K9 mark can recruit heterochromatin protein 1 (HP1), which correlates with closed chromatin. However, the H3K9 and HP1 marks are not exclusive to transcriptionally silent chromatin but can serve as a platform for diverse processes (21).
Epigenetic silencing encompasses several steps: initiation, maintenance, and reactivation. In the initiation step, the chromatin region must be targeted for modification, while for the maintenance step the modification must be sustained, which includes the transfer of these markings to newly assembled chromatin during S-phase. If the maintenance step is interrupted, reactivation can occur. For example, the activity of the HDAC family members (classes 1 and 2) can be blocked by chemical compounds, the HDAC inhibitors (HDIs) (6, 13). Treatment of cells with HDIs results in widespread histone acetylation and the activation of a small subset of genes, presumably corresponding to those for which silencing is maintained by HDAC complexes; however, HDI treatment can also result in the repression of specific genes (6). Most HDIs presently in use show little specificity with respect to class 1 and class 2 HDAC family members (6, 13). Nevertheless, some HDIs have shown promise in cancer therapy and may act partly through the reactivation of silent tumor suppressor genes (6, 36, 37). HDIs are also capable of reactivating silent HIV type 1 DNA and may be useful as part of a strategy to eliminate cells that harbor latent proviruses (31, 58).
HeLa cells can support the early steps of infection of a variety of retrovirus-based vectors derived from heterologous host species. We have used pseudotyped avian sarcoma virus (ASV)-based vectors that encode a green fluorescent protein (GFP) reporter gene to monitor the early steps of infection in human HeLa cells. As the cell-to-cell spread of this vector is restricted in human cells (1), GFP expression is limited to the initially infected cell. Using this system, we have found that integrated GFP retroviral reporter genes, under the control of several different promoters, are subject to high-frequency epigenetic repression and silencing (28). Furthermore, the repressed and silent GFP reporter genes could be activated with trichostatin A (TSA), an HDI. As TSA produced only a transient increase in GFP expression in some cells, a multistep cell-sorting strategy allowed us to enrich for GFP-negative [GFP(−)] cells that harbored TSA-inducible silent retroviral vectors (28). These cells maintain their GFP− phenotype in long-term culture, but when they are challenged with HDIs, robust GFP reactivation is observed. These findings were consistent with an earlier study in which we demonstrated, using chromatin immunoprecipitation (ChIP), that the class 1 HDACs, HDAC1 and HDAC2, as well as Daxx, a transcriptional repressor known to associate with HDACs (25), were present in a complex with the ASV DNA early after infection (20). The Daxx/HDAC complex appears to be recruited to viral DNA through interaction with the ASV integrase protein. We proposed that the Daxx/HDAC complexes have a role in initiating epigenetic silencing in this system, as part of an antiviral response (20, 28).
Our previous studies provided a strong rationale for screening for specific host proteins that maintain retroviral silencing in this model system. Our approach has been to use small interfering RNAs (siRNAs) to knock down candidate proteins and measure GFP reactivation. The results show that knockdown of HDAC1, Daxx, or HP1γ is sufficient for robust retroviral reporter gene reactivation; furthermore, expression of putative viral countermeasure proteins that inhibit HDACs and Daxx also resulted in reactivation. These studies identify roles for specific cellular proteins in the maintenance of retroviral epigenetic silencing and support a model whereby the cellular Daxx and HDAC proteins mediate both initiation and maintenance of retroviral silencing as part of an antiviral response. The results also validate the use of an siRNA-based approach to identify specific components of corepressor complexes and indicate that siRNAs can be used as reagents to interrupt the maintenance of epigenetic silencing.
MATERIALS AND METHODS
Cells.
HeLa cell populations containing silent GFP genes were described previously (28). TI-C cell clones were isolated by cell sorting as described previously (28).
Analysis of GFP expression.
GFP expression was quantitated by FACScan as described previously (20, 28).
Western blot analysis.
Western blotting was performed by standard methods as described previously (20). Anti-HP1α (MAB3446), anti-HP1β (MAB3448), anti-HP1γ (MAB3450), and anti-GAPDH (AP124P) were purchased from Chemicon, Temecula, CA. Anti-NF-κB p50 (ab7971), anti-Dicer (ab13502), anti-HDAC1 (ab19845), and anti-HDAC2 (ab16032) were purchased from Abcam, Cambridge, MA. Anti-Daxx (D7810) was purchased from Sigma-Aldrich, St. Louis, MO. Goat anti-rabbit (31462; Pierce, Rockford, IL) and goat anti-mouse (AP124P; Chemicon) peroxidase-conjugated second antibodies and enhanced chemiluminescence reagents (Pierce) were used according to the manufacturers’ instructions.
siRNAs.
All siRNA SMARTpools and single siDuplexes were purchased from Dharmacon (Lafayette, CO). DharmaFECT 1 transfection reagent (T-2001-02) was used according to the manufacturer's protocols. The following siRNAs were used: siCONTROL GAPDH duplex (D-001140-01), siCONTROL RISC-free siRNA #1 (D-001220-01), and siCONTROL nontargeting siRNA #1 (D-001210-01). The following siRNA SMARTpools were used: HP1α/CBX1 (M-009716-00), HP1β/CBX3 (M-010033-00), HP1γ/CBX5 (M-004296-01), Dicer1 (M-003483-00), HDAC1 (M-003493-02), HDAC2 (M-094936-00), HDAC3 (M-003496-00), HDAC4 (M-003497-02), and Daxx (M-004420-00).
The following single siRNAs were used: NFκB1 (D-003520-01/02/03/05), Daxx (D-004420-01/02/03/04), HDAC1 (D-003493-01/02/04/09), H3.3A (D-011684-04), H3.3B (D-012051), and HIRA (D-013610-02/04).
Quantitative RT-PCR (qRT-PCR).
RNA was quantified using the Agilent 2100 BioAnalyzer in combination with an RNA 6000 Nano LabChip. RNA was reverse transcribed using the Moloney murine leukemia virus reverse transcriptase (RT) (Ambion, Austin TX) and a mixture of anchored oligo(dT) and random decamers. Aliquots of cDNAs were used for PCR. Real-time PCR assays were run using an ABI 7900 HT instrument. The primers and probes were designed using Primer Express version 1.5 software from Applied Biosystems and synthesized by the Fox Chase Cancer Center Fannie Rippel Biotechnology Facility. The probes were 5′-6FAM and 3′-BHQ1 labeled. PCR master mix from Eurogentec was used for PCR. Cycling conditions were 95°C for 15 min followed by 40 (two-step) cycles (95°C for 15 s and 60°C for 60 s). PolR2F was used as the reference gene. The 2−ΔΔCT method (CT, threshold cycle) was used to calculate relative changes in expression. For each sample, the values are averages and standard deviations of data from two PCRs performed with two amounts (100 and 20 ng) of total RNA in the RT reaction. The following primers and probe sequences were used: for Daxx, 5′-AGGGCCATTAGGAAACAGCTA (forward), 5′-AGGGTACATATCTTTTTCCCATTCTT (reverse), and 5′-TGGAAAGGCAAAGGTCAGTGCATGA (probe); for HDAC1, 5′-TGAGGACGAAGACGACCCT (forward), 5′-CTCACAGGCAATTCGTTTGTC (reverse), and 5′-CAAGCGCATCTCGATCTGCTCCTC (probe); for HDAC2, 5′-CTTTCCTGGCACAGGAGACTT (forward), 5′-CACATTGGAAAATTGACAGCATAGT (reverse), and 5′-AGGGATATTGGTGCTGGAAAAGGCAA (probe); for HDAC3, 5′-GCTCCTCACTAATGGCCTCTTC (forward), 5′-GGTGGTTATACTGTCCGAAATGTT (reverse), and 5′-AGCAGCGATGTCTCATATGTCCAGCA (probe); and for HDAC4, 5′-TTGAGCGTGAGCAAGATCCT (forward), 5′-GACGCTAGGGTCGCTGTAGAA (reverse), and 5′-CGTGGACTGGGACGTGCACCA (probe).
dnHP1 expression vector.
The construction and use of a retroviral vector encoding a dominant negative form of HP1 (dnHP1) was described previously (HP1βΔN) (60).
Plasmids and transfection.
The immediate early 1 (IE1) and IE2 expression plasmids (44) were provided by Tom Shenk, the Gam1 wild-type (wt) and mutant expression plasmids (9) were provided by Susan Chiocca, and the pp71 wt and mutant expression plasmids were provided by Robert Kalejta. Transfections were carried out using Lipofectamine or Lipofectamine 2000 (Invitrogen, Carlsbad CA) as described by the supplier.
Construction and transfection of an HDAC1 siRNA-resistant expression plasmid.
An HDAC1 expression plasmid was purchased from Origene (Rockville, MD). The QuikChange mutagenesis kit (Stratagene, La Jolla, CA) was used to introduce silent changes in the HDAC1 codons to create an siRNA-resistant site for HDAC1 siRNA 01. The oligodeoxynucleotides used for mutagenesis were 5′-GGATACGGAGATCCCTAATGAGCTCCCCTACAATGACTACTTTG-3′ and 5′-CAAAGTAGTCATTGTAGGGGAGCTCATTAGGGATCTCCGTATCC-3′. The wt and resistant plasmids were used to transfect HeLa TI-C cells in the presence of HDAC1 siRNA 01 by use of the Dharmacon DharmaFECT Duo transfection reagent as described by the supplier.
ChIP.
ChIP reactions from TI-C cells were performed using the Upstate/Millipore EZ ChIP kit. Anti-Daxx antibody was obtained from Santa Cruz (sc-7152), and the immunoglobulin G negative control antibody was provided with the EZ ChIP kit. For PCRs, 2 μl of purified DNA from precipitated chromatin was amplified by PCR with the following primers: CMV-GFP (positions −306 to +20 surrounding the transcriptional start site) (5′-CTT ATG GGA CTT TCC TAC TTG-3′ [forward] and 5′-TCC TCG CCC TTG CTC ACC ATG-3′ [reverse]) and β-actin coding region (positions 68 to 327) (5′-CTC ACC ATG GAT GAT GAT ATC GC-3′ [forward] and 5′-ATT TTC TCC ATG TCG TCC CAG TTG-3′ [reverse]) (29). The PCR products were analyzed on 2% agarose gels. For the quantitation of PCR products, gels were stained with Syto 60 (Invitrogen) and analyzed using the Odyssey imaging system (LI-COR).
RESULTS
Silent retroviruses are reactivated by HDAC1 and Daxx siRNAs.
We previously isolated a subset of HeLa cells that contain silent retroviral GFP reporter genes that can be reactivated by treatment with a variety of HDIs, including TSA (28). We have designated these cells TSA inducible (TI) and we have derived versions harboring silent GFP retroviral reporter genes under the control of the human cytomegalovirus (hCMV) IE (TI-C) or ASV (TI-L) long terminal repeat (LTR) (TI-L) promoters (28). Such cells represented a significant fraction of the infected culture, as the ratio of GFP+ cells to TI-C cells was approximately 2:1 (28).
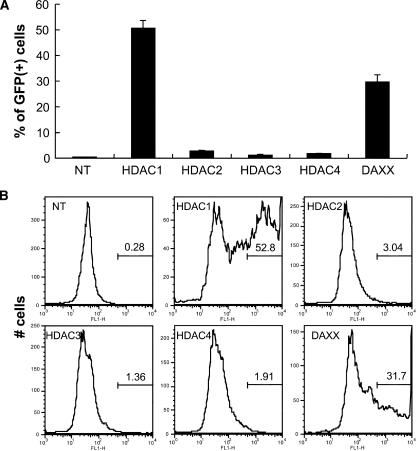
TSA has broad activity against class 1 (widely expressed) and class 2 (primarily tissue-specific) HDACs. We considered HDAC1 and HDAC2 (class 1) to be strong candidates for meditating silencing, as they were detected in complexes with ASV viral DNA in HeLa cells early after infection (20). Expression of the class 2 HDAC4 is typically more restricted to specific tissues; however, we previously detected HDAC4 in HeLa cells (20). Our initial experiments were designed to use siRNAs to identify roles for one or more HDACs in silencing maintenance, as siRNA-mediated knockdown of HDACs might phenocopy HDI activity. TI-C cells were treated with siRNA “SMARTpools” (Dharmacon Corp.) (100 nM) comprising a mixture of four siRNAs that target single mRNAs. Our initial tests included siRNAs specific for HDAC1, HDAC2, HDAC3, and HDAC4. As shown in Fig. 1A, treatment of the TI-C population with an siRNA pool that targets HDAC1 resulted in the appearance of a significant fraction of cells that expressed GFP, as measured by fluorescence-activated cell sorter (FACS) analysis. In contrast, siRNA pools that target HDAC2, HDAC3, and HDAC4 mRNA had no significant effect. As shown in the FACS profiles in Fig. 1B, treatment with the HDAC1 siRNA pool resulted in the appearance of cells with very high GFP fluorescence intensities. The HDAC2 siRNA pool consistently produced a small increase in the number of GFP-expressing cells, but the GFP intensity in this small fraction was just above the background level (Fig. 1B). Our previous studies indicated a role for Daxx in the initiation of retroviral silencing in this system (20). Transfection of the TI-C cell population with the Daxx siRNA pool also resulted in robust reactivation of GFP (Fig. 1A and B), although it was not as pronounced as observed with the HDAC1 siRNA (Fig. 1B).
FIG. 1.
Knockdown of HDAC1 or Daxx results in reactivation of the silent GFP reporter. (A) HeLa TI-C cells (28) were transfected with the indicated siRNA SMARTpools (100 nM) (Dharmacon) and incubated for 96 h. GFP expression was analyzed by FACS. NT, not transfected. (B) Histograms of GFP intensities (x axis) from the experiment shown in panel A. Percentages of GFP-positive cells are indicated by the numerical values. Autoscaling was used to portray the distribution of GFP intensities.
We previously found that the TI-C cells used here oscillate between responsive and nonresponsive states in terms of reactivation by HDIs (28). It is likely that this phenomenon also contributes to the incomplete reactivation observed with HDAC1 and Daxx siRNAs.
Validation and biological relevance of siRNA-mediated reactivation.
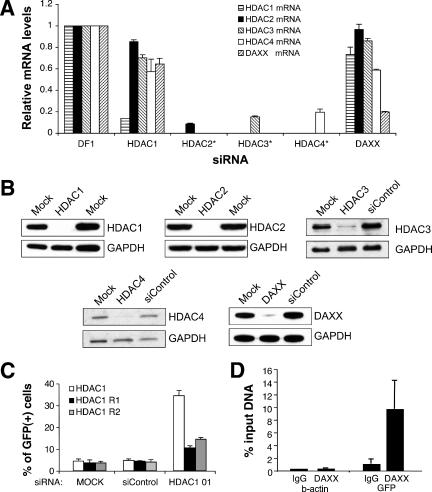
To confirm the specificities of the reactivation shown in Fig. 1, we measured target mRNA levels by qRT-PCR (Fig. 2A). The results confirm that treatment with an siRNA pool that targets HDAC1 mRNA resulted in a substantial reduction in HDAC1 mRNA levels but not in Daxx mRNA levels. siRNAs directed against HDAC2, -3, and -4 mRNAs were effective at reducing the levels of their respective target mRNAs. Daxx siRNA treatment resulted in a significant reduction in Daxx mRNA, with only a negligible effect on the HDAC1 mRNA level. Western blotting confirmed that the siRNA pools produced significant reductions in the amounts of the corresponding proteins in all cases (Fig. 2B). Thus, the lack of reactivation by HDAC2, -3, and -4 siRNA pools could not be attributed to ineffectiveness of these siRNAs.
FIG. 2.
Determination of specificity of siRNA knockdown. (A) qRT-PCR analysis of target mRNAs. For each target mRNA measurement, the values were normalized to a control which was treated with transfection reagent only (DharmaFECT 1 [DF1]). For HDAC1 and Daxx siRNA treatments, the levels of HDAC1, HDAC2, HDAC3, HDAC4, and Daxx mRNAs were measured. For HDAC2, HDAC3, and HDAC4 siRNAs, only the cognate target mRNA levels were measured (*). (B) Assessment of knockdown by Western blotting. TI-C cells were treated with 100 nM of siRNAs indicated above the panel and cells were processed for Western blotting after 72 h. GAPDH antibody was used to monitor recovery. Mock siRNA treatments were performed in duplicate. (C) Transfection of a plasmid encoding an siRNA-resistant form of HDAC1 mRNA. Silent mutations that destroy the HDAC1 siRNA 01 annealing site were introduced into an HDAC1 expression plasmid, as described in Materials and Methods. Mutant plasmids prepared in duplicate (R1, R2) or a wt control plasmid was introduced into TI-C cells along with the HDAC1 siRNA 01. GFP expression was monitored by FACS. As shown, HDAC 01 siRNA was capable of stimulating GFP reactivation after transfection of the wt HDAC1 plasmid. In contrast, the siRNA-resistant plasmids were able to repress GFP expression in the presence of the siRNA. (D) Localization of Daxx at the GFP promoter. ChIP analysis was carried out as described in Materials and Methods. Two primer sets were used, targeting the silent viral GFP promoter region or the active cellular β-actin gene. Experiments shown are representative, and the Daxx results are averages of triplicate immunoprecipitations. IgG, immunoglobulin G.
We also considered the possibility that “off-target” effects of siRNA pools might lead to GFP reactivation through the knockdown of an unintended mRNA target. As off-target effects are frequently sequence based (4, 11), analysis of several single siRNAs was carried out. In these experiments, the TI-C population was treated with each of the four individual siRNAs (25 nM concentration) from the HDAC1 and Daxx pools. Three individual siRNAs from the HDAC1 pool (01, 02, and 09) produced both efficient knockdown of HDAC1 and GFP reactivation (data not shown). As a final confirmation of specificity, an expression plasmid encoding an siRNA-resistant form of HDAC1 siRNA 01 was constructed, and we demonstrated that the introduction of this plasmid could repress the GFP reporter in the presence of HDAC1 siRNA 01 (Fig. 2C). All four of the individual Daxx siRNAs produced a level of reactivation that was similar to what was seen for the pool, also eliminating the possibility of off-target effects (data not shown).
To assess a direct role for Daxx in the maintenance of silencing at the retroviral loci in TI-C cells, we used ChIP. As shown in Fig. 2D, Daxx was detected in the hCMV IE promoter/enhanced GFP transcriptional start site region of the silent viral loci but was not detected at the active β-actin cellular locus. Taken together, the siRNA and ChIP results indicate that Daxx plays a direct role in the long-term maintenance of silencing.
Specific candidate and control siRNAs do not reactivate silent GFP retroviral reporter genes.
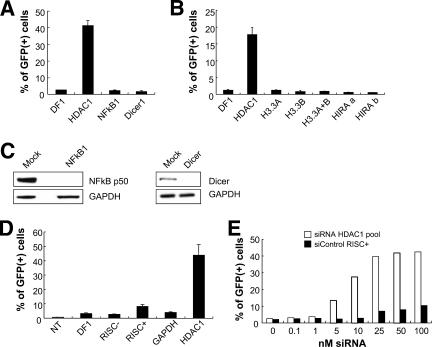
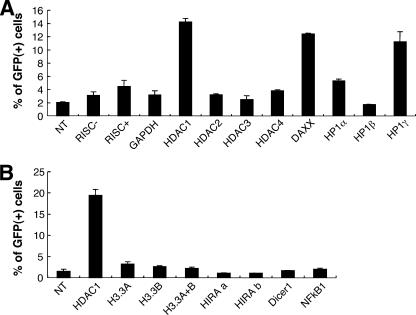
We tested the effects of other siRNA pools which might have the potential to produce broad effects on HDAC1-mediated silencing (NF-κB) (55), chromatin structure (histone chaperone HIRA and histone H3.3), and microRNA processing (Dicer). For example, human Dicer1 regulates many genes via its role in microRNA processing (14) and also by production of siRNAs that can direct heterochromatin formation (18, 27). We found that the knockdown of NF-κB or Dicer did not result in retroviral reporter gene reactivation (Fig. 3A and C). The introduction of siRNA pools directed against the histone variants H3.3A and H3.3B or the histone chaperone protein HIRA also failed to reactivate the silent GFP reporter under these conditions (Fig. 3B).
FIG. 3.
Knockdown of several candidate proteins or treatment with various control siRNAs fails to reactivate the silent GFP reporter. (A and B) HeLa TI-C cells were treated with the indicated siRNAs and the percentages of GFP-positive cells were determined by FACS at 96 h posttransfection. Single siRNAs were used for H3.3A, H3.3B, and HIRA. Two independent single siRNAs (designated a and b) were tested for HIRA. DF1, DharmaFECT 1 transfection reagent. (C) TI-C cells were treated with 100 nM siRNAs as indicated above the panels and cells were processed for Western blotting after 72 h. (D) Treatment and analysis with the indicated siRNAs was as for panel A. Negative control siRNAs RISC−, RISC+, and GAPDH were analyzed. (E) The HDAC1 siRNA SMARTpool was titrated to determine the lowest effective concentration versus the negative control siRNA RISC+. Analysis was as for panel A.
We also tested specifically designed negative control siRNAs (Dharmacon Corp.) that do not target cellular mRNA sequences (nontargeting) but are either able or unable to assemble into the RISC complex that mediates target mRNA degradation. The nontargeting siRNA can reveal the potential for off-target effects that are mediated through the RISC complex (designated the RISC+ control). The “RISC-free” siRNA is modified to prevent assembly into the RISC complex and serves as a control for the treatment of cells with the transfection reagent in conjunction with a functionally inert RNA payload (designated RISC− control). At 100 nM concentrations, the RISC+ control siRNA showed some low level of reactivation compared to the RISC− or GAPDH siRNA controls (Fig. 3D). As RISC-dependent off-target effects are typically more prominent at high concentrations of individual siRNAs (e.g., 100 nM) (11), this observation gave us an opportunity to more carefully address the effects of siRNA concentration with respect to specific versus off-target effects on GFP reactivation. Our standard conditions utilize siRNA pools at a 100 nM final concentration, with each of four siRNAs being present at 25 nM. Parallel titrations of the RISC+ control and the HDAC1 siRNA pool revealed that significant reactivation by the HDAC1 siRNA pool could be observed at concentrations (≥10 nM) where the RISC+ had no effect above what was seen for the transfection reagent alone (Fig. 3E). This titration also revealed that the HDAC1 siRNA pool could reactivate GFP at a low concentration (25 nM), characteristic of specific siRNA targeting. From the experiments shown in Fig. 3, we conclude that several nonspecific controls, or siRNAs that are predicted to have broad effects on gene expression, do not promote reactivation; furthermore, we could not identify a role for human Dicer1 in the maintenance of silencing in this system.
In summary, the results of the experiments shown in Fig. 1 through 3 validate our experimental design, namely, to use siRNAs to interrogate GFP-silent cells to identify factors mediating silencing. We conclude from these experiments that HDAC1 and Daxx have specific roles in the maintenance of retroviral silencing in this system.
Evidence for position-independent roles for HDAC1 and Daxx in retroviral reporter gene silencing.
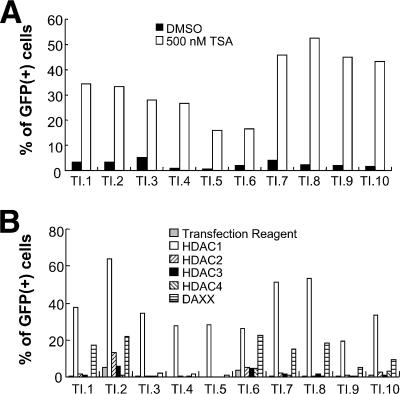
To determine if the integration sites of the retroviral DNA are determinants for the participation of specific host factors that control epigenetic silencing, we examined a series of TI-C cell clones. These clones were derived from a pool of cells in which the average GFP copy number was ca. 1 as measured by quantitative real-time PCR (28). As shown in Fig. 4A, treatment of each of these cell clones with TSA induced GFP reactivation, but to various degrees. Challenge with the HDAC1 siRNA pool also resulted in reactivation in all clones, and the Daxx siRNA pool produced a measurable reactivation in 7 of the 10 clones tested (Fig. 4B, clones 1, 2, 6, 7, 8, 9, and 10). As with the TI-C cell population, transfection with the HDAC1 siRNA pool phenocopied the TSA treatment (compare Fig. 4A and B). Based on analyses of these clones, we conclude that HDAC1 and, in most cases, Daxx are required for the maintenance of silencing at independent loci, but the integration site may modulate the extent of reactivation.
FIG. 4.
Analysis of TI-C silent cell clones. (A) Clones were treated with TSA or a dimethyl sulfoxide (DMSO) control, and GFP was monitored by FACS after 24 h. (B) Cell clones were transfected with the indicated siRNAs, and GFP reactivation was monitored by FACS after 96 h. Representative results are shown.
Clones 2 and 6 also showed modest levels of reactivation after treatment with HDAC2 and HDAC3 siRNA pools; however, the mean fluorescence intensities of GFP positive cells were significantly lower than that produced by the HDAC1 siRNA pool (not shown). Furthermore, clones 2 and 6 also showed exaggerated responses to the transfection reagent alone, compared to other clones. Clone 2 was also prone to spontaneous or stress-induced GFP reactivation (data not shown). These results also indicate that the integration locus can affect the reactivation properties.
HP1 plays a role in retroviral gene silencing.
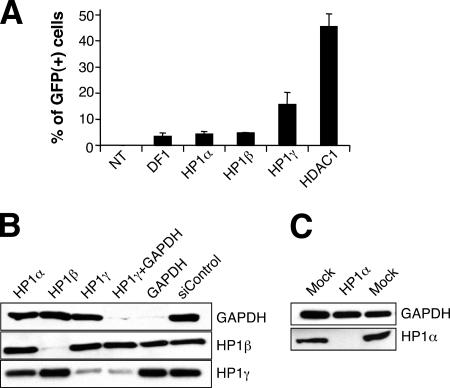
The three isoforms of HP1, designated α, β, and γ, have been implicated in a variety of processes, including the maintenance of epigenetic gene silencing (21). To investigate the possible role of these proteins in retroviral reporter gene silencing, we transfected HeLa TI-C cells with HP1α, β, and γ siRNA pools. As shown in Fig. 5A, the HP1γ siRNA pool induced significant GFP reactivation, whereas the HP1α and HP1β siRNA pools had no effect compared to what was seen for transfection agent alone. Although transfection of the HP1γ siRNA pool resulted in a smaller percentage of GFP-expressing cells than transfection of the HDAC1 siRNA pool (Fig. 5A), the GFP intensity in the activated cells was high (data not shown).
FIG. 5.
Analysis of the role of HP1 isoforms in silencing maintenance. (A) HeLa TI-C cells were transfected with the indicated HP1 isoform siRNA SMARTpools, and GFP reactivation was monitored by FACS analysis after 96 h. Abbreviations: NT, not transfected; DF1, DharmaFECT 1 transfection reagent. (B and C) Western blot analyses of siRNA knockdown of the HP1 family of proteins. Cells were transfected with the siRNAs indicated above the panels. The detection of HP1α required loading of 10-fold more protein.
The HP1β and HP1γ isoforms were found to be highly abundant in HeLa cells, and we were able to confirm siRNA-specific knockdown of both proteins (Fig. 5B). The amount of HP1α was very low in untransfected HeLa cells, but knockdown was confirmed using more-sensitive Western blotting conditions (Fig. 5C).
To address the possibility that residual amounts of HP1γ might account for the only limited reactivation produced by the HP1γ siRNA pool, we cotransfected GAPDH and HP1γ siRNA pools in order to internally monitor the transfection efficiency as measured by GAPDH knockdown. As expected, transfection of the GAPDH siRNA pool resulted in knockdown of GAPDH and had no effect on HP1β and HP1γ levels (Fig. 5B). In GAPDH-HP1γ-cotransfected cells, we could detect residual levels of HP1γ under conditions in which GAPDH was nearly undetectable. We conclude that either the HP1γ siRNA is pool less potent or a long-lived form of HP1γ protein persists. Thus, the limited reactivation in response to HP1γ siRNAs could be due to the presence of residual HP1γ protein.
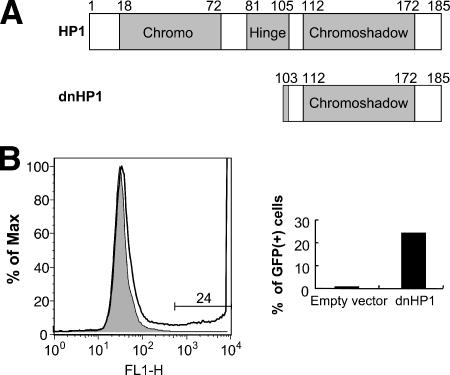
To independently assess a role for HP1 isoforms in silencing, we used a dominant negative form (dnHP1) (60). The dnHP1 was constructed by deleting the chromodomain from HP1β, leaving only the chromoshadow multimerization domain (Fig. 6A). The dnHP1 form was introduced into TI-C cells with a retroviral vector, followed by the selection of transduced cells by use of puromycin. A dramatic reactivation of the GFP gene was observed in a large subset of these cells (6B, right), and the GFP intensity was very high (Fig. 6B, left). Parallel puromycin selection of cells transduced with an empty vector failed to induce GFP expression (Fig. 6B, left). Based on the siRNA results (Fig. 5A), it is likely that the relevant target of inhibition by dnHP1 is HP1γ. We therefore conclude that HP1γ contributes to the maintenance of retroviral reporter gene silencing in this system.
FIG. 6.
Expression of a dnHP1 reactivates silent GFP. (A) Map of dnHP1. Chromo, chromodomain. (B) A retroviral vector encoding a dnHP1 (60) was used to infect HeLa TI-C cells. Cells were placed under selection with puromycin and were monitored for reactivation of GFP expression by FACS. The FACS profile obtained at 5 days postinfection shows GFP expression in cells selected with the dnHP1 expression vector (no fill) versus the empty vector (filled). The expression of dnHP1 produced a population of GFP-positive cells (24%), some of which were very bright and appear off scale in the graph. A representative experiment is shown.
Virus-encoded inhibitors of HDACs and Daxx can reactivate the silent GFP retroviral reporter gene.
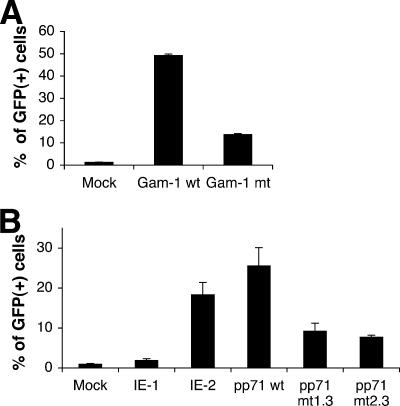
Several viral proteins are known to bind to and inhibit HDACs (9, 44). These proteins may act as countermeasures to protect viral genomes from repression by HDACs, consistent with a role for HDACs in an antiviral response. The avian adenovirus protein Gam1 has been demonstrated to inhibit human HDAC1 (9), while the hCMV proteins IE1 and IE2 inhibit HDAC3 (44). As shown in Fig. 7A, transfection of an expression plasmid that encodes the Gam1 protein resulted in a dramatic reactivation of the silent GFP retroviral reporter in the TI-C cell population. A mutant plasmid that encodes a protein with diminished capacity to inhibit HDAC1 showed less of an effect. Expression of the wt and mutant Gam-1 proteins was confirmed by Western blotting (data not shown). We also expressed Gam1 in TI-C cell clone 3, in which GFP was reactivated only by HDAC1 siRNA (Fig. 4B). Again, expression of Gam1, but not of the Gam1 mutant, resulted in strong retroviral reporter gene reactivation with this clone (data not shown). As Gam1 expression phenocopies the effect of HDAC1 siRNA, this experiment provides independent confirmation that inhibition of HDAC1 is sufficient for reactivation.
FIG. 7.
Reactivation of silent GFP by viral proteins. (A and B) HeLa TI-C cells were transfected with expression plasmids encoding the indicated proteins and GFP reactivation was monitored after 48 h by FACS analysis.
Expression of hCMV IE2 but not IE1 resulted in strong retroviral reporter gene reactivation in clone 3 (Fig. 7B). The expression of both proteins was confirmed by Western blotting (data not shown). Both IE1 and IE2 are reported to form complexes with HDAC3 and inhibit its activity (44), although broader HDAC inhibitory specificities of these proteins have not been explored. Again, as the siRNA experiments identified a role for HDAC1 but not HDAC3 in silencing maintenance in our system, these findings indicate that IE2 may inhibit HDAC1 as well as HDAC3. Further studies will be required to obtain support for this interpretation. Although Gam1, IE1, and IE2 may have diverse and complex functions beyond HDAC inhibition (for an example, see reference 5), our system has apparently provided a means to detect their HDAC-inhibitory activity.
Several studies have identified a role for Daxx in the repression of hCMV gene expression (7, 33, 46). Furthermore, hCMV encodes a protein, pp71, that inhibits Daxx by targeting it for proteasome-mediated degradation (49). The fact that hCMV encodes a Daxx “countermeasure” supports a model whereby Daxx mediates an antiviral response that is overcome by pp71 (20, 49). Our original findings suggested a role for Daxx in the initiation of viral transcriptional repression (20). The ability of Daxx siRNAs to reactivate silent retroviral DNA in long-term-passage cells implicates a role for Daxx in the maintenance of silencing. As a further test of this interpretation, we transfected TI-C cells with a plasmid encoding hCMV pp71. As shown in Fig. 7B, the transfection of plasmids encoding wt pp71, but not of two mutant forms of this protein (24), resulted in a robust reactivation of GFP expression, thus providing independent confirmation of a role for Daxx in the maintenance of silencing of integrated retroviral DNA in this system.
The retroviral reporter gene promoter is not the major determinant of silencing.
In the experiments described above, the silent, TSA-sensitive GFP gene was under the control of the hCMV IE promoter (TI-C cells). To determine if the results that we obtained were dependent on this promoter, we tested another cell population in which the silent GFP reporter gene is driven by the native retroviral LTR promoter (TI-L cells) and for which rechallenge with HDIs also results in GFP reactivation (28). As shown in Fig. 8, when this cell population was treated with the collection of siRNA pools described above, only HDAC1, Daxx, and HP1γ siRNAs produced reactivation significantly above background levels. The overall detection of the GFP response is reduced compared to what was seen for the TI-C cells, owing to the weaker LTR promoter (28). We obtained similar results with a third cell population, in which the silent reporter was under the control of the cellular EF1-α promoter (28) (data not shown). From these experiments, we conclude that the reporter gene promoter is not the major determinant in eliciting the activities of a signature constellation of factors that participate in silencing maintenance.
FIG. 8.
Analysis of HeLa cells harboring silent GFP under the control of the ASV LTR. (A and B) TI-L cells were transfected with the indicated siRNA SMARTpools and GFP expression was measured after 96 h by FACS analysis. NT, not transfected; two independent single siRNAs tested for HIRA are designated HIRA a and b.
DISCUSSION
Our present study has focused on the functional identification of host factors that maintain retroviral silencing and further testing of the hypothesis that silencing is a component of an antiviral response. We reasoned that siRNA-mediated knockdown of individual host proteins that maintain retroviral silencing would result in reactivation and that this approach could be used to identify specific silencing factors. siRNA-based methods are quite powerful but are prone to a variety of artifacts, including off-target effects (4, 11). Furthermore, siRNA transfection or target knockdown could produce secondary effects, such as a cellular stress response. As retroviruses and transposable elements can be reactivated by various forms of cellular stress, it seemed possible that “off-target” or nonspecific responses to siRNA transfection might produce a high background in the assay. As such, numerous controls were used to unequivocally demonstrate specificity. Our initial attempt to apply and validate the use of the siRNA methodology began with the prediction that the knockdown of one or more HDACs would phenocopy the effects of HDIs. We show that challenge of a GFP-silent cell population (or clones derived thereof) with siRNAs targeted to HDAC1 resulted in significant and specific reactivation of the GFP retroviral reporter gene. We describe rigorous control experiments, including the use of an siRNA-resistant HDAC1 mutant. As the HDAC1 siRNA and HDIs produce the same phenotype, we conclude that HDAC1 is the relevant target for HDIs in this system. Remarkably, there seems to be no redundancy in HDAC-mediated silencing in this system. This finding indicates that the silencing of viral vectors could be reversed by highly specific siRNA reagents.
We previously identified a role for the transcriptional repressor Daxx in the initiation of retroviral silencing and hypothesized that this protein participated in an antiviral response (20). We also found that the association of HDACs with retroviral DNA was dependent on Daxx, consistent with an overall model whereby the repressive activities of Daxx are mediated by HDAC binding partners (25). Subsequent studies by others have shown that Daxx also represses hCMV early gene expression (7, 33, 46, 49). An antiviral role for Daxx has now been supported by the finding that the hCMV pp71 protein acts as a viral countermeasure that targets Daxx for proteasomal degradation (49). The avian adenovirus Gam1 and hCMV IE1/IE2 proteins inhibit HDACs and may also act as viral countermeasures to overcome HDAC-mediated repression (9, 44). Here, we have shown that expression of pp71, Gam1, or IE2 results in GFP reactivation, consistent with their proposed roles as Daxx and HDAC inhibitors. We note that Daxx siRNA or Daxx inhibitor pp71 expression resulted in the reactivation of silent GFP in long-term-passage cells. This finding revealed a role for Daxx in the maintenance of silencing, in addition to the earlier identified role in the initiation of silencing. However, in several cell clones, the knockdown of Daxx did not result in detectable reactivation (Fig. 4B). It is possible that the relevant HDAC1 activity in these clones is associated with a different binding partner or complex.
Based on our previous studies (20) and those of others, it seems likely that the roles of HDAC1 and Daxx in retroviral silencing are direct, i.e., that these factors interact with integrated viral chromatin. Here, we provide evidence that Daxx is physically associated with the viral locus during the long-term maintenance of silencing (Fig. 2D). With respect to HDAC1, we detected rapid histone H4 hyperacetylation and the concomitant induction of GFP mRNA within several hours after TSA treatment of TI-C cells (data not shown). Therefore, the effects of TSA appear to be direct, and the relevant target of inhibition by TSA is likely HDAC1 resident at the silent viral locus. There is additional evidence to support a direct role for HDAC activity in silencing maintenance. It is now clear that HDACs play a role in the repression of a variety of retroviruses (10, 23, 28, 38, 47, 54, 55, 58), large DNA viruses (9, 42, 44), and viral transgenes (8), as measured by reversal of silencing by HDI treatment, and such treatment can produce a transition from a hypo- to a hyperacetylated histone state in the vicinity of the viral promoter (44). Although the simplest model is that HDACs maintain silencing at the viral loci via deacetylation of histone tails, the acetylation-deacetylation cycle is a common regulatory mechanism of cellular processes, and HDACs have broad substrate activity on nonhistone substrates, including transcription factors (19). However, the fact that HDAC1 is required to maintain the silencing of two different promoters (hCMV and the ASV LTR) suggests that the critical substrate(s) is a general factor (e.g., histones) rather than a specific factor (e.g., transcription factors). Irrespective of the precise mechanism of action, we have shown that siRNAs provide a highly specific means of reversing epigenetic silencing.
Our results provide some insight into general models for the initiation and maintenance of retroviral silencing. Examination of cell clones harboring silent retroviruses revealed that, in each case, silencing was controlled by HDAC1, indicating that this factor can mediate silencing at numerous integration sites (Fig. 4D). Our previous studies showed that ca. 40% of ASV integration events occur in protein-encoding genes (43), yet HDAC-mediated silencing and repression occurred at high frequency (28). It is unlikely, therefore, that the HDAC-mediated effects are limited to rare integration events into preexisting hypoacetylated heterochromatin. Relevant studies using HDIs have been interpreted to mean that HATs and HDACs are unable to access heterochromatin in the absence of DNA replication (reviewed in reference 53). In independent studies, we have observed that TSA-mediated reactivation of silent retroviral reporter genes is DNA replication independent (28; also data not shown). This behavior therefore provides further support for the idea that the location of these silent proviruses is not restricted to preexisting heterochromatin sites. Lastly, we do not believe that levels of repressive factors (e.g., HDAC1) in individual cells determine the frequency of silencing in our system, as a matched HIV-based GFP vector was uniformly resistant to silencing (28). Taken together, our results support a model whereby repressive factors are able to nucleate on viral chromatin independently of the integration site (Fig. 4B) (28, 56), although their repressive activities are not fully penetrant and may be modulated by the integration site (28).
The studies described here focus on, but are not limited to, retroviral constructs in which the silent GFP gene is under the control of the strong hCMV IE promoter. This promoter is capable of driving high-intensity GFP expression and thus provides an excellent on-off dynamic range (Fig. 1B). These characteristics initially led us to identify cells harboring silent viral DNA (28). We also reasoned that repression of this strong promoter likely signified roles for potent silencing factors. After identifying these factors (HDAC1, etc.) by use of the hCMV IE promoter system, we went on to ask if they were promoter specific by testing the silent GFP genes under the control of the native viral LTR (Fig. 8) and the EF1-α cellular promoter (28; also data not shown). We tentatively conclude that the promoters are not a major determinant in shaping the constellation of repressive factors that we have identified. It is possible that viral sequences, including the native retroviral LTR, which is retained in all of viral constructs, provide a common determinant.
The HP1 family members typically concentrate in silent pericentric heterochromatin via binding to a specific histone modification associated with repressive chromatin, H3K9 methylation. This modification is written and erased by activities of histone methyltransferases and demethylases, respectively (50). The recognition of H3K9 methylation by HP1 is mediated by the HP1 chromodomain, while the multimerization of HP1, which is believed to drive chromatin compaction, is mediated by the HP1 chromoshadow domain (Fig. 6A). These properties of HP1 provide a strong paradigm for how histone code modifications may drive heterochromatin formation and silencing. HP1α and HP1β localize to heterochromatin, while HP1γ is found in both heterochromatin and euchromatin (39). We found that knockdown of the HP1γ but not the HP1α or HP1β isoform was sufficient for reactivation. Assuming that reactivation is the direct result of HP1γ depletion, our results suggest that HP1γ contributes to retroviral gene silencing at numerous loci, possibly within euchromatin. Consistent with this interpretation, HP1 proteins are known to mediate the silencing of specific euchromatic genes as part of sequence-specific repressor complexes (45).
In addition to roles for HP1 and H3K9 methylation in heterochromatin formation/maintenance, several studies have identified an association of heterochromatic markers with a subset of transcribed genes. These findings indicate that heterochromatin can also serve as a platform for various activities, including transcription (21). More-recent studies have indicated that HP1γ itself is regulated by a modification subcode; specifically, phosphorylation of HP1γ Ser83 is associated with actively transcribed genes (34). Our findings are more consistent with the classical view that HP1γ participates in gene silencing. The fact that the HP1γ isoform mediates silencing in our system is also consistent with our current hypothesis, namely, that the chromosomal location of silent or partially repressed retroviruses is not limited to constitutive heterochromatin (28). More-definitive studies, including chromosomal mapping of integration sites and identification of the forms of HP1 that are physically associated with these integrated viral genomes, are required to test this model. We also considered that our cell-sorting strategy to isolate GFP-silent cells may have introduced a bias in terms of the constellation of factors that maintain silencing (i.e., HDAC1, HP1γ). Although this bias cannot be ruled out, our previous study (28) indicated that HDAC-mediated repression and silencing occurred at a very high frequency in this system. Thus, the selected cells used in this study likely do not represent a rare subset. Furthermore, our findings are consistent with those in other systems that indicate roles for HDACs (10, 20, 23, 28, 38, 47, 54, 55, 58), as well as HP1γ (15, 35, 56), in retroviral silencing. Therefore, we suggest that the function identified for HP1γ in HIV type 1 DNA latency (15, 35) may signify a more generic role.
In summary, we describe compelling evidence for roles for specific host factors in retroviral reporter gene silencing. Furthermore, the factors we have identified are not uniquely dedicated to the silencing of retroviruses and may even represent a more general system to silence foreign DNA. This work has also established that siRNAs provide a powerful tool to dissect the function of host proteins in this process. Reversal of epigenetic silencing by HDIs and other compounds may be a useful component of both cancer (6, 36, 37) and HIV (31) therapies. It will be important to develop more-specific therapeutics to inhibit individual HDACs and other mediators of silencing to avoid broad effects on the epigenetic state of treated cells. Our findings indicate that such approaches may be possible.
Acknowledgments
This work was supported by National Institutes of Health grants CA71515 and CA06927 and by an appropriation from the Commonwealth of Pennsylvania. R.A.K. was funded by grants from the Pennsylvania Department of Health and by National Institutes of Health grant NS053666. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
We acknowledge the use of the Fox Chase Cancer Center Flow Cytometry and Cell Sorting Facility. We are also grateful to Emmanuelle Nicolas from the Fox Chase Cancer Center Fannie E. Rippel Biochemistry and Biotechnology Facility for carrying out qRT-PCR experiments. We also thank Paul Cairns, Jared Evans, and Glenn Rall for critical comments on the manuscript and Tom Shenk, Susan Chiocca, and Rob Kalejta for providing expression plasmids. We are grateful to Marie Estes for assistance in preparing the manuscript.
The contents of this work are solely our responsibility and do not necessarily represent the official views of the National Cancer Institute or any other sponsoring organization.
Footnotes
Published ahead of print on 19 December 2007.
REFERENCES
- 1.Barsov, E. V., and S. H. Hughes. 1996. Gene transfer into mammalian cells by a Rous sarcoma virus-based retroviral vector with the host range of the amphotropic murine leukemia virus. J. Virol. 703922-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, B. E., A. Meissner, and E. S. Lander. 2007. The mammalian epigenome. Cell 128669-681. [DOI] [PubMed] [Google Scholar]
- 4.Birmingham, A., E. M. Anderson, A. Reynolds, D. Ilsley-Tyree, D. Leake, Y. Fedorov, S. Baskerville, E. Maksimova, K. Robinson, J. Karpilow, W. S. Marshall, and A. Khvorova. 2006. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 3199-204. [DOI] [PubMed] [Google Scholar]
- 5.Boggio, R., R. Colombo, R. T. Hay, G. F. Draetta, and S. Chiocca. 2004. A mechanism for inhibiting the SUMO pathway. Mol. Cell 16549-561. [DOI] [PubMed] [Google Scholar]
- 6.Bolden, J. E., M. J. Peart, and R. W. Johnstone. 2006. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 5769-784. [DOI] [PubMed] [Google Scholar]
- 7.Cantrell, S. R., and W. A. Bresnahan. 2006. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 806188-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, W. Y., and T. M. Townes. 2000. Molecular mechanism for silencing virally transduced genes involves histone deacetylation and chromatin condensation. Proc. Natl. Acad. Sci. USA 97377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiocca, S., V. Kurtev, R. Colombo, R. Boggio, M. T. Sciurpi, G. Brosch, C. Seiser, G. F. Draetta, and M. Cotten. 2002. Histone deacetylase 1 inactivation by an adenovirus early gene product. Curr. Biol. 12594-598. [DOI] [PubMed] [Google Scholar]
- 10.Coull, J. J., F. Romerio, J. M. Sun, J. L. Volker, K. M. Galvin, J. R. Davie, Y. Shi, U. Hansen, and D. M. Margolis. 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 746790-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullen, B. R. 2006. Enhancing and confirming the specificity of RNAi experiments. Nat. Methods 3677-681. [DOI] [PubMed] [Google Scholar]
- 12.Dalle, B., J. E. Rubin, O. Alkan, T. Sukonnik, P. Pasceri, S. Yao, R. Pawliuk, P. Leboulch, and J. Ellis. 2005. eGFP reporter genes silence LCRbeta-globin transgene expression via CpG dinucleotides. Mol. Ther. 11591-599. [DOI] [PubMed] [Google Scholar]
- 13.de Ruijter, A. J., A. H. van Gennip, H. N. Caron, S. Kemp, and A. B. van Kuilenburg. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370737-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du, T., and P. D. Zamore. 2005. microPrimer: the biogenesis and function of microRNA. Development 1324645-4652. [DOI] [PubMed] [Google Scholar]
- 15.du Chene, I., E. Basyuk, Y. L. Lin, R. Triboulet, A. Knezevich, C. Chable-Bessia, C. Mettling, V. Baillat, J. Reynes, P. Corbeau, E. Bertrand, A. Marcello, S. Emiliani, R. Kiernan, and M. Benkirane. 2007. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis, J. 2005. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 161241-1246. [DOI] [PubMed] [Google Scholar]
- 17.Ellis, J., and S. Yao. 2005. Retrovirus silencing and vector design: relevance to normal and cancer stem cells? Curr. Gene Ther. 5367-373. [DOI] [PubMed] [Google Scholar]
- 18.Fukagawa, T., M. Nogami, M. Yoshikawa, M. Ikeno, T. Okazaki, Y. Takami, T. Nakayama, and M. Oshimura. 2004. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat. Cell Biol. 6784-791. [DOI] [PubMed] [Google Scholar]
- 19.Glozak, M. A., N. Sengupta, X. Zhang, and E. Seto. 2005. Acetylation and deacetylation of non-histone proteins. Gene 36315-23. [DOI] [PubMed] [Google Scholar]
- 20.Greger, J. G., R. A. Katz, A. M. Ishov, G. G. Maul, and A. M. Skalka. 2005. The cellular protein Daxx interacts with avian sarcoma virus integrase and viral DNA to repress viral transcription. J. Virol. 794610-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grewal, S. I., and S. Jia. 2007. Heterochromatin revisited. Nat. Rev. Genet. 835-46. [DOI] [PubMed] [Google Scholar]
- 22.Han, Y., M. Wind-Rotolo, H. C. Yang, J. D. Siliciano, and R. F. Siliciano. 2007. Experimental approaches to the study of HIV-1 latency. Nat. Rev. Microbiol. 595-106. [DOI] [PubMed] [Google Scholar]
- 23.He, G., and D. M. Margolis. 2002. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol. Cell. Biol. 222965-2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann, H., H. Sindre, and T. Stamminger. 2002. Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 765769-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollenbach, A. D., C. J. McPherson, E. J. Mientjes, R. Iyengar, and G. Grosveld. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 1153319-3330. [DOI] [PubMed] [Google Scholar]
- 26.Jaenisch, R., and A. Bird. 2003. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33(Suppl.)245-254. [DOI] [PubMed] [Google Scholar]
- 27.Kanellopoulou, C., S. A. Muljo, A. L. Kung, S. Ganesan, R. Drapkin, T. Jenuwein, D. M. Livingston, and K. Rajewsky. 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19489-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katz, R. A., E. Jack-Scott, A. Narezkina, I. Palagin, P. Boimel, J. Kulkosky, E. Nicolas, J. G. Greger, and A. M. Skalka. 2007. High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms. J. Virol. 812592-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, J. H., H. Li, and M. R. Stallcup. 2003. CoCoA, a nuclear receptor coactivator which acts through an N-terminal activation domain of p160 coactivators. Mol. Cell 121537-1549. [DOI] [PubMed] [Google Scholar]
- 30.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 31.Lehrman, G., I. B. Hogue, S. Palmer, C. Jennings, C. A. Spina, A. Wiegand, A. L. Landay, R. W. Coombs, D. D. Richman, J. W. Mellors, J. M. Coffin, R. J. Bosch, and D. M. Margolis. 2005. Depletion of latent HIV-1 infection in vivo: a proof-of-concept study. Lancet 366549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewinski, M. K., D. Bisgrove, P. Shinn, H. Chen, C. Hoffmann, S. Hannenhalli, E. Verdin, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2005. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 796610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieberman, P. M. 2006. Chromatin regulation of virus infection. Trends Microbiol. 14132-140. [DOI] [PubMed] [Google Scholar]
- 34.Lomberk, G., D. Bensi, M. E. Fernandez-Zapico, and R. Urrutia. 2006. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 8407-415. [DOI] [PubMed] [Google Scholar]
- 35.Marban, C., S. Suzanne, F. Dequiedt, S. de Walque, L. Redel, C. Van Lint, D. Aunis, and O. Rohr. 2007. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 26412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marks, P. A. 2007. Discovery and development of SAHA as an anticancer agent. Oncogene 261351-1356. [DOI] [PubMed] [Google Scholar]
- 37.Marks, P. A., and R. Breslow. 2007. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2584-90. [DOI] [PubMed] [Google Scholar]
- 38.Merezak, C., M. Reichert, C. Van Lint, P. Kerkhofs, D. Portetelle, L. Willems, and R. Kettmann. 2002. Inhibition of histone deacetylases induces bovine leukemia virus expression in vitro and in vivo. J. Virol. 765034-5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minc, E., J. C. Courvalin, and B. Buendia. 2000. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet. Cell Genet. 90279-284. [DOI] [PubMed] [Google Scholar]
- 40.Mizutani, T., T. Ito, M. Nishina, N. Yamamichi, A. Watanabe, and H. Iba. 2002. Maintenance of integrated proviral gene expression requires Brm, a catalytic subunit of SWI/SNF complex. J. Biol. Chem. 27715859-15864. [DOI] [PubMed] [Google Scholar]
- 41.Mok, H. P., and A. M. Lever. 2007. Chromatin, gene silencing and HIV latency. Genome Biol. 8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 211112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narezkina, A., K. D. Taganov, S. Litwin, R. Stoyanova, J. Hayashi, C. Seeger, A. M. Skalka, and R. A. Katz. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 7811656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nevels, M., C. Paulus, and T. Shenk. 2004. Human cytomegalovirus immediate-early 1 protein facilitates viral replication by antagonizing histone deacetylation. Proc. Natl. Acad. Sci. USA 10117234-17239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412561-565. [DOI] [PubMed] [Google Scholar]
- 46.Preston, C. M., and M. J. Nicholl. 2006. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 871113-1121. [DOI] [PubMed] [Google Scholar]
- 47.Quivy, V., S. De Walque, and C. Van Lint. 2007. Chromatin-associated regulation of HIV-1 transcription: implications for the development of therapeutic strategies. Subcell. Biochem. 41371-396. [PubMed] [Google Scholar]
- 48.Ruthenburg, A. J., C. D. Allis, and J. Wysocka. 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 2515-30. [DOI] [PubMed] [Google Scholar]
- 49.Saffert, R. T., and R. F. Kalejta. 2006. Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 803863-3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi, Y., and J. R. Whetstine. 2007. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell 251-14. [DOI] [PubMed] [Google Scholar]
- 51.Svoboda, J., J. Hejnar, J. Geryk, D. Elleder, and Z. Vernerova. 2000. Retroviruses in foreign species and the problem of provirus silencing. Gene 261181-188. [DOI] [PubMed] [Google Scholar]
- 52.Swindle, C. S., and C. A. Klug. 2002. Mechanisms that regulate silencing of gene expression from retroviral vectors. J. Hematother. Stem Cell Res. 11449-456. [DOI] [PubMed] [Google Scholar]
- 53.Taddei, A., D. Roche, W. A. Bickmore, and G. Almouzni. 2005. The effects of histone deacetylase inhibitors on heterochromatin: implications for anticancer therapy? EMBO Rep. 6520-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Lint, C., V. Quivy, D. Demonte, A. Chariot, C. Vanhulle, S. de Walque, G. Gaudray, E. Veithen, V. Bours, J. Piette, and A. Burny. 2004. Molecular mechanisms involved in HIV-1 transcriptional latency and reactivation: implications for the development of therapeutic strategies. Bull. Mem. Acad. R. Med. Belg. 159176-189. [PubMed] [Google Scholar]
- 55.Williams, S. A., L. F. Chen, H. Kwon, C. M. Ruiz-Jarabo, E. Verdin, and W. C. Greene. 2006. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 25139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolf, D., and S. P. Goff. 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 13146-57. [DOI] [PubMed] [Google Scholar]
- 57.Yao, S., T. Sukonnik, T. Kean, R. R. Bharadwaj, P. Pasceri, and J. Ellis. 2004. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol. Ther. 1027-36. [DOI] [PubMed] [Google Scholar]
- 58.Ylisastigui, L., N. M. Archin, G. Lehrman, R. J. Bosch, and D. M. Margolis. 2004. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 181101-1108. [DOI] [PubMed] [Google Scholar]
- 59.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13335-340. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, R., S. T. Liu, W. Chen, M. Bonner, J. Pehrson, T. J. Yen, and P. D. Adams. 2007. HP1 proteins are essential for a dynamic nuclear response that rescues the function of perturbed heterochromatin in primary human cells. Mol. Cell. Biol. 27949-962. [DOI] [PMC free article] [PubMed] [Google Scholar]