Abstract
The untranslated leader of the human immunodeficiency virus type 1 (HIV-1) RNA genome encodes essential sequence and structural motifs that control various replication steps. The 5′ splice site or splice donor (SD) is embedded in a semistable hairpin, but the function of this structure is unknown. We stabilized this SD hairpin by creating an additional base pair and demonstrated a severe HIV-1 replication defect. A splicing defect was apparent in RNA analyses of virus-infected cells and cells transfected with appropriate reporter constructs. We selected multiple virus revertants in search for interesting second-site escape pathways. Most revertants acquired an additional mutation that modulated the stability of the mutant SD hairpin. One revertant acquired a single nucleotide change in the upstream DIS hairpin. We demonstrate that a novel SD site is created by this upstream mutation, which obviously reduces the number of leader nucleotides that are included in spliced HIV-1 transcripts. These results suggest a novel role of RNA structure in the regulation of HIV-1 splicing.
The human immunodeficiency virus type 1 (HIV-1) proviral DNA genome expresses a primary transcript of 9 kb that serves not only as genomic RNA for progeny virus but also as the mRNA that encodes the viral Gag and Gag-Pol proteins. Successful infection and production of new infectious viruses require the balanced expression of seven additional viral proteins. To achieve this proteomic diversity, the primary transcript is alternatively and incompletely spliced, and nuclear export of the unspliced transcript is regulated (21, 34-37). The process of HIV-1 RNA splicing is highly orchestrated. Several sequence motifs within the RNA are required for recognition by the cellular spliceosome, including the 5′ splice site or splice donor (SD), a branch point, and a 3′ splice site or splice acceptor (SA). HIV-1 uses multiple alternative 5′ and 3′ splice sites to generate more than 40 spliced mRNA species (32, 36, 38). Spliced mRNAs can be divided into two classes, namely, multiply spliced (∼1.8 kb) and singly spliced (∼4 kb) RNAs (Fig. 1A). In the early phase of HIV-1 gene expression, only the completely spliced mRNAs are transported to the cytoplasm for translation of the Tat, Rev, and Nef proteins. As the Rev protein accumulates, nuclear export of the singly and unspliced mRNAs is facilitated (27, 31). These mRNAs express the Vif, Vpr, Vpu, and Env proteins and the Gag and Gag-Pol polyproteins, respectively. For cellular gene expression, only intronless RNAs are exported from the nucleus (24). To circumvent this intron-dependent export block, both singly spliced and unspliced HIV-1 RNAs harbor the Rev-responsive element (RRE) in the env intron (Fig. 1A). Rev binds to the RRE and mediates nuclear export of the RRE-containing RNAs by delivering them to the Crm1-dependent pathway (14, 19, 20). To allow the expression of all HIV-1 transcripts, splicing of the primary transcript must be inefficient, allowing Rev to facilitate nuclear export before splicing is completed. Inefficient splicing of the HIV-1 primary transcript has been attributed to suboptimal 3′ SAs and noncanonical branch-point sequences (3, 18, 25, 34, 36, 38, 40). Furthermore, splice enhancers and silencers throughout the viral genome have been implicated in splicing regulation (12, 17, 41).
FIG. 1.
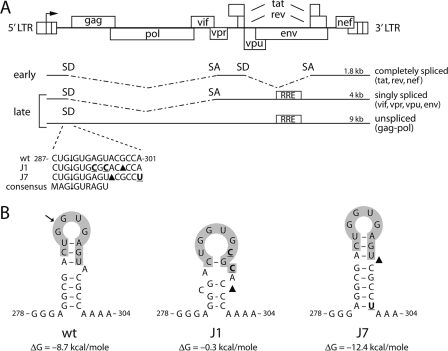
HIV-1 SD mutants. (A) The HIV-1 DNA genome and the three classes of spliced mRNAs are shown. The 5′ and 3′ LTRs are divided into three segments (U3, R, and U5). Transcription starts at the U3-R border (arrow). SD and SA sites are shown in a simplified scheme. The RRE is required to export the unspliced and singly spliced mRNAs from the nucleus to the cytoplasm. All spliced mRNAs are spliced at the 5′ major SD. The wt and mutant (J1 and J7) sequences of the major 5′ SD site are shown. Mutated nucleotides are indicated in bold and underlined, and deletions are indicated with black triangles. ↓, cleavage site. The consensus sequence is indicated underneath, with M indicating an A or C nucleotide and R indicating a purine. (B) The J1 and J7 mutations affect the SD hairpin structure. ΔG values were determined with the Mfold program and are indicated underneath the RNA structure. The J1 hairpin is unlikely to fold, since its ΔG value is close to zero.
We recently became interested in the HIV-1 5′ major SD, which is used in all splicing events. Mutation of the 5′ SD blocks all downstream splicing events (9). The 5′ major SD fits the consensus sequence MAG↓GURAGU (in which M is either C or A) in eight of nine positions and has been described as an efficient SD (34). The 5′ major SD is positioned in a semistable hairpin structure of the untranslated leader RNA (Fig. 1B). We previously mutated this SD hairpin as part of an extended mutational analysis of the leader RNA structure (2). The SD mutants exhibited a rather unusual phenotype, suggesting that the hairpin structure may affect RNA splicing. In this study, we tested the hypothesis that the local SD structure regulates HIV-1 splicing efficiency. The replication of SD mutant viruses was analyzed, and interesting revertant viruses were selected. A detailed splicing analysis of these mutant and revertant HIV-1 genomes and reporter constructs indicates that the activity of the 5′ SD can be suppressed by stable local RNA structure.
MATERIALS AND METHODS
DNA constructs.
The introduction of the J1 and J7 mutations into pLAI-R37 and pLTR-gag-flag-luc was previously described (2). pLAI-R37 is a derivative of the full-length infectious clone pLAI, containing a deletion in the 3′ R region (7). The pLTR-gag-flag-luc plasmid contains the HIV-1 5′ long terminal repeat (LTR) promoter region, the complete leader RNA, nucleotides 1 to 75 of the Gag coding sequence, an in-frame Flag coding sequence, and the firefly luciferase open reading frame. The fusion protein contains the N-terminal 25 Gag amino acids followed by the Flag peptide (amino acids DYKDDDDKD) and the firefly luciferase protein. The pRL-CMV plasmid contains the Renilla luciferase reporter gene under the control of the cytomegalovirus (CMV) promoter (Promega). pcDNA3-Tat (42) expresses the HIV-1 LAI Tat protein under the control of the CMV promoter and is a derivative of the pcDNA3 vector (Invitrogen). Plasmid DNA isolation was performed with a Qiagen plasmid isolation kit according to the manufacturer's protocol (Qiagen, Chatsworth, CA).
CA-p24 ELISA.
Culture supernatant was heat inactivated at 56°C for 30 min in the presence of 0.05% Empigen-BB (Calbiochem, La Jolla, CA). The CA-p24 concentration was determined by a twin-site enzyme-linked immunosorbent assay (ELISA), with D7320 (Biochrom, Berlin, Germany) as the capture antibody and alkaline phosphatase-conjugated anti-p24 monoclonal antibody (EH12-AP) as the detection antibody. Detection was done with a Lumiphos Plus system (Lumigen, MI) in a LUMIstar Galaxy (BMG Labtechnologies, Offenburg, Germany) luminescence reader. Recombinant CA-p24 expressed in a baculovirus system was used as the reference standard.
Virus production.
C33A cells, a human cervix carcinoma cell line, were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS), nonessential amino acids (Invitrogen), 20 mM glucose, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C and 5% CO2. Virus production was determined for C33A cell cultures (24-well plate) transfected with 1 μg proviral DNA by the calcium phosphate method. CA-p24 was measured by ELISA 2 days after transfection, both in the culture medium and intracellularly. Cells were lysed in phosphate-buffered saline (PBS) containing 1% Empigen-BB.
Virus replication and evolution studies.
The SupT1 T-cell line was cultured in RPMI 1640 medium supplemented with 10% (vol/vol) FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C and 5% CO2. SupT1 T cells (10 × 106) were transfected with 250 ng proviral DNA by electroporation (250 V, 975 μF), using a Bio-Rad Gene Pulser II instrument (Bio-Rad, Hercules, CA), or were infected with viruses produced in C33A cells (equivalent to 1 ng CA-p24). Cells were split 1 to 10 twice a week. CA-p24 levels in the culture medium were determined by ELISA. The protocol for virus evolution by prolonged cell-free passage of virus on fresh, uninfected SupT1 cells was described previously (30). Isolation of total cellular DNA was performed by proteinase K treatment (4). The LTR-leader region was PCR amplified with primers T7-1 and TA014 (2). The PCR products were sequenced directly, thus providing the population sequence of the viral quasispecies, and subsequently used for cloning purposes. The PCR product with the J7 mutations and the C267U reversion was termed J7R.
The J7R PCR product, containing the leader-gag-flag sequences, was cloned into the pCRII-TOPO vector (Invitrogen). The HindIII-NcoI fragment was subsequently cloned into pLTR-gag-flag-luc. In addition, the HindIII-ClaI fragment was cloned into Blue-5′LTR digested with the same restriction enzymes, rendering Blue-J7R-LTR. This pBluescript-derived construct (29) contains the XbaI-ClaI fragment of the infectious pLAI clone, including the 5′ LTR promoter sequence, the full-length leader sequence, and part of the Gag open reading frame (positions −454 to +376 relative to the transcriptional start site at +1). The J7R XbaI-ClaI fragment was subsequently cloned into pLAI-R37. The resulting constructs were named pLTR-gag-flag-luc-J7R and pLAI-J7R. All constructs were verified by restriction enzyme digestion and BigDye Terminator sequencing (Applied Biosystems, Foster City, CA) on an automatic sequencer (Applied Biosystems 377 DNA sequencer).
Luciferase expression.
Reporter gene expression driven by the wild-type (wt) and mutant leader RNAs was analyzed in C33A cells. The cells (24-well plates) were transfected with 1 μg calcium phosphate-precipitated luciferase constructs in the presence of 5 ng pcDNA3-Tat or pcDNA3 (16, 42). pRL-CMV (5 ng) expressing Renilla luciferase was cotransfected as an internal control. Cells were washed with PBS 2 days after transfection and lysed in passive lysis buffer provided by a dual-luciferase reporter assay system (Promega). The firefly and Renilla luciferase activities were determined according to the manufacturer's protocol. Firefly luciferase activity was normalized to Renilla luciferase activity.
Single-cycle infection.
TZM-bl cells (43, 44) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, nonessential amino acids (Invitrogen), 20 mM glucose, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5% CO2. For transient transfection assays, approximately 5 × 104 cells were seeded into 24-well plates to 40 to 60% confluence. TZM-bl cells were infected with equal amounts of virus produced in C33A cells (10 ng CA-p24) in the presence of 80 μg/ml DEAE-dextran. At 2 days postinfection, cells were washed with PBS and lysed in passive lysis buffer (Promega). Firefly luciferase in cell lysates was measured using assay reagents from Promega.
Splice site analysis.
C33A cells were transfected with 1 μg of a wt or mutant luciferase construct in the presence of 50 ng pcDNA3-Tat or pcDNA3 (16, 42). Cells were washed with PBS 2 days after transfection, and total RNA was isolated by the method of Boom et al. (10). Total RNA was used as a template for reverse transcription (RT) at 60°C (ThermoScript RT-PCR system; Invitrogen), using the reverse oligonucleotide primer TA113 (GCATACGACGATTCTGTG), which is complementary to nucleotides (nt) 666 to 683 of the HIV gag-Flag-luc mRNA (nt 226 to 243 of the luciferase open reading frame).
C33A cells were transfected with 1 μg of a wt or mutant HIV-1 molecular clone. At 2 days posttransfection, cells were harvested and washed with PBS. Total RNA was isolated according to the Boom method and used for RT, using a Tat intron primer complementary to nt 5725 to 5703 of the HIV-1 LAI genome. The luciferase and HIV-1 cDNAs were subsequently amplified in a standard PCR, with a forward oligonucleotide primer that corresponds to nt 1 to 18 of the HIV-1 leader downstream of a T7 promoter sequence. The products were analyzed in a 1.5% ethidium bromide-containing agarose gel and cloned into the pCRII-TOPO vector (Invitrogen) for sequence analysis.
RNA secondary structure prediction.
Computer-assisted RNA secondary structure predictions were performed using the Mfold, version 3.0, algorithm (33, 46) offered by the MBCMR Mfold server (http://mfold.burnet.edu.au/). Standard settings were used for all folding jobs (37°C and 1.0 M NaCl). Folding was performed with sequences comprising nt 1 to 368 of the genomic RNA sequence of the wt and mutant HIV-1 leader RNAs.
RESULTS
Mutation of HIV-1 SD hairpin structure.
The 5′ major SD is positioned in a semistable stem-loop structure (SD hairpin) in the HIV-1 leader RNA (Fig. 1). The SD is used for the generation of all spliced viral RNAs. Previous studies showed that certain mutations in the SD hairpin (J1 and J7) (Fig. 1B) affect expression from an HIV-1 LTR-leader luciferase reporter gene (2). The J1 mutations change two nucleotides of the consensus splice site sequence (Fig. 1A). In addition, these mutations destabilize the SD hairpin by disrupting one C-G and two A-U base pairs (Fig. 1B). The J7 mutations leave the consensus sequence intact and stabilize the SD hairpin because of a deletion of the bulged A nucleotide and an extension of the stem with an additional A-U base pair (Fig. 1). The J1 and J7 mutations may affect mRNA splicing through different mechanisms, as the J1 mutations destroy the SD consensus sequence, whereas the J7 mutations decrease the accessibility of the SD consensus sequence.
Impaired replication of SD mutant viruses.
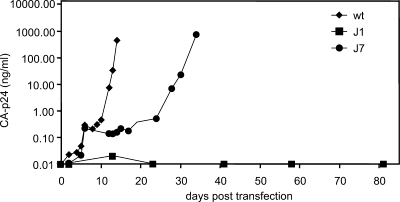
We analyzed the effect of these mutations on virus replication. Molecular clones with the wt and mutant leader sequences were transfected into the SupT1 T-cell line, and viral replication was monitored for several months (Fig. 2). The J1 mutant virus did not replicate at all, even when large amounts of input DNA were used for transfection (results not shown). The effect of the J7 mutations on virus replication was not as severe but was still very significant. Replication of the J7 virus was much delayed compared to that of the wt virus. Infection assays with an equal amount of input virus gave similar results (results not shown).
FIG. 2.
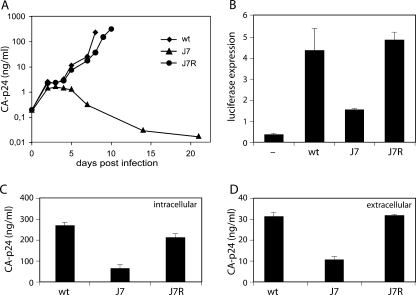
Replication of wt and SD mutant viruses. SupT1 cells were transfected with 250 ng of wt or mutant molecular clones. Virus production was measured in the culture medium by CA-p24 ELISA at several days posttransfection.
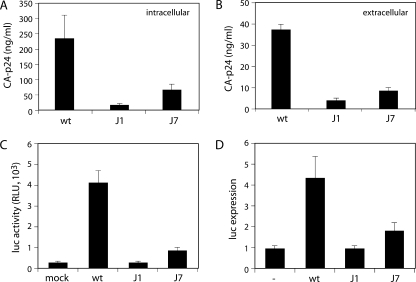
Subsequently, we determined which part of the replication cycle, the production phase or the entry/integration phase, was affected by the SD mutations. The production phase was analyzed by monitoring virus production in nonpermissive C33A cells transfected with the wt and mutant molecular clones. CA-p24 production was quantified as a measure of virus production in cell lysates and the culture medium (Fig. 3A and B, respectively). Virus gene expression was greatly decreased for the J1 and J7 mutants. The decrease in protein production of the J1 mutant was more severe than that of the J7 mutant. The production of Env protein was similarly decreased by the SD mutations (results not shown). These results indicate that virus production is impaired by the J1 and J7 mutations due to decreased protein production.
FIG. 3.
Characterization of wt and SD mutant viruses. (A and B) SD mutations decrease CA-p24 expression intracellularly (A) and extracellularly (B). C33A cells were transfected with wt or mutant proviral clones and lysed at 2 days posttransfection, and CA-p24 was determined by ELISA in the lysates and culture medium. (C) Infectivity of SD mutant viruses is severely decreased. TZM-bl indicator cells that express luciferase from a 5′ LTR promoter were infected with the indicated viruses. Luciferase activity was measured at 2 days postinfection. RLU, relative light units. (D) Tat production is greatly reduced by SD mutations. Tat protein production was determined in cotransfection assays with the indicated wt or mutant molecular clones and an LTR-luciferase expression construct. The basal expression level was determined in the absence of proviral DNA (lane labeled with a dash).
To analyze the entry/integration phase of the viral life cycle, equal amounts of wt and mutant viruses were used to infect TZM-Bl cells. These cells express the HIV-1 receptors and carry an LTR-driven luciferase transgene. Viruses that successfully complete entry, RT, and integration produce the Tat protein, which trans-activates the LTR-luciferase transgene. In the absence of virus, Tat is not produced and the basal LTR promoter activity drives luciferase expression (Fig. 3C, mock). Infection by wt HIV-1 increased luciferase expression approximately 40-fold (Fig. 3C). The J1 and J7 mutants showed defects in luciferase induction. The J1 mutant did not stimulate luciferase expression at all, whereas the J7 mutant upregulated luciferase expression sevenfold (Fig. 3C).
These combined results suggest that the J1 and J7 mutations cause defects in both phases of the viral life cycle. However, they could also be explained by one defect in RNA splicing that affects both assays. If splicing is inhibited, the amounts of tat and rev mRNAs will decrease, resulting in reduced levels of Tat and Rev proteins. Consequently, Tat-induced upregulation of viral gene expression in C33A cells and luciferase expression in HeLa TZM-bl cells will be reduced. In addition, Rev levels may be too low to facilitate the nuclear export of the unspliced and singly spliced mRNAs, thus abrogating the expression of Gag, Gag-Pol, and Env in C33A cells. As a direct test for the amount of Tat produced from the J1 and J7 proviral genomes, we cotransfected the molecular clones with an LTR-luciferase construct (Fig. 3D). Luciferase production in the absence of molecular clones represents the basal LTR promoter activity (lane labeled with a dash). The wt HIV-1 DNA upregulated luciferase expression severalfold, in a dose-dependent manner. Under linear assay conditions, we observed a fourfold stimulation. The J1 molecular clone failed to upregulate luciferase expression and therefore did not seem to express a detectable level of Tat protein. The J7 construct showed significantly decreased luciferase induction. These results are in agreement with the proposed gene expression defect, possibly due to an effect on splicing caused by mutation of the SD signal or structure.
Evolution of HIV-1 mutants with a stabilized SD hairpin.
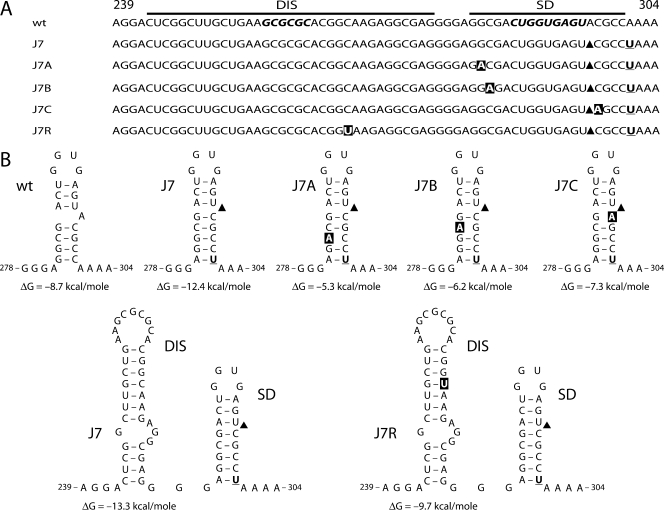
To further understand the effect of the SD mutations on RNA splicing, we performed evolution studies with the J1 and J7 viruses. We started infections by transfection of SupT1 T cells with large amounts of the proviral clones and prolonged six independent cultures for each construct for a considerable time. If virus replication was observed, viruses were passaged at the peak of infection onto fresh uninfected cells. The leader region of the passaged viruses was sequenced to check for the presence of the original mutations and the occurrence of second-site reversions. All attempts to obtain a replicating J1 virus variant were unsuccessful. In contrast, all J7-infected cultures yielded quickly replicating viruses. Interestingly, the J7 viruses maintained the original SD mutations, but additional mutations were acquired during the course of the evolution experiment (Fig. 4A). These adaptations were divided into two classes, namely, mutations located within the SD hairpin (J7A, -B, and -C) and a true second-site mutation in the upstream region that encodes the RNA dimerization signal, or DIS hairpin (J7R). The J7A reversion was observed in three of six cultures, suggesting a preferential escape route, which may be triggered by the relatively easy G-to-A transition (6). None of the acquired mutations affects the SD consensus sequence, and J7R does not affect the DIS palindrome that is used in RNA dimerization. We analyzed the effect of the reversions on the RNA secondary structure with the use of Mfold software (Fig. 4B). The J7A, -B, and -C reversions destabilize the mutant SD hairpin, such that wt stability is restored. These results strongly suggest that the stabilized J7 SD hairpin causes a drop in viral fitness, which is restored in the revertants. Interestingly, the upstream DIS mutation in J7R does not change the stabilized SD hairpin and only slightly reduces the stability of the DIS hairpin. No simple mechanistic explanation for the J7R reversion is apparent, and we therefore decided to focus on this intriguing second-site repair pathway.
FIG. 4.
Forced evolution of the J7 SD mutant yields two classes of virus revertants. (A) Six cultures of SupT1 cells were transfected with the J7 proviral clone. Over time, replicating viruses were observed in all cultures, and viruses were passaged onto fresh cells at the peak of infection. In addition, chromosomal DNA was isolated from infected cells at the time of passage, and the leader region of the integrated proviral genome was sequenced, of which only the DIS-SD region is depicted. The J7 mutations (indicated in bold and underlined; black triangles indicate the deletion) remained present. Nucleotide changes acquired during evolution are shown in white in black boxes. Nucleotide positions are indicated. The J7A mutation (G283A) was selected in three cultures. (B) Effects of J7 variants on DIS and SD hairpins. The J7A, J7B, and J7C mutations destabilize the SD hairpin by disrupting a base pair. The ΔG values are indicated below the RNA structures. The J7R reversion leaves the stabilized SD hairpin intact but slightly destabilizes the DIS hairpin by altering a G-C to a G-U base pair. The ΔG values of the wt and J7R DIS hairpins are indicated.
The DIS mutation restores protein expression and virus replication.
We tested whether the J7R reversion in the DIS hairpin was indeed responsible for improved replication of the J7 mutant. We introduced the mutation into the original J7 virus and infected the SupT1 T-cell line. Indeed, the mutation greatly improved the replication of the J7 mutant (Fig. 5A). The J7R revertant showed a slight replication delay compared to the wt, and a similar small defect was observed in the TZM-bl infection assay (results not shown). Tat production was restored to wt levels in a cotransfection assay with the molecular clone and an LTR-luciferase reporter (Fig. 5B). Virus production measured by intra- and extracellular CA-p24 protein expression was also restored to wt levels (Fig. 5C and D, respectively), and Env protein production was largely restored (results not shown). The J7R mutation clearly rescues the detrimental effect of SD hairpin stabilization on viral replication and protein expression.
FIG. 5.
The J7R mutation restores protein production and virus replication. (A) Replication of wt, J7, and J7R mutant viruses. SupT1 cells were infected with equal amounts of viruses. CA-p24 production was measured in the culture medium at several days postinfection. (B) Tat expression is restored to wt levels by the J7R mutation. See the legend to Fig. 3D for details. (C and D) CA-p24 expression in C33A cells (C) and in culture medium (D) at 2 days posttransfection.
Surprisingly, the J7R mutation does not directly affect the stability of the extended SD hairpin and only moderately affects the stability of the DIS hairpin. It is possible that DIS hairpin destabilization has an indirect effect on the folding and accessibility of the SD. Mfold analyses do not support this hypothesis. In addition, the stability of the DIS hairpin regulates the LDI/BMH riboswitch of the HIV-1 leader RNA (23). The SD region is folded differently in the long distance interaction (LDI) and branched multiple hairpin (BMH) conformations, and the equilibrium may therefore also influence pre-mRNA splicing (1). However, in vitro RNA folding and dimerization assays do not support this possibility (results not shown).
The DIS mutation creates an alternative SD.
The results thus far show that SD mutations impair viral gene expression. To establish that this inhibition is caused by an RNA splicing defect, we determined the RNA splicing efficiencies of luciferase constructs (Fig. 6A). We transfected C33A cells with the wt, mutant, and revertant constructs in the presence or absence of a Tat expression vector (Fig. 6B). Luciferase expression was slightly but consistently improved by the J7 mutations in both the absence and presence of Tat, as previously described (2). Most importantly, the J7R mutant restored the wt expression level in both the absence and presence of Tat.
FIG. 6.
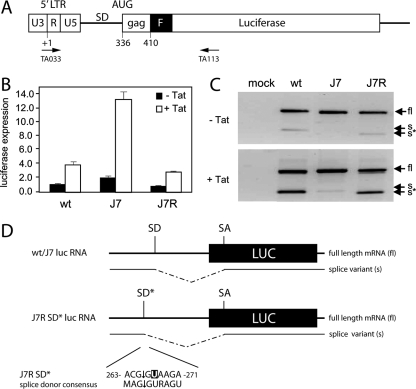
Effect of J7R mutation on gene expression. (A) Luciferase reporter. See the legend to Fig. 1 for details. Primers used for RT-PCR analysis are indicated. (B) wt and mutant HIV-1 leader RNA-driven luciferase expression. Luciferase constructs were transfected into C33A cells, and pcDNA3 (white bars) or pcDNA3-Tat (black bars) was cotransfected. pRL-CMV was cotransfected as an internal control. Transfections were performed in triplicate. The graph shows normalized firefly luciferase activities. Similar results were obtained in three independent experiments. (C) Analysis of luciferase mRNA expressed by the wt and mutant reporter constructs. Transfection assays were performed as described for panel B. Total RNA was isolated at 2 days posttransfection and reverse transcribed with primer TA113. The cDNA was subsequently amplified with the TA033 and TA113 primers. The PCR products were applied to agarose gels. Arrows indicate PCR products derived from the unspliced, full-length (fl), and spliced (s) luciferase mRNAs. The J7R lane shows a shorter splice product (s*). In the presence of Tat, the ratio between fl and s or s* products is altered. A control reaction in which reverse transcriptase was omitted from the RT-PCR did not yield any amplified products (results not shown). (D) The J7R mutation results in the formation of an SD* site that results in splicing at nt 265 of the leader RNA. The fl, s, and s* RT-PCR products were cloned and sequenced. All wt and J7 mutant s products were spliced at the 5′ major SD, whereas the J7R mutant s* was spliced at the alternative major splice site (SD*) in the DIS region (nt 265). The sequence surrounding the J7R mutation constitutes an SD consensus sequence. For both s and s* mRNAs, the 3′ SA site is located at nt 569 of the luciferase transcript.
To determine whether altered levels of RNA splicing are responsible for these changes in gene expression, total RNA was isolated from transfected cells and reverse transcribed. The cDNA products were amplified with a standard PCR. Since the luciferase sequence does not carry a known 3′ SA, we used reverse primers that are complementary to different locations in the luciferase mRNA to identify aberrant splice products. The primer TA113 (Fig. 6A) detected both unspliced and spliced transcripts (Fig. 6C). The splicing event was confirmed by cDNA sequencing, indicating that the HIV-1 5′ SD is joined to a cryptic 3′ SA at position 570 in the luciferase mRNA (Fig. 6D). This observation supports our hypothesis that splicing occurs on the luciferase mRNA and consequently affects protein expression. The J7 sample did not yield the spliced product, further supporting our hypothesis that stabilization of the SD hairpin prevents RNA splicing, which triggers increased luciferase expression. The J7R mutation corrected the splicing defect, consistent with reduced luciferase expression (Fig. 6B). However, close examination of the migration pattern shows that the J7R splice product is smaller than the wt product. Strikingly, sequencing of this PCR product indicated that the cryptic luciferase 3′ SA was not linked to the major 5′ SD but to an upstream DIS sequence near the C267U reversion mutation. Indeed, nt 263 to 271 of the J7R leader RNA (ACG↓GUAAGA) constitute an SD consensus sequence (MAG↓GURAGU). Apparently, stabilization of the SD hairpin causes a splicing defect that is restored by creation of an alternative SD* in the upstream DIS region.
The alternative SD* is used as a major SD in the revertant virus.
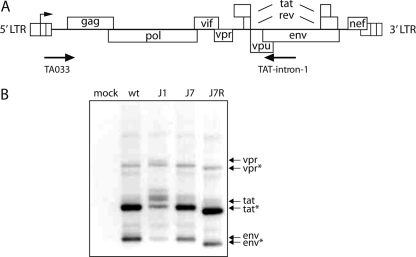
Finally, we determined whether the alternative 5′ SD* is used for the generation of spliced mRNAs in HIV-1. Proviral constructs were transfected into C33A cells, and total RNA was isolated and subjected to RT with an env primer (Fig. 7A). This reaction will generate cDNAs of the 4-kb singly spliced RNAs. The cDNAs were amplified by PCR, and the products were analyzed in an agarose gel (Fig. 7B). The PCR products from the wt samples were as expected and were verified by sequencing as vpr, tat, and env RNAs. The J1 mutant with a defective SD was used as a negative control. Hardly any splice products and a few aberrant PCR products were detected. The J7 mutant yielded the same three products as the wt, but with reduced intensities. The J7R mutation seemed to restore the intensities of the splice products, but they migrated faster than the wt PCR products, and sequence analysis confirmed that splicing connected the alternative SD* to the canonical 3′ SA sites.
FIG. 7.
The alternative SD* is used as a 5′ splice site in J7R viral RNA. (A) The HIV-1 DNA genome and the positions of oligonucleotide primers used for RT-PCR are shown. The Tat-intron-1 primer is located in the env region. RT-PCR is able to detect all spliced mRNAs of the 4-kb class (singly spliced; also see Fig. 1). (B) Identification of wt and mutant splice variants. C33A cells were transfected with proviral constructs. Total RNA was isolated at 2 days posttransfection and reverse transcribed. The cDNA was amplified and analyzed via agarose gel electrophoresis. The J7R mutation resulted in a population of smaller mRNAs than those observed for the wt and J7 mutant viruses. Sequence analysis indicated that the alternative SD* was used for splicing in all clones (eight of eight clones analyzed).
DISCUSSION
Splicing of the HIV-1 RNA genome is highly orchestrated to yield the proper ratio of more than 40 alternatively spliced mRNAs and the unspliced RNA. The major 5′ SD in the untranslated leader RNA is used in all spliced mRNAs. Because the 5′ SD is part of a semistable RNA hairpin structure, we set out to test if a stable RNA structure can modulate the splicing efficiency. Stabilization of the SD hairpin results in a severe virus replication defect that is caused by reduced expression of the Tat and Rev proteins, which are needed for transcriptional activation of viral gene expression and expression of the unspliced and singly spliced mRNAs, respectively. A similar phenotype was observed for a virus with a mutated SD sequence. We were able to select revertant viruses that maintained the original mutations but acquired additional mutations that weakened the stabilized SD hairpin. We can thus exclude the possibility that the SD structure mutant is impaired in replication because of changes in the nucleotide sequence. These results demonstrate that RNA structure can be used to suppress a splice signal, and we propose that the SD hairpin regulates splicing efficiency. This adds another regulatory mechanism to the extremely coordinated process of HIV-1 splicing.
To directly test this scenario of splicing regulation in the wt virus by RNA structure, we cloned the observed second-site reversion mutations in the context of the wt virus to destabilize the SD hairpin. Because no significant replication defect was observed (results not shown), we also combined these mutations to more dramatically destabilize the SD hairpin. These attempts did not induce a clear replication defect, suggesting that the wt SD hairpin does not play a pivotal role in virus replication, at least in cell culture infections. More sensitive experiments, e.g., competition assays and infection of primary cells, are required to fully address this issue. The SD mutations were also cloned into the luciferase expression cassette, and preliminary results indicate small effects on luciferase activity that are in agreement with enhanced RNA splicing. However, more detailed experimentation, including the generation and analysis of more mutants, is required to confirm these findings.
We selected one unique virus revertant that maintained the stabilized SD hairpin but acquired a single nucleotide substitution in the upstream DIS hairpin. We demonstrated that a new SD was created, with seven of nine matches with the consensus SD sequence. The substitution created a GU dinucleotide in the SD* site, which is nearly invariant in all SDs and greatly determines splicing efficiency (13, 36). The creation of a new SD site provides independent evidence for our conclusion that splicing is hampered by placement of the SD in an excessively stable hairpin structure. Apparently, the DIS hairpin is not sufficiently stable to suppress this new SD site. Two factors may contribute to this. First, the reversion mutation weakens the DIS hairpin by replacing a G-C with a G-U base pair. In fact, Mfold analysis indicates folding of an alternative structure in which the SD* is accessible for the splicing machinery. Second, the revertant leader RNA may have shifted toward the LDI conformation rather than the DIS hairpin-containing BMH conformation (1, 23). However, in vitro RNA dimerization assays do not support this idea (results not shown).
This evolutionary escape provides another example of the extreme possibilities of HIV-1 adaptation, even in the conserved leader region. A new SD site can be created by a single nucleotide change, apparently without destroying the natural function of the DIS hairpin in RNA dimerization, although this function is less important than previously thought (8, 22). In addition, usage of the new SD* excludes 24 nt in the spliced mRNAs, which apparently does not result in a drastic effect on virus replication. There are additional indications that HIV-1 can easily overcome a mutational attack on the SD site. Two groups mutated the 5′ major SD and reported the usage of cryptic SD sites 4 and 50 nt further downstream (11, 36). Furthermore, analysis of aberrant splice products of the J1 mutant (Fig. 7B) indicated the usage of multiple alternative SD sites at positions 265, 339, and 349.
We propose that RNA structure can downmodulate a strong SD sequence. How would this operate in mechanistic terms? It is likely that the initial step of splicing is blocked by RNA structure, that is, binding of U1 snRNA to the SD. Thus, RNA structure is likely to restrict the accessibility of the SD sequence, but this should be verified with biochemical assays. HIV-1 needs to prevent constitutive splicing in order to allow the expression of the singly spliced and unspliced RNAs. To do so, suppression of the first RNA splicing step, that is, the binding of U1 snRNP, seems an attractive approach. It has previously been shown that destruction of the 5′ major SD abolishes all downstream splicing events (9). Our results suggest that local RNA structure mediates this SD suppression. A similar RNA structure phenomenon was described for the cellular SMN and Tau genes (26, 39). Tau is involved in frontotemporal dementia and parkinsonism associated with chromosome 17 (FTDP17). Tau proteins are expressed as two isoforms. The ratio between both isoforms is 1:1 in the normal cerebral cortex of human adults. Some FTDP17 patients harbor mutations that destabilize an RNA structure at the 5′ SD of exon 10 (SD10). Consequently, splicing is enhanced at this position, which results in a shifted ratio of Tau isoforms and subsequent disease development. Binding of U1 snRNP to SD10 was indeed increased for the mutant mRNAs, suggesting that the decreased hairpin stability increased the accessibility of the SD10 consensus sequence for the U1 snRNP (26). In addition, RNA structure was shown to affect SA usage in the HIV-1 genome (25). The SA3 site is required for Tat production and is located in an RNA stem-loop structure, similar to the 5′ major SD. RNA structure was proposed to modulate the pre-mRNA interaction with the splicing apparatus. Apparently, RNA structure can be used to control the recognition of RNA sequence motifs. In our laboratory, we have observed this type of regulation for several RNA functions in the HIV-1 life cycle, such as polyadenylation and reverse transcription (4, 5, 15, 28, 45).
Acknowledgments
We thank Rogier Sanders, Fokla Zorgdrager, and Bep Klaver for technical support and Wim van Est for artwork.
RNA studies in the laboratory of Ben Berkhout are supported by NWO-CW (TOP grant) and ZonMw (Vici program).
Footnotes
Published ahead of print on 26 December 2007.
REFERENCES
- 1.Abbink, T. E. M., and B. Berkhout. 2003. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the gag start codon. J. Biol. Chem. 27811601-11611. [DOI] [PubMed] [Google Scholar]
- 2.Abbink, T. E. M., M. Ooms, P. C. J. Haasnoot, and B. Berkhout. 2005. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry 449058-9066. [DOI] [PubMed] [Google Scholar]
- 3.Amendt, B. A., Z.-H. Si, and C. M. Stoltzfus. 1995. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol. Cell. Biol. 154606-4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beerens, N., and B. Berkhout. 2002. The tRNA primer activation signal in the HIV-1 genome is important for initiation and processive elongation of reverse transcription. J. Virol. 762329-2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beerens, N., F. Groot, and B. Berkhout. 2001. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 27631247-31256. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout, B., A. T. Das, and N. Beerens. 2001. HIV-1 RNA editing, hypermutation and error-prone reverse transcription. Science 2927. [DOI] [PubMed] [Google Scholar]
- 7.Berkhout, B., J. van Wamel, and B. Klaver. 1995. Requirements for DNA strand transfer during reverse transcription in mutant HIV-1 virions. J. Mol. Biol. 25259-69. [DOI] [PubMed] [Google Scholar]
- 8.Berkhout, B., and J. L. B. van Wamel. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 706723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bohne, J., H. Wodrich, and H. G. Krausslich. 2005. Splicing of human immunodeficiency virus RNA is position-dependent suggesting sequential removal of introns from the 5′ end. Nucleic Acids Res. 33825-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-van Dillen, and J. Van der Noordaa. 1990. A rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borg, K. T., J. P. Favaro, S. J. Arrigo, and M. Schmidt. 1999. Activation of a cryptic splice donor in human immunodeficiency virus type-1. J. Biomed. Sci. 645-52. [DOI] [PubMed] [Google Scholar]
- 12.Caputi, M., A. Mayeda, A. R. Krainer, and A. M. Zahler. 1999. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 184060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartegni, L., S. L. Chew, and A. R. Krainer. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat. Rev. Genet. 3285-298. [DOI] [PubMed] [Google Scholar]
- 14.Daly, T. J., K. S. Cook, G. S. Gray, T. E. Maione, and J. R. Rusche. 1989. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 342816-819. [DOI] [PubMed] [Google Scholar]
- 15.Das, A. T., B. Klaver, and B. Berkhout. 1999. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J. Virol. 7381-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, A. T., B. Klaver, B. I. F. Klasens, J. L. B. van Wamel, and B. Berkhout. 1997. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 712346-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domsic, J. K., Y. Wang, A. Mayeda, A. R. Krainer, and C. M. Stoltzfus. 2003. Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol. Cell. Biol. 238762-8772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dyhr-Mikkelsen, H., and J. Kjems. 1995. Inefficient spliceosome assembly and abnormal branch site selection in splicing of an HIV-1 transcript in vitro. J. Biol. Chem. 27024060-24066. [DOI] [PubMed] [Google Scholar]
- 19.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82475-483. [DOI] [PubMed] [Google Scholar]
- 20.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 901051-1060. [DOI] [PubMed] [Google Scholar]
- 21.Frankel, A. D., and J. A. Young. 1998. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 671-25. [DOI] [PubMed] [Google Scholar]
- 22.Hill, M. K., M. Shehu-Xhilaga, S. M. Campbell, P. Poumbourios, S. M. Crowe, and J. Mak. 2003. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J. Virol. 778329-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huthoff, H., and B. Berkhout. 2001. Two alternating structures for the HIV-1 leader RNA. RNA 7143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izaurralde, E., and I. W. Mattaj. 1995. RNA export. Cell 81153-159. [DOI] [PubMed] [Google Scholar]
- 25.Jacquenet, S., D. Ropers, P. S. Bilodeau, L. Damier, A. Mougin, C. M. Stoltzfus, and C. Branlant. 2001. Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res. 29464-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang, Z., J. Cote, J. M. Kwon, A. M. Goate, and J. Y. Wu. 2000. Aberrant splicing of tau pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell. Biol. 204036-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, S., R. Byrn, J. Groopman, and D. J. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 633708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klasens, B. I. F., A. T. Das, and B. Berkhout. 1998. Inhibition of polyadenylation by stable RNA secondary structure. Nucleic Acids Res. 261870-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klaver, B., and B. Berkhout. 1994. Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J. Virol. 683830-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klaver, B., and B. Berkhout. 1994. Evolution of a disrupted TAR RNA hairpin structure in the HIV-1 virus. EMBO J. 132650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klotman, M. E., S. Kim, A. Buchbinder, A. DeRossi, D. Baltimore, and F. Wong-Staal. 1991. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc. Natl. Acad. Sci. USA 885011-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutzelberger, M., L. S. Reinert, A. T. Das, B. Berkhout, and J. Kjems. 2006. A novel splice donor site in the gag-pol gene is required for HIV-1 RNA stability. J. Biol. Chem. 28118644-18651. [DOI] [PubMed] [Google Scholar]
- 33.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288911-940. [DOI] [PubMed] [Google Scholar]
- 34.O'Reilly, M. M., M. T. McNally, and K. L. Beemon. 1995. Two strong 5′ splice sites and competing, suboptimal 3′ splice sites involved in alternative splicing of human immunodeficiency virus type 1 RNA. Virology 213373-385. [DOI] [PubMed] [Google Scholar]
- 35.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52491-532. [DOI] [PubMed] [Google Scholar]
- 36.Purcell, D. F. J., and M. A. Martin. 1993. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 676365-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabson, A. B., and B. J. Graves. 1997. Synthesis and processing of viral RNA, p. 205-262. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 38.Schwartz, S., B. K. Felber, D. M. Benko, E. M. Fenyo, and G. N. Pavlakis. 1990. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J. Virol. 642519-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Si, Z., B. A. Amendt, and C. M. Stoltzfus. 1997. Splicing efficiency of human immunodeficiency virus type 1 tat RNA is determined by both a suboptimal 3′ splice site and a 10 nucleotide exon splicing silencer element located within tat exon 2. Nucleic Acids Res. 25861-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh, N. N., R. N. Singh, and E. J. Androphy. 2007. Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 35371-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staffa, A., and A. Cochrane. 1994. The tat/rev intron of human immunodeficiency virus type 1 is inefficiently spliced because of suboptimal signals in the 3′ splice site. J. Virol. 683071-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tange, T. O., C. K. Damgaard, S. Guth, J. Valcarcel, and J. Kjems. 2001. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 205748-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verhoef, K., M. Koper, and B. Berkhout. 1997. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology 237228-236. [DOI] [PubMed] [Google Scholar]
- 44.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422307-312. [DOI] [PubMed] [Google Scholar]
- 46.Westerhout, E. M., M. Ooms, M. Vink, A. T. Das, and B. Berkhout. 2005. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 33796-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuker, M., and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.