Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 facilitates the expression of both intronless viral ORF59 genes and intron-containing viral K8 and K8.1 genes (V. Majerciak, N. Pripuzova, J. P. McCoy, S. J. Gao, and Z. M. Zheng, J. Virol. 81:1062-1071, 2007). In this study, we showed that disruption of ORF57 in a KSHV genome led to increased accumulation of ORF50 and K8 pre-mRNAs and reduced expression of ORF50 and K-bZIP proteins but had no effect on latency-associated nuclear antigen (LANA). Cotransfection of ORF57 and K8β cDNA, which retains a suboptimal intron of K8 pre-mRNA due to alternative splicing, promoted RNA splicing of K8β and production of K8α (K-bZIP). Although Epstein-Barr virus EB2, a closely related homolog of ORF57, had a similar activity in the cotransfection assays, herpes simplex virus type 1 ICP27 was inactive. This enhancement of RNA splicing by ORF57 correlates with the intact N-terminal nuclear localization signal motifs of ORF57 and takes place in the absence of other viral proteins. In activated KSHV-infected B cells, KSHV ORF57 partially colocalizes with splicing factors in nuclear speckles and assembles into spliceosomal complexes in association with low-abundance viral ORF50 and K8 pre-mRNAs and essential splicing components. The association of ORF57 with snRNAs occurs by ORF57-Sm protein interaction. We also found that ORF57 binds K8β pre-mRNAs in vitro in the presence of nuclear extracts. Collectively our data indicate that KSHV ORF57 functions as a novel splicing factor in the spliceosome-mediated splicing of viral RNA transcripts.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, is a human gammaherpesvirus that is closely related to Epstein-Barr virus (EBV) and herpesvirus saimiri (HVS) (7, 41, 45). KSHV infection is associated with all forms of Kaposi's sarcoma, primary effusion lymphoma or body cavity-based B-cell lymphoma, and multicentric Castleman disease (20, 53, 55, 62). Latent KSHV infection in Kaposi's sarcoma tissues and B-cell lines features the restricted expression of only five viral genes (13, 74). The lytic KSHV infection produces progeny virus from infected cells and can be induced by chemicals such as tetradecanoyl phorbol acetate (43), butyrate (39), or valproic acid (25) or by hypoxia (12) in primary effusion lymphoma-derived B cells with latent KSHV infection. Chemical induction in latently infected cells initiates the expression of a viral transactivator, ORF50, which is essential for the switch from KSHV latency to the lytic phase (32, 57).
KSHV ORF57 (mRNA transcript accumulation [MTA]), which is transactivated by ORF50 (31, 63), encodes a viral early nuclear protein of 455 amino acid (aa) residues (15, 24) that is homologous to herpes simplex virus (HSV) ICP27 (IE63), EBV EB2 (SM), and HVS ORF57. KSHV ORF57 promotes the expression of ORF56 and ORF59 genes at the posttranscriptional level (24, 34, 35), but how ORF57 functions is poorly understood. Previous reports indicated that HSV ICP27 mediates viral intronless RNA export (49), inhibits RNA splicing of host cell transcripts (5, 18, 30), and selectively stabilizes some labile mRNAs containing AU-rich instability elements in their 3′ untranslated regions (4, 11). A recent study also suggested that KSHV ORF57 mediates the nuclear export of viral RNAs in a CRM1-independent manner, presumably through its interaction with the cellular export factor Aly/REF (36), which is similar to the properties of ORF57 homologs in other members of the herpesvirus family (21, 26, 64). However, Aly/REF binding by KSHV ORF57 in vivo appears to have no effect on ORF57-mediated enhancement of KSHV ORF59 expression (34). Our recent studies showed that infecting cells with an ORF57-disrupted KSHV genome prevents the expression of K8α and K8.1 (33), two viral split genes with multiple introns and exons, thus indicating that KSHV ORF57 also regulates the expression of intron-containing viral genes. Here, we provide evidence that KSHV ORF57 can function as a viral splicing factor to promote splicing of K8 transcripts and, ultimately, K8α (K-bZIP) protein production.
MATERIALS AND METHODS
Cells and KSHV induction.
A KSHV+/EBV+ B-cell line, JSC-1 (6), was maintained in RPMI 1640 medium. Human HEK-293 cells, HeLa cells, and stable Bac36 cells containing either a wild-type KSHV genome (Bac36-wt) or an ORF57-knockout KSHV genome (Bac36-Δ57) (33) were grown in Dulbecco's modified Eagle's medium. Both culture media contained 10% fetal bovine serum and were supplemented with 2 mM l-glutamine, 100 units penicillin/ml, and 100 μg streptomycin/ml. To induce the expression of KSHV lytic genes, JSC-1 cells or stable Bac36 cells were cultured in the presence of sodium butyrate at a final concentration of 3 mM for 24 h or with valproic acid (VA) at a final concentration of 1 mM for 72 h.
Plasmids.
All plasmids used in this study were previously described, as follows: pVM7 (FLAG-tagged, full-length ORF57) (35); pVM24 (FLAG-ORF57 aa 1 to 251) and its mutant derivatives pVM45 (mNLS1), pVM46 (mNLS2), pVM47 (mNLS3), pVM48 (mNLS1 + 2), pVM49 (mNLS1 + 3), pVM50 (mNLS2 + 3), pKY15 (FLAG-ICP27), and pGS113 (myc-EBV EB2) (34); pST3 (K8β cDNA) and pKY3 (K8β cDNA with optimized intron 5′ and 3′ splice sites) (68); and pZMZ70 (green fluorescent protein [GFP]-human papillomavirus type 16 [HPV16] E6) and pTMF11 (GFP-human β-globin) (73).
Transient transfection.
HEK-293 cells (5 × 105 in a 6-cm petri dish) and HeLa cells (1 × 107 in a 10-cm petri dish) were transfected, respectively, with approximately 2 and 4 μg of plasmid DNA using Lipofectamine 2000. Protein samples in radioimmunoprecipitation assay buffer (Boston BioProducts, Ashland, MA) were prepared at 24 h (HEK-293) or 48 h (HeLa) after transfection and used for various assays. Total cell RNA was also prepared from separate transfections for reverse transcription-PCR (RT-PCR) or Northern blotting.
Immunofluorescence staining.
JSC-1 cells with or without butyrate induction were washed twice with phosphate-buffered saline, spotted onto polylysine-treated glass coverslips, and fixed with 4% paraformaldehyde. Stable Bac36 cells were grown on coverslips and induced with or without butyrate before fixation. Immunofluorescence staining was performed as described previously (33). The primary antibodies rabbit polyclonal anti-ORF57 antibody (33) and mouse monoclonal anti-SC35 (Sigma, St. Louis, MO), antinucleophosmin (NPM/B23; Zymed Laboratories Inc, South San Francisco, CA), anti-K8 (Promab, Albany, CA), and anti-ORF50 antibodies were used, together with fluorescein isothiocyanate-conjugated anti-rabbit or tetramethyl rhodamine isothiocyanate-conjugated anti-mouse antibodies (Sigma). Confocal fluorescence images were collected using a Zeiss LSM510 META laser-scanning microscope equipped with a 63× Plan-Apochromat (numerical aperture, 1.4) oil immersion objective lens, with an optical slice thickness of 1.0 μm, an x-y pixel sampling of 0.1 μm, and a Z-step size of 0.2 μm. Individual optical slices were saved in TIFF format, and Adobe Photoshop 6.0 software (Adobe Systems, San Jose, CA) was used to process the images into composite figures. Colocalization analysis was performed using the colocalization module of Imaris v 4.0 software (Bitplane).
UV cross-linking and IP of ORF57-associated RNAs and proteins.
KSHV-infected JSC-1 cells with or without butyrate induction or HEK-293 cells with ORF57 overexpression in PBS (Ca2+ and Mg2+ free) were used for UV cross-linking and immunoprecipitation (IP) to detect ORF57-associated RNAs and proteins as described previously (34). After digestion with proteinase K, the RNA in the IP pull-downs was extracted, treated with DNase I, and analyzed by RT-PCR or Northern blotting. The proteins in the IP pull-downs were analyzed by Western blotting.
RT-PCR and Northern blot analysis.
Total cell RNA treated with DNase I was reverse transcribed and amplified by PCR using different primer sets (Table 1) for detection of a specific RNA. The RT-PCR products were gel purified and verified by sequencing.
TABLE 1.
Primer pairs for RT-PCR analyses of viral or cellular RNA transcripts
| Oligonucleotide | Positiona | Gene | Sequence (5′→3′) |
|---|---|---|---|
| oST1 | 75182 (s) | K8 | TAATACGACTCACTATAGGG/ACCACCAAGAGGACCACACATTTC |
| oST3 | 75838 (as) | K8 | GTACTCACCCC/CACACAAAGTCTGGCATGGTTCTCCC |
| oKY27 | 71575 (s) | ORF50 | ACTGTCCAGGCAGCCACA |
| oKY28 | 72699 (as) | ORF50 | AGGTCACTGGGATCGTAGATTG |
| oKY30 | 127846 (as) | LANA | AGAGCAGCAGCTTGGTCCG |
| oST14 | 127264 (s) | LANA | GTCCCGACCTCAGGCGCATTCC |
| oKY46 | 126877 (s) | LANA | CTGGAGACTGCGTGGGTGG |
| oKY47 | 9 (s) | U1 | CTGGCAGGGGAGATACCATG |
| oKY48 | 163 (as) | U1 | AGGGGAAAGCGCGAACGCA |
| oKY49 | 13 (s) | U2 | CCTTTTGGCTAAGATCAAGTGTAG |
| oKY50 | 183 (as) | U2 | CACCGTTCCTGGAGGTACTG |
| oZMZ326 | 1 (s) | U6 | CATATCTCGC/GTGCTCGCTTCGGCAGCACA |
| oST197 | 106 (as) | U6 | AAAATATGGAACGCTTCACGA |
| oZMZ269 | 225 (s) | GAPDH | GTCATCAATGGAAATCCCATCACC |
| oZMZ270 | 502 (as) | GAPDH | TGAGTCCTTCCACGATACCAAA |
| oZMZ237 | 106 (s) | E6 | GTTTCAGGACCCACAGGAGC |
| oZMZ222 | 562 (as) | E6 | GTACTCACCCC/TGATTACAGCTGGGTTTC |
| oZMZ318 | 1b | β-Globin | ATCTGACTCGAG/CTTACATTTGCTTCTGACACAAC |
| oZMZ106 | 359b | β-Globin | GGGTTGCCCATAACAGCATCAGG |
| oKY32 | 2809 (s) | Survivin | GCATGGGTGCCCCGACGTTG |
| oKY33 | 12060 (as) | Survivin | GCTCCGGCCAGAGGCCTCAA |
| oKY34 | 51 (s) | IL-6 | AGGAGCCCAGCTATGAACTC |
| oKY35 | 410 (as) | IL-6 | GTCGAGGATGTACCGAATTTG |
s, sense; as, antisense.
Based on Promega pSP64 HβΔ6.
Northern blot analyses of small nuclear RNAs (snRNAs) from IP pull-downs were conducted as described previously (35) with antisense RNA probes prepared by in vitro transcription from various plasmids containing U1, U2, U4, U5, and U6 snRNAs.
Western blot analysis.
Protein samples were separated on a 4% to 12% bis-Tris gel (Invitrogen) and immunoblotted as described previously (68) with rabbit polyclonal anti-ORF57 antibody or mouse monoclonal anti-K8α (K-bZIP), anti-FLAG M2 (Sigma), anti-SR mAb104 (ATCC, Manassas, VA), anti-c-Myc (Sigma), anti-Sm (Y12 clone; Lab Vision Co., Fremont, CA), anti-SF2/ASF (Zymed Laboratories, Inc.), anti-U2AF35, and anti-human β-tubulin (BD Pharmingen, San Diego, CA).
In vitro RNA splicing.
In vitro splicing assays were carried out in a 12.5-μl reaction volume with 20 fmol of 32P-labeled pre-mRNA transcripts as described previously (37, 38, 72) in the presence of HeLa or HEK-293 nuclear extracts (NE) or cytoplasmic S100 with or without addition of SF2/ASF expressed in bacteria (75) or FLAG-tagged ORF57 expressed in baculovirus (34).
RESULTS
KSHV ORF57 is involved in the expression of K-bZIP and ORF50 and distributes in the nuclear speckles in KSHV-infected B cells.
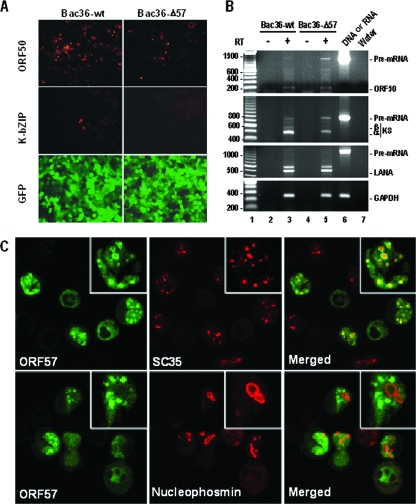
Recently, we demonstrated that a mutant KSHV genome with a disrupted ORF57 (Bac36-Δ57) is unable to express a subset of viral lytic genes and to produce infectious virions (33). Our present results also show that the ORF57-disrupted mutant KSHV genome exhibited reduced expression of ORF50, a viral transactivator, in addition to lacking viral K-bZIP (K8α) protein expression during viral lytic induction (Fig. 1A). To determine the mechanism through which ORF57 influences lytic gene expression, we extracted total cell RNA from Bac36-wt- and Bac36-Δ57-stably infected cells 24 h after butyrate or 72 h after VA induction and examined viral RNA splicing of ORF50, K8, and latency-associated nuclear antigen (LANA) by RT-PCR. We found that ORF50, K8, and LANA RNAs expressed from Bac36-wt underwent RNA splicing, with the majority of detected RT-PCR products being spliced mRNA. However, expression of ORF50 and K8 RNAs from Bac36-Δ57 led to substantial (∼6- and ∼4-fold, respectively) accumulation of the respective pre-mRNAs (Fig. 1B), whereas expression of LANA from Bac36-Δ57 was not altered compared to that from Bac36-wt, suggesting a defect in RNA splicing of both ORF50 and K8, but not LANA, pre-mRNAs. The similar results were also confirmed with VA-induced the two Bac36 cells (data not shown). Using confocal fluorescence microscopy to examine the distribution of KSHV ORF57, we found that the majority (∼ 96%) of total splicing factor SC35 in nuclear speckle domains colocalized with ORF57 in KSHV-infected JSC-1 cells after butyrate induction, but only ∼ 23% of total ORF57 was present in speckle domains, where it colocalizes with SC35, indicating that a partial, but significant, proportion of total ORF57 colocalizes with SC35. In contrast, very little (<1%) of nucleolar nucleophosmin colocalized with ORF57. Likewise, very little (<3%) of total ORF57 colocalized with nucleophosmin (Fig. 1C). The localization in nuclear speckle domains, which function as storage sites for splicing factors (27), suggests that one of KSHV ORF57's activities may be to directly or indirectly facilitate RNA splicing of intron-containing viral pre-mRNAs.
FIG. 1.
KSHV ORF57 regulates the expression of viral intron-containing genes and distributes in nuclear splicing speckles in KSHV-infected B cells. (A) KSHV ORF57 is important for production of viral ORF50 and K-bZIP proteins. HEK-293 cells stably transfected with Bac36-wt or Bac36-Δ57 DNA were induced by sodium butyrate for lytic infection and stained with anti-ORF50 or anti-K8 (K-bZIP) antibody. GFP expression from Bac36-wt and Bac36-Δ57 genomes in the HEK-293 cells was imaged directly from the fixed cells. (B) Accumulation of ORF50 and K8 pre-mRNAs in butyrate-induced stable Bac36-Δ57 cells. Total RNA extracted 24 h after butyrate induction from HEK-293 cells stably transfected with Bac36-wt or Bac36-Δ57 was analyzed by RT-PCR for the expression of KSHV ORF50, K8, and LANA RNAs using a pair of gene-specific exon primers (Table 1). The primer pair oKY30 and oKY46 was used for LANA detection. Cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA served as the RNA loading control. Lane 6 indicates that Bac36 DNA in ORF50, K8, and LANA detection served as controls in each assay. (C) Distribution of KSHV ORF57 in nuclear speckles in KSHV-infected JSC-1 cells. Butyrate-activated JSC-1 cells were stained with rabbit anti-ORF57, mouse monoclonal anti-SC35, or antinucleophosmin and were then imaged by using fluorescein isothiocyanate-labeled anti-rabbit immunoglobulin G (green) or tetramethyl rhodamine isothiocyanate-labeled anti-mouse immunoglobulin G (red). Insets show enlarged representative cells.
KSHV ORF57 promotes K-bZIP production from K8β cDNA by facilitating viral RNA splicing.
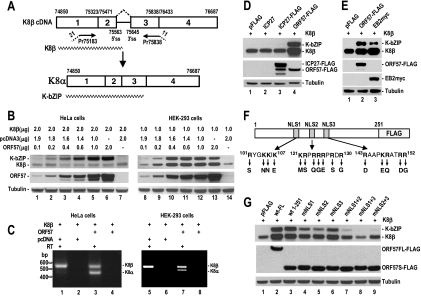
To examine whether ORF57 facilitates viral RNA splicing, we analyzed the expression of K8β cDNA in the presence or absence of KSHV ORF57 in cotransfected HeLa cells and HEK-293 cells. The K8β cDNA corresponds to K8β mRNA, a partially spliced intermediate of K8 pre-mRNA that retains a suboptimal intron with weak 5′ and 3′ splice sites (Fig. 2A) (68). When the intron in K8β RNA remains unspliced, the resulting mRNA harbors a stop codon within the intron but does not undergo nonsense-mediated decay (68). Consequently, K8β mRNA encodes a truncated protein with the same N terminus as K-bZIP (K8α) protein but missing a C-terminal leucine zipper domain (68). Surprisingly, the cotransfection assays showed that ORF57 promotes K-bZIP protein production from K8β cDNA in a dose-dependent manner (Fig. 2A and B), more dramatically in HeLa cells. RT-PCR analysis of total cell RNA extracted from the cotransfected cells showed that the enhanced expression of K-bZIP protein from K8β cDNA by ORF57 was mainly due to enhanced production of K8α mRNA from K8β pre-mRNAs (Fig. 2C), with an enhanced K8β/K8α ratio of 1:1 in HeLa cells and 2.5:1 in HEK-293 cells. HSV-1 ICP27 did not function in the same capacity (Fig. 2D); however, EBV EB2, a more closely related homolog of ORF57, functioned in a similar manner as ORF57 by promoting K8β mRNA splicing and K-bZIP production (Fig. 2E), indicating that both KSHV ORF57 and EBV EB2 promote splicing of an intron downstream of a large exon (565 nucleotide [nt]). In addition, three nuclear localization signals (NLSs) in the ORF57 N terminus (34) must be intact for this function. Introducing point mutations to disrupt one or two NLSs in an N-terminal fragment of ORF57 protein that retains ∼90% of the activity of full-length ORF57 in promoting ORF59 expression (34) reduced or completely prevented K-bZIP production from K8β pre-mRNA in HEK-293 cell cotransfection assays (Fig. 2F and G), despite the proper localization of the mutant ORF57 protein in the nucleus (34).
FIG. 2.
KSHV ORF57 promotes the expression of K-bZIP protein by enhancing K8β RNA splicing. (A) Structures of the full-length KSHV K8β cDNA and K8α RNA, with spliced exons (numbered boxes) and introns (line) illustrated. K8β is an alternative spliced product of K8 pre-mRNA and retains a suboptimal intron (intron 2). The numbers above the cDNA or RNA are the nucleotide positions in the KSHV genome (GenBank accession number U75689 [45]). Splice junctions of intron 1 and intron 3 are indicated in K8β cDNA as 75323/75471 and 75838/76433, respectively. K8β encodes a truncated protein (jagged line) of 190 aa. Splicing of the suboptimal intron from K8β RNA leads to the production of K8α mRNA, which encodes a K-bZIP protein (jagged line) of 238 aa. (B) KSHV ORF57 stimulates K-bZIP expression from K8β cDNA in a dose-dependent manner. HeLa and HEK-293 cells were cotransfected with a KSHV K8β cDNA expression vector, pST3, and a FLAG-tagged KSHV ORF57 expression vector, pVM7, or an empty vector, pcDNA3. The protein samples were prepared 24 or 48 h after transfection and analyzed by Western blotting. (C) KSHV ORF57 promotes RNA splicing of K8β pre-mRNAs. Total RNA from HeLa or HEK-293 cells cotransfected as described above was prepared 48 h after transfection and analyzed for K8 by RT-PCR. (D and E) KSHV ORF57 functionally resembles EBV EB2 protein but differs in activity from HSV ICP27 with respect to its effect on splicing of K8β RNA. HSV ICP27 with or without a FLAG tag was compared with FLAG-tagged KSHV ORF57 and Myc-tagged EBV EB2 by contransfection of HEK-293 cells with K8β cDNA as described above for K8 expression. Protein samples prepared 24 h after cotransfection were analyzed by Western blotting. Lower bands in lane 3 (D) are presumed degradation products of ICP27-FLAG. (F) Diagram of three NLSs in a truncated, FLAG-tagged ORF57 and various mutants in which the indicated residues were replaced with neutral or acidic residues. (G) NLSs of ORF57 play an important role in ORF57-mediated enhancement of K8β RNA splicing. The protein samples prepared 24 h after cotransfection of HEK-293 cells with K8β cDNA plus full-length ORF57 (ORF57FL), ORF57 aa 1 to 251 (ORF57S), or ORF57 aa 1 to 251 with one or two NLS mutations were analyzed by Western blotting for K8 and ORF57 expression. Tubulin from each sample in panels B, D, E, and G served as a sample loading control.
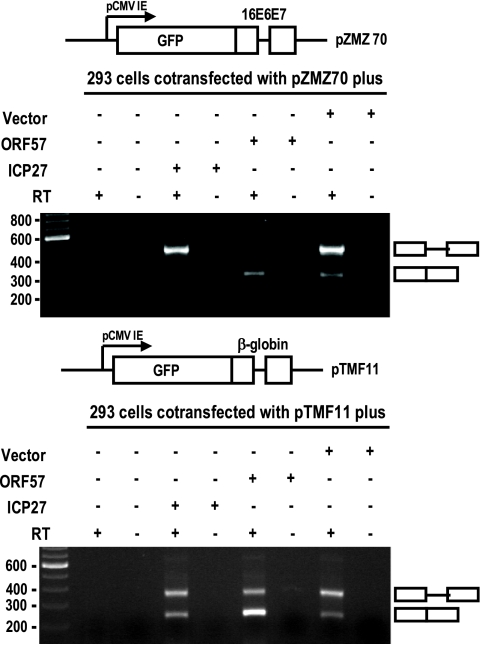
Similar experiments were also conducted by cotransfection of HEK-293 cells with ORF57 or ICP27, together with HPV16 E6E7 or human β-globin expression plasmids. The latter two plasmids have a GFP sequence fused to exon 1 that results in deficient RNA splicing due to a very large exon 1 (>900 nt) (73). We found that KSHV ORF57, but not ICP27, also facilitated splicing of both HPV16 E6E7 and β-globin pre-mRNAs with an enlarged exon 1 (Fig. 3), indicating that the splicing enhancement function of ORF57 is not limited to KSHV K8β pre-mRNAs.
FIG. 3.
KSHV ORF57 promotes splicing of other pre-mRNAs with an enlarged exon 1. HPV16 E6E7 (pZMZ70) or human β-globin (pTMF11) inserted downstream of GFP in a pEGFP expression vector (73) was expressed in HEK-293 cells by transient cotransfection with ORF57 (pVM7), ICP27 (pKY15), or an empty FLAG vector. Total cell RNA was prepared 48 h posttransfection and examined by RT-PCR with E6E7- or β-globin-specific primers (Table 1).
KSHV ORF57 preferentially interacts with intron-containing viral RNAs.
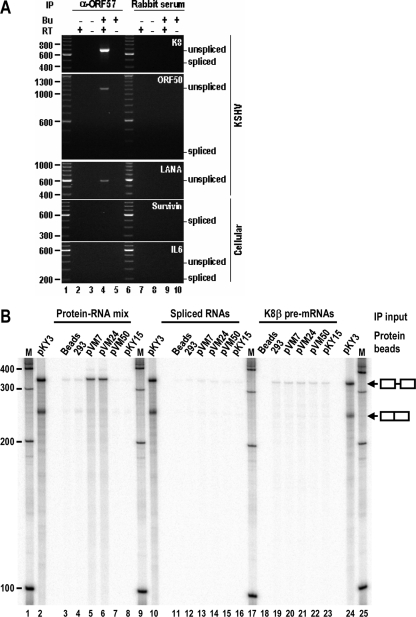
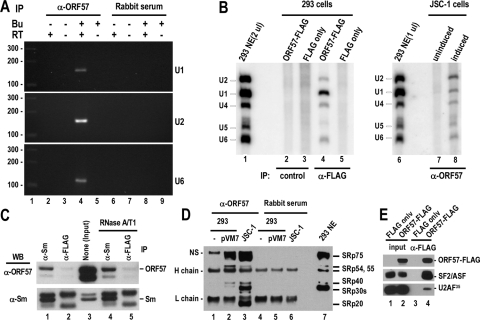
To understand the mechanism by which ORF57 promotes viral RNA splicing, we isolated ORF57-RNA complexes from butyrate-induced JSC-1 cells by in vivo UV cross-linking in combination with IP techniques. This approach covalently cross-links ORF57 bound to RNA, and the covalent adducts are then isolated by anti-ORF57 IP. The ORF57-bound RNA species was subsequently examined, and we found that KSHV ORF57 selectively interacts with low-abundance pre-mRNAs of K8, ORF50, and LANA but not cellular survivin and interleukin-6 (IL-6) during lytic virus infection (Fig. 4A). We noticed that this difference in preferential selection of viral RNAs by ORF57 was not due to the relative abundance of the viral versus cellular RNAs (data not shown). Notably, all of the RNA molecules isolated from the immunoprecipitated complexes contained introns as verified by sequencing. To confirm this observation, we carried out in vitro K8β pre-mRNA splicing and ORF57 binding assays. K8β (pKY3) RNA was subjected to in vitro splicing, and the resulting products were used for the ORF57 binding assays, which were carried out as follows. FLAG-tagged ORF57 proteins expressed in HEK-293 cells were immobilized on agarose beads and incubated either with a whole K8β (pKY3) RNA splicing reaction mixture (in the presence of HEK-293 cell nuclear extract), with the isolated K8β (pKY3) RNA splicing products (no extract), or with the K8β pre-mRNAs. Subsequently, the RNA-protein complexes that remained bound to the beads after extensive washes were resolved using an 8% denaturing polyacrylamide gel. As shown in Fig. 4B, the full-length (pVM7) and N-terminal (pVM24) ORF57 selectively bound K8β pre-mRNA in the presence of other cellular proteins (lanes 5 and 6), whereas the N-terminal ORF57 with point mutations in the NLS2 and NLS3 motifs (pVM50) lost this selective binding affinity (lane 7), and the similar protein ICP27 (pKY15) failed to bind K8β pre-mRNA (lane 8). Interestingly, ORF57 was unable to bind either unspliced K8β pre-mRNAs or spliced K8β mRNA in the absence of other cellular proteins (compare lanes 13, 14, 20, and 21 to lanes 5 and 6). Together, these results indicate that RNA introns are needed for the interaction of ORF57 with a pre-mRNA in the presence of other cellular proteins.
FIG. 4.
KSHV ORF57 preferentially interacts with viral pre-mRNAs. (A) KSHV ORF57 selectively binds to intron-containing viral pre-mRNAs in JSC-1 cells. KSHV ORF57-RNA complexes in JSC-1 cells with or without butyrate (Bu) activation were cross-linked by UV irradiation and immunoprecipitated with anti-ORF57 antibody or nonimmune rabbit serum. RNA extracted from the protein-RNA immunoprecipitates was examined by RT-PCR for the presence of a specific RNA using a pair of gene-specific exon primers (Table 1). The primer pair oST14 and oKY30 was used for LANA detection. (B) KSHV ORF57 selectively binds to K8β pre-mRNAs in the presence of HEK-293 nuclear extract in vitro. FLAG-tagged ORF57 in a full-length (pVM7) or truncated (aa 1 to 251, pVM24) version or the truncated version with point mutations in NLS2 and NLS3 motif (pVM50) purified from HEK-293 cells at 24 h posttransfection was compared to FLAG-tagged ICP27 (pKY15) for RNA binding in vitro. Approximately 200 μg of total protein from each transfection was immobilized on anti-FLAG-conjugated beads before RNA binding. An optimized, 32P-labeled K8β pre-mRNA (pKY3) (68) transcribed in vitro was spliced for 2 h at 30°C in the presence of HEK-293 nuclear extracts (72). The whole splicing reactions (lanes 3 to 8) or RNAs extracted from the splicing reactions (lanes 11 to 16) were then compared with K8β pre-mRNA (lanes 18 to 23) in RNA binding assays with each protein. Lanes 2, 10, and 24 were the spliced K8β RNA products (pKY3) resolved in an 8% denaturing polyacrylamide gel.
KSHV ORF57 interacts with essential components of the spliceosome in vivo.
Considering that ORF57 localizes to nuclear speckles and selectively binds RNA intron regions, we hypothesized that ORF57 is likely assembled into spliceosomal complexes to facilitate splicing of viral pre-mRNA. To test this hypothesis, we isolated ORF57-RNA complexes from butyrate-induced JSC-1 cells by in vivo UV cross-linking in combination with anti-ORF57 IP. The RNAs present in the anti-ORF57 IP pull-down fractions were examined for the presence of cellular spliceosomal snRNAs using RT-PCR (Fig. 5A) or Northern blot analysis (Fig. 5B). Indeed, we found KSHV ORF57 in association with U1, U2, U4, U5, and U6 snRNAs, all of which are essential components of spliceosomes, during KSHV lytic induction (Fig. 5A and B, right panel), but not with cellular ADAMST-1 RNA (data not shown). This association with snRNAs occurred in the absence of other viral proteins, since transfection of ORF57 alone was sufficient for interaction of ORF57 with various U snRNAs (Fig. 5B, left panel). This interaction was mediated through an ORF57-Sm protein interaction (Fig. 5C), as determined by using anti-Sm IP assays in the presence or absence of RNase A/T1 (Fig. 5C). ORF57 was present in the anti-Sm pull-downs under both conditions. Moreover, we also found that ORF57 was associated with various cellular serine-arginine-rich (SR) proteins in both butyrate-induced JSC-1 cells and transiently transfected HEK-293 cells, as demonstrated by mAb104, anti-U2AF35, and anti-SF2/ASF immunoblotting in IP-Western blot assays (Fig. 5D and E). Together, these results demonstrate that ORF57 interacts with the cellular splicing machinery, most likely in spliceosomal complexes.
FIG. 5.
KSHV ORF57 interacts with snRNAs and splicing factors. (A) Determination of ORF57-snRNA interactions by RT-PCR. UV-cross-linked ORF57-RNA complexes were immunoprecipitated from JSC-1 cells with or without butyrate (Bu) activation, using rabbit anti-ORF57 antibody or preimmune serum. The snRNAs in the immunoprecipitated ORF57-RNA complexes were detected by RT-PCR. (B) Determination of ORF57-snRNA interactions by Northern blot analysis. The snRNAs in the immunoprecipitated ORF57-RNA complexes obtained from ORF57-transfected HEK-293 cells or JSC-1 cells with or without butyrate activation were examined by Northern blotting using a pool of U1, U2, U4, U5, and U6 probes. Total RNA from HEK-293 nuclear extract served as control. (C) ORF57 interacts with snRNAs through Sm proteins. Protein-protein complexes immunoprecipitated from butyrate-activated JSC-1 cells with monoclonal anti-Sm or anti-FLAG antibodies (a control antibody), with or without RNase A/T1 treatment, were immunoblotted with rabbit anti-ORF57 or anti-Sm antibodies. IP, antibodies used for IP; WB, antibodies used for Western blotting. Two bands in anti-FLAG IP (lower panel) are nonspecific. (D and E) ORF57 interacts with SR proteins and U2AF35. Cellular splicing factors in ORF57-FLAG (pVM7)-expressing HEK-293 cells (D and E) or in butyrate-activated JSC-1 cells (D) were immunoprecipitated with anti-ORF57 antibody and then immunoblotted with a monoclonal anti-SR antibody, mAb104 (D), or immunoprecipitated with anti-FLAG antibody and then immunoblotted with anti-SF2/ASF, anti-U2AF35, or anti-ORF57 antibody (E). NS, nonspecific; 293 NE, HEK-293 nuclear extract (serving as SR protein controls) (D).
Recombinant KSHV ORF57 does not behave like a classical SR protein in S100 complementation assays.
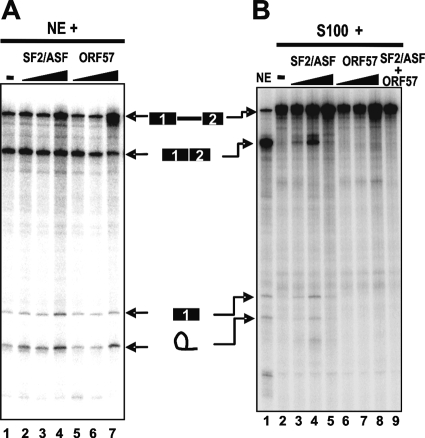
Because ORF57 binds RNA and has several RS/SR dipeptide repeats, we tested whether it might function similarly to SR proteins, which promote spliceosome assembly and splicing in S100 complementation assays (38). We compared the ability of ORF57 to facilitate splicing of various pre-mRNA substrates with that of the prototypical SR protein, SF2/ASF, in nuclear extract and S100 complementation in vitro splicing assays. Baculovirus-expressed FLAG-tagged ORF57 had no effect on splicing of human β-globin (Fig. 6A), BPV-1 late, and Drosophila Ftz pre-mRNAs and failed to facilitate splicing of KSHV K8β pre-mRNA (data not shown) in all in vitro splicing reactions in the presence of HeLa NE. Moreover, FLAG-tagged ORF57, in contrast to SF2/ASF, did not complement, at all doses examined, splicing of human β pre-mRNA in SR protein-deficient S100 extract (Fig. 6B, compare lanes 3 to 5 with lanes 6 to 8). Because SF2/ASF at 15 pmol sequestrates the splicing machinery in the assay (Fig. 6B, lane 5) as predicted, an effective 8-pmol dose of SF2/ASF (Fig. 6B, lane 4) was mixed with the same amount of recombinant ORF57 in the assay to examine its synergic effect. In fact, we found that ORF57 interfered with the function of SF2/ASF when both proteins at this dose were present together (Fig. 6B, lane 9). Our results indicate that the recombinant KSHV ORF57 that lacks a normal set of posttranslational modifications does not function like an SR protein, despite the presence of four RS or SR dipepetides in its N-terminal half (34).
FIG. 6.
Effect of SF2/ASF and ORF57 on in vitro splicing of human β-globin pre-mRNA. (A) Human β-globin pre-mRNA was incubated in NE along with increasing amounts (2, 8, and 15 pmol) of recombinant SF2/ASF (lanes 2 to 4) or ORF57 (lanes 5 to 7). (B) S100-complementation splicing assays of human β-globin pre-mRNA with 2, 8, or 15 pmol of either SF2/ASF (lanes 3 to 5), ORF57 (lanes 6 to 8), or a mixture (8 pmol each) of both (lane 9). The mobilities of the pre-mRNA and splicing products are indicated between the two splicing gels.
DISCUSSION
In this study, we demonstrated that KSHV ORF57 functions as a viral splicing factor that promotes splicing of viral lytic pre-mRNAs. Expression of an ORF57-null KSHV genome resulted in deficient viral RNA splicing, leading to accumulation of ORF50 and K8 pre-mRNAs and production of considerably reduced amounts of ORF50 and K-bZIP proteins. However, ORF57 appears to have no effect on LANA expression during lytic virus infection, consistent with LANA expression independent of ORF57 during viral latent infection. Cotransfection of ORF57 and K8β cDNA demonstrated that ORF57 promotes splicing of K8β RNAs and production of K8α (K-bZIP) proteins in a dose-dependent manner. This novel finding is in sharp contrast to what is known about the functions of ORF57 homologs, which, in general, suppress splicing of intron-containing cellular pre-mRNAs (5, 18, 30, 47, 48) and facilitate RNA export of intronless viral transcripts (8, 21, 46).
Although the accumulation of mRNA transcripts is a known, evolutionarily conserved function of ORF57 and related homologs, the mechanism by which KSHV ORF57 promotes mRNA accumulation remains largely unknown. Numerous studies, including our own, have described diverse roles for the ORF57 homologs in posttranscriptional regulation of gene expression. KSHV ORF57 is phylogenetically and functionally more closely related to EBV EB2 than the other homologs. Interestingly, we found that both KSHV ORF57 and EBV EB2, but not HSV ICP27, enhance RNA splicing of K8β pre-mRNAs, colocalize with SC35 (9), and promote the expression of KSHV ORF59 (34). A separate study found the EBV EB2 protein to be as efficient as KSHV ORF57 at enhancing expression of the KSHV PAN, mCP, and ORF9 genes (17). However, KSHV ORF57 and EBV EB2 were unable to substitute for each other in rescuing virus production from the corresponding ORF57- or EB2-null genome (17). KSHV ORF57 also differs in its nuclear localization pattern from HVS ORF57, another homologous protein in the gammaherpesvirus subfamily. Although both proteins distribute in the nuclear speckles (this study and reference 10), KSHV ORF57 in the present study was not found much so in the nucleolus, in contrast to HVS ORF57 (3).
The KSHV ORF50 and K8 genes in early viral lytic infection express, respectively, a tricistronic and a bicistronic pre-mRNA, which overlap with each other and undergo alternative splicing to generate two major spliced mRNA isoforms, α and β, by inclusion (β) or exclusion (α) of an intron (K8 intron 2 or ORF50 intron 3) at nt 75563 to 75645. This intron is suboptimal, with a low binding affinity of its 5′ splice site to U1 snRNA and of its 3′ splice site to U2AF (68). Despite the suboptimal features of the K8 intron 2, this intron was completely spliced out of the majority of the K8 pre-mRNAs to produce a fully spliced K8α message, which encodes a K-bZIP protein important for viral gene transcription (22, 28, 42, 63), DNA replication (1, 29, 65), and cell cycle arrest (23, 66, 67). However, retention of this intron in K8β mRNA results in the production of a truncated protein that plays a dominant-negative role in fully spliced K8α expression, K-bZIP-mediated induction of p21 and p53, and K-bZIP-CDK2 interactions (68). Our previous studies, aimed at investigating what promotes the splicing of K8 intron 2 (ORF50 intron 3), demonstrated that splicing of this K8 intron is triggered by exon definition crossing over exon 3 (58, 68, 71). In the present study, we identified KSHV ORF57 as a viral splicing factor responsible for splicing of this intron, uncovering an important mechanism for how KSHV regulates the expression of its own genes during lytic viral infection. Although it remains to be determined why KSHV ORF57 in promotion of K8β splicing and K-bZIP production is more effective in HeLa cells than in HEK-293 cells, this might reflect a difference between the two cell lines in the expression levels of various cellular splicing components. It is perhaps not surprising that a viral splicing factor is needed during KSHV infection, because the KSHV genome carries more than 30 split genes whose expression requires extensive RNA splicing. This represents more than 30% of the total number of known genes in the KSHV genome (35, 59, 70). Among the split genes, many have a rather small intron downstream of an oversized exon, making them distinct from most mammalian pre-mRNAs, which consist of small exons and large introns. We postulated that an unknown mechanism might be needed for efficient exon definition (2, 56, 69, 73) leading to splicing of these viral RNAs by the host splicing machinery. Facilitation of viral RNA splicing by a viral protein over the cellular splicing machinery during lytic viral infection provides an advantage for KSHV to accelerate viral gene expression and commandeer the cellular controls of its RNA processing. During the course of preparation of this paper, Tormanen et al. (60) reported that an adenovirus protein, L4-33K, also functions as a virus-encoded RNA splicing factor that preferentially activates splicing of viral transcripts with a weak 3′ splice site. The finding that KSHV ORF57 is associated selectively with low-abundance, intron-containing viral pre-mRNAs and with essential splicing components of the spliceosome suggests that ORF57 might be involved in the expression of other viral or cellular split genes containing introns with suboptimal splice sites along with an oversized upstream exon. Consistently, KSHV LANA pre-mRNA contains a small exon upstream of a large intron with an optimal splice site, and its interaction with KSHV ORF57 during viral lytic induction does not result in an increased RNA splicing as we observed in this study. This description of ORF57 function in RNA splicing is in contrast to that for HSV ICP27, whose major functions are to suppress cellular RNA splicing (5, 18, 30) and to promote intronless viral RNA export (8, 49). However, this contrast in protein function seems logical, as only five of ∼90 HSV genes contain introns (44).
Although KSHV ORF57 is shown here to be a potent viral splicing factor that can promote splicing even in the absence of other viral proteins, we do not yet know how it facilitates splicing. Its activity is distinct from that of SR proteins, judging from in vitro splicing assays. Tagged KSHV ORF57 expressed in baculovirus or mammalian cells did not bind to RNAs with high affinity, in the absence of cellular proteins (reference 34 and this study). KSHV ORF57 has four RS or SR repeats in the N-terminal half of the protein (34), but the length and position of the RS repeats are atypical for an SR protein. Our observation that KSHV ORF57 coimmunoprecipitates with various spliceosomal snRNAs via interaction with Sm proteins (40, 54, 61), with cellular splicing factors including U2AF (16, 51, 52) and SR proteins (14, 19, 50), and selectively with low-abundance, intron-containing pre-mRNAs suggests that KSHV ORF57 plays a crucial auxiliary role in viral RNA splicing by mediating protein-protein interactions with various cellular splicing factors.
Acknowledgments
We thank Rozanne Sandri-Goldin for providing the pCMV-ICP27 plasmid (p320), Sankar Swaminathan for the EBV EB2myc plasmid (pGS113), Sam Gunderson for the U1 to U6 expression plasmids, Tom Maniatis for anti-U2AF35, Zuo Zhang for SF2/ASF, and Muzammel Haque and Koichi Yamanishi for anti-ORF50 antibody. We also thank Rozanne Sandri-Goldin, Donald Ganem, and George Miller for critical reading of the manuscript.
E.A. and A.R.K. were supported by grant CA13106 from the National Cancer Institute. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 9 January 2008.
REFERENCES
- 1.AuCoin, D. P., K. S. Colletti, S. A. Cei, I. Papouskova, M. Tarrant, and G. S. Pari. 2004. Amplification of the Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 lytic origin of DNA replication is dependent upon a cis-acting AT-rich region and an ORF50 response element and the trans-acting factors ORF50 (K-Rta) and K8 (K-bZIP). Virology 318542-555. [DOI] [PubMed] [Google Scholar]
- 2.Berget, S. M. 1995. Exon recognition in vertebrate splicing. J. Biol. Chem. 2702411-2414. [DOI] [PubMed] [Google Scholar]
- 3.Boyne, J. R., and A. Whitehouse. 2006. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc. Natl. Acad. Sci. USA 10315190-15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, C. R., M. S. Nakamura, J. D. Mosca, G. S. Hayward, S. E. Straus, and L. P. Perera. 1995. Herpes simplex virus trans-regulatory protein ICP27 stabilizes and binds to 3′ ends of labile mRNA. J. Virol. 697187-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant, H. E., S. E. Wadd, A. I. Lamond, S. J. Silverstein, and J. B. Clements. 2001. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol. 754376-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon, J. S., D. Ciufo, A. L. Hawkins, C. A. Griffin, M. J. Borowitz, G. S. Hayward, and R. F. Ambinder. 2000. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi's sarcoma herpesvirus-containing supernatant. J. Virol. 7410187-10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 2661865-1869. [DOI] [PubMed] [Google Scholar]
- 8.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 7612877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., G. Liao, M. Fujimuro, O. J. Semmes, and S. D. Hayward. 2001. Properties of two EBV Mta nuclear export signal sequences. Virology 288119-128. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, M., D. J. Goodwin, K. T. Hall, A. J. Stevenson, D. M. Meredith, A. F. Markham, and A. Whitehouse. 1999. The gene product encoded by ORF 57 of herpesvirus saimiri regulates the redistribution of the splicing factor SC-35. J. Gen. Virol. 801311-1316. [DOI] [PubMed] [Google Scholar]
- 11.Corcoran, J. A., W. L. Hsu, and J. R. Smiley. 2006. Herpes simplex virus ICP27 is required for virus-induced stabilization of the ARE-containing IEX-1 mRNA encoded by the human IER3 gene. J. Virol. 809720-9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, D. A., A. S. Rinderknecht, J. P. Zoeteweij, Y. Aoki, E. L. Read-Connole, G. Tosato, A. Blauvelt, and R. Yarchoan. 2001. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood 973244-3250. [DOI] [PubMed] [Google Scholar]
- 13.Fakhari, F. D., and D. P. Dittmer. 2002. Charting latency transcripts in Kaposi's sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J. Virol. 766213-6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graveley, B. R. 2000. Sorting out the complexity of SR protein functions. RNA. 61197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta, A. K., V. Ruvolo, C. Patterson, and S. Swaminathan. 2000. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J. Virol. 741038-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guth, S., T. O. Tange, E. Kellenberger, and J. Valcarcel. 2001. Dual function for U2AF(35) in AG-dependent pre-mRNA splicing. Mol. Cell. Biol. 217673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han, Z., and S. Swaminathan. 2006. Kaposi's sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J. Virol. 805251-5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 687790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hastings, M. L., and A. R. Krainer. 2001. Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13302-309. [DOI] [PubMed] [Google Scholar]
- 20.Hayward, G. S. 2003. Initiation of angiogenic Kaposi's sarcoma lesions. Cancer Cell 31-3. [DOI] [PubMed] [Google Scholar]
- 21.Hiriart, E., L. Bardouillet, E. Manet, H. Gruffat, F. Penin, R. Montserret, G. Farjot, and A. Sergeant. 2003. A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J. Biol. Chem. 27837790-37798. [DOI] [PubMed] [Google Scholar]
- 22.Izumiya, Y., S. F. Lin, T. Ellison, L. Y. Chen, C. Izumiya, P. Luciw, and H. J. Kung. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 771441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izumiya, Y., S. F. Lin, T. J. Ellison, A. M. Levy, G. L. Mayeur, C. Izumiya, and H. J. Kung. 2003. Cell cycle regulation by Kaposi's sarcoma-associated herpesvirus K-bZIP: direct interaction with cyclin-CDK2 and induction of G1 growth arrest. J. Virol. 779652-9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 743586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klass, C. M., L. T. Krug, V. P. Pozharskaya, and M. K. Offermann. 2005. The targeting of primary effusion lymphoma cells for apoptosis by inducing lytic replication of human herpesvirus 8 while blocking virus production. Blood 1054028-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 205769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamond, A. I., and D. L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4605-612. [DOI] [PubMed] [Google Scholar]
- 28.Liao, W., Y. Tang, S. F. Lin, H. J. Kung, and C. Z. Giam. 2003. K-bZIP of Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 (KSHV/HHV-8) binds KSHV/HHV-8 Rta and represses Rta-mediated transactivation. J. Virol. 773809-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, C. L., H. Li, Y. Wang, F. X. Zhu, S. Kudchodkar, and Y. Yuan. 2003. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J. Virol. 775578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindberg, A., and J. P. Kreivi. 2002. Splicing inhibition at the level of spliceosome assembly in the presence of herpes simplex virus protein ICP27. Virology 294189-198. [DOI] [PubMed] [Google Scholar]
- 31.Lukac, D. M., L. Garibyan, J. R. Kirshner, D. Palmeri, and D. Ganem. 2001. DNA binding by Kaposi's sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J. Virol. 756786-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 739348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majerciak, V., N. Pripuzova, J. P. McCoy, S. J. Gao, and Z. M. Zheng. 2007. Targeted disruption of KSHV ORF57 in the viral genome is detrimental for the expression of ORF59, K8α, and K8.1 and the production of infectious virus. J. Virol. 811062-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Majerciak, V., K. Yamanegi, S. H. Nie, and Z. M. Zheng. 2006. Structural and functional analyses of Kaposi's sarcoma-associated herpesvirus (KSHV) ORF57 nuclear localization signals in living cells. J. Biol. Chem. 28128365-28378. [DOI] [PubMed] [Google Scholar]
- 35.Majerciak, V., K. Yamanegi, and Z. M. Zheng. 2006. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J. Virol. 8011968-11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malik, P., D. J. Blackbourn, and J. B. Clements. 2004. The evolutionarily conserved Kaposi's sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J. Biol. Chem. 27933001-33011. [DOI] [PubMed] [Google Scholar]
- 37.Mayeda, A., and A. R. Krainer. 1999. Mammalian in vitro splicing assays. Methods Mol. Biol. 118315-321. [DOI] [PubMed] [Google Scholar]
- 38.Mayeda, A., and A. R. Krainer. 1999. Preparation of HeLa cell nuclear and cytosolic S100 extracts for in vitro splicing. Methods Mol. Biol. 118309-314. [DOI] [PubMed] [Google Scholar]
- 39.Miller, G., M. O. Rigsby, L. Heston, E. Grogan, R. Sun, C. Metroka, J. A. Levy, S. J. Gao, Y. Chang, and P. Moore. 1996. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N. Engl. J. Med. 3341292-1297. [DOI] [PubMed] [Google Scholar]
- 40.Nagai, K., Y. Muto, D. A. Pomeranz Krummel, C. Kambach, T. Ignjatovic, S. Walke, and A. Kuglstatter. 2001. Structure and assembly of the spliceosomal snRNPs. Biochem. Soc. Trans. 2915-26. [DOI] [PubMed] [Google Scholar]
- 41.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 714187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, J., T. Seo, S. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 7411977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2342-346. [DOI] [PubMed] [Google Scholar]
- 44.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 45.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl Acad. Sci.USA 9314862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruvolo, V., A. K. Gupta, and S. Swaminathan. 2001. Epstein-Barr virus SM protein interacts with mRNA in vivo and mediates a gene-specific increase in cytoplasmic mRNA. J. Virol. 756033-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruvolo, V., L. Sun, K. Howard, S. Sung, H. J. Delecluse, W. Hammerschmidt, and S. Swaminathan. 2004. Functional analysis of Epstein-Barr virus SM protein: identification of amino acids essential for structure, transactivation, splicing inhibition, and virion production. J. Virol. 78340-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruvolo, V., E. Wang, S. Boyle, and S. Swaminathan. 1998. The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc. Natl. Acad. Sci. USA 958852-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanford, J. R., J. Ellis, and J. F. Caceres. 2005. Multiple roles of arginine/serine-rich splicing factors in RNA processing. Biochem. Soc. Trans. 33443-446. [DOI] [PubMed] [Google Scholar]
- 51.Sickmier, E. A., K. E. Frato, H. Shen, S. R. Paranawithana, M. R. Green, and C. L. Kielkopf. 2006. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell 2349-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soares, L. M., K. Zanier, C. Mackereth, M. Sattler, and J. Valcarcel. 2006. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science 3121961-1965. [DOI] [PubMed] [Google Scholar]
- 53.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, and F. Sigaux. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 861276-1280. [PubMed] [Google Scholar]
- 54.Stark, H., P. Dube, R. Luhrmann, and B. Kastner. 2001. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature 409539-542. [DOI] [PubMed] [Google Scholar]
- 55.Staskus, K. A., R. Sun, G. Miller, P. Racz, A. Jaslowski, C. Metroka, H. Brett-Smith, and A. T. Haase. 1999. Cellular tropism and viral interleukin-6 expression distinguish human herpesvirus 8 involvement in Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. J. Virol. 734181-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sterner, D. A., T. Carlo, and S. M. Berget. 1996. Architectural limits on split genes. Proc. Natl. Acad. Sci. USA 9315081-15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 9510866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang, S., and Z. M. Zheng. 2002. Kaposi's sarcoma-associated herpesvirus K8 exon 3 contains three 5′-splice sites and harbors a K8.1 transcription start site. J. Biol. Chem. 27714547-14556. [DOI] [PubMed] [Google Scholar]
- 59.Taylor, J. L., H. N. Bennett, B. A. Snyder, P. S. Moore, and Y. Chang. 2005. Transcriptional analysis of latent and inducible Kaposi's sarcoma-associated herpesvirus transcripts in the K4 to K7 region. J. Virol. 7915099-15106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tormanen, H., E. Backstrom, A. Carlsson, and G. Akusjarvi. 2006. L4-33K, an adenovirus-encoded alternative RNA splicing factor. J. Biol. Chem. 28136510-36517. [DOI] [PubMed] [Google Scholar]
- 61.Urlaub, H., V. A. Raker, S. Kostka, and R. Luhrmann. 2001. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 20187-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Viejo-Borbolla, A., M. Ottinger, and T. F. Schulz. 2003. Human herpesvirus 8: biology and role in the pathogenesis of Kaposi's sarcoma and other AIDS-related malignancies. Curr. Infect. Dis. Rep. 5169-175. [DOI] [PubMed] [Google Scholar]
- 63.Wang, S. E., F. Y. Wu, Y. Yu, and G. S. Hayward. 2003. CCAAT/enhancer-binding protein-alpha is induced during the early stages of Kaposi's sarcoma-associated herpesvirus (KSHV) lytic cycle reactivation and together with the KSHV replication and transcription activator (RTA) cooperatively stimulates the viral RTA, MTA, and PAN promoters. J. Virol. 779590-9612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams, B. J., J. R. Boyne, D. J. Goodwin, L. Roaden, G. M. Hautbergue, S. A. Wilson, and A. Whitehouse. 2005. The prototype gamma-2 herpesvirus nucleocytoplasmic shuttling protein, ORF 57, transports viral RNA through the cellular mRNA export pathway. Biochem. J. 387295-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, F. Y., J. H. Ahn, D. J. Alcendor, W. J. Jang, J. Xiao, S. D. Hayward, and G. S. Hayward. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 751487-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, F. Y., Q. Q. Tang, H. Chen, C. ApRhys, C. Farrell, J. Chen, M. Fujimuro, M. D. Lane, and G. S. Hayward. 2002. Lytic replication-associated protein (RAP) encoded by Kaposi sarcoma-associated herpesvirus causes p21CIP-1-mediated G1 cell cycle arrest through CCAAT/enhancer-binding protein-alpha. Proc. Natl. Acad. Sci. USA 9910683-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu, F. Y., S. E. Wang, Q. Q. Tang, M. Fujimuro, C. J. Chiou, Q. Zheng, H. Chen, S. D. Hayward, M. D. Lane, and G. S. Hayward. 2003. Cell cycle arrest by Kaposi's sarcoma-associated herpesvirus replication-associated protein is mediated at both the transcriptional and posttranslational levels by binding to CCAAT/enhancer-binding protein alpha and p21(CIP-1). J. Virol. 778893-8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yamanegi, K., S. Tang, and Z. M. Zheng. 2005. Kaposi's sarcoma-associated herpesvirus K8β is derived from a spliced intermediate of K8 pre-mRNA and antagonizes K8α (K-bZIP) to induce p21 and p53 and blocks K8α-CDK2 interaction. J. Virol. 7914207-14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, M. Q. 1998. Statistical features of human exons and their flanking regions. Hum. Mol. Genet. 7919-932. [DOI] [PubMed] [Google Scholar]
- 70.Zheng, Z. M. 2003. Split genes and their expression in Kaposi's sarcoma-associated herpesvirus. Rev. Med. Virol. 13173-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng, Z. M. 2004. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 11278-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng, Z. M., and C. C. Baker. 2000. Parameters that affect in vitro splicing of bovine papillomavirus type 1 late pre-mRNAs. J. Virol. Methods 85203-214. [DOI] [PubMed] [Google Scholar]
- 73.Zheng, Z. M., M. Tao, K. Yamanegi, S. Bodaghi, and W. Xiao. 2004. Splicing of a Cap-proximal human papillomavirus 16 E6E7 intron promotes E7 expression, but can be restrained by distance of the intron from its RNA 5′ cap. J. Mol. Biol. 3371091-1108. [DOI] [PubMed] [Google Scholar]
- 74.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 936641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu, J., and A. R. Krainer. 2000. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 143166-3178. [DOI] [PMC free article] [PubMed] [Google Scholar]