Abstract
Sulfur-containing compounds play an important role in plant stress defense; however, only a little is known about the molecular mechanisms of regulation of sulfate assimilation by stress. Using known Arabidopsis (Arabidopsis thaliana) mutants in signaling pathways, we analyzed regulation of the key enzyme of sulfate assimilation, adenosine 5′-phosphosulfate reductase (APR), by salt stress. APR activity and mRNA levels of all three APR isoforms increased 3-fold in roots after 5 h of treatment with 150 mm NaCl. The regulation of APR was not affected in mutants deficient in abscisic acid (ABA) synthesis and treatment of the plants with ABA did not affect the mRNA levels of APR isoforms, showing that APR is regulated by salt stress in an ABA-independent manner. In mutants deficient in jasmonate, salicylate, or ethylene signaling, APR mRNA levels were increased upon salt exposure similar to wild-type plants. Surprisingly, however, APR enzyme activity was not affected by salt in these plants. The same result was obtained in mutants affected in cytokinin and auxin signaling. Signaling via gibberellic acid, on the other hand, turned out to be essential for the increase in APR mRNA by salt treatment. These results demonstrate an extensive posttranscriptional regulation of plant APR and reveal that the sulfate assimilation pathway is controlled by a complex network of multiple signals on different regulatory levels.
During their lifetime, plants are exposed to a variety of biotic and abiotic stresses. The common feature of the stresses is the generation of reactive oxygen species (ROS), which are potentially damaging to cell structures and components. Detoxification of ROS is essential to limit oxidative stress and a key mechanism for this is the ascorbate-glutathione cycle, which plays a pivotal role in defense (Noctor and Foyer, 1998). Therefore, glutathione synthesis is often induced under stress conditions, including salt stress, and the capacity for its synthesis is correlated with tolerance to various stresses (Noctor et al., 1998; Kocsy et al., 2001, 2004; Ruiz and Blumwald, 2002; Mittova et al., 2003).
Glutathione (GSH) is a tripeptide composed of Glu, Cys, and Gly. Its synthesis is primarily dependent on availability of the constituent amino acids (Strohm et al., 1995). Because Cys is the final product of assimilatory sulfate reduction, there is a tight link between the demand for GSH and the rate of sulfate reduction (Lappartient and Touraine, 1997; Vauclare et al., 2002; Kopriva and Rennenberg, 2004). Sulfate assimilation provides reduced sulfur for synthesis of the amino acids Cys and Met, many coenzymes and prosthetic groups (such as iron-sulfur centers, thiamine, lipoic acid, etc.), and a variety of secondary compounds. Sulfate taken up into plant cells has to be activated first by adenylation to adenosine 5′-phosphosulfate (APS) catalyzed by ATP sulfurylase. APS is reduced to sulfite by APS reductase (APR), the electrons are provided by GSH, and sulfite is subsequently reduced to sulfide by ferredoxin-dependent sulfite reductase. Sulfide is incorporated into the amino acid skeleton of O-acetyl-Ser, synthesized from Ser and acetyl-CoA by Ser acetyltransferase, by O-acetyl-Ser (thiol) lyase (OASTL) to form Cys, which is the primary donor of reduced sulfur for all subsequent biosynthetic reactions (for review, see Leustek et al., 2000; Kopriva, 2006).
Sulfate assimilation is highly regulated in a demand-driven manner (Lappartient and Touraine, 1996; Leustek et al., 2000; Kopriva and Rennenberg, 2004; Kopriva, 2006). The pathway is induced when GSH concentration is reduced by high demand (e.g. in detoxification of heavy metals or protection against cold and osmotic stress; Brunner et al., 1995; Lee and Leustek, 1999; Kocsy et al., 2004), by blocking its synthesis with enzyme inhibitors (Hartmann et al., 2004) or during sulfate starvation (e.g. Nikiforova et al., 2003). On the other hand, a surplus of reduced sulfur compounds by fumigation with H2S or by feeding thiols represses the pathway (Westerman et al., 2001; Vauclare et al., 2002). The key regulatory steps of sulfate assimilation are the transport of sulfate into the cells and the reduction of APS to sulfite by APR (Vauclare et al., 2002). Until now, however, we had a very limited knowledge of the molecular regulation of sulfate assimilation. The recently discovered sulfur-responsive SURE element and SLIM1 transcriptional regulator are the first cis- and trans-factors associated with regulation of the pathway in the promoter of the high-affinity sulfate transporters SULTR1;1 and SULTR1;2, respectively (Maruyama-Nakashita et al., 2005, 2006). Both sulfate transporters and APR seem to be regulated at the transcriptional level because, on most occasions, changes in activity correlate with changes in mRNA levels. In Arabidopsis (Arabidopsis thaliana), APR activity and the mRNA levels for its three isoforms undergo a diurnal rhythm with the highest level at the beginning of the light period (Kopriva et al., 1999). They are reduced during nitrogen deficiency (Koprivova et al., 2000) and by treatment with thiols (Vauclare et al., 2002), and increase upon treatment with O-acetyl-Ser and carbohydrates (Hesse et al., 2003). In addition, a posttranslational regulation of APR was shown to be responsible for rapid modulation of activity during oxidative stress (Bick et al., 2001).
We are interested in the molecular mechanism of regulation of sulfate assimilation. Here, we describe a genetic approach to identify components in the regulation of APR by salt stress. To address the regulation in its whole complexity, we determined not only mRNA levels, but also APR protein accumulation and enzyme activity. We found that APR is regulated at different levels by a complex network of multiple signals. We provide evidence for a novel translational regulation of APR and for involvement of GAs in transcriptional regulation of the corresponding genes.
RESULTS
Regulation of APR by Salt Stress
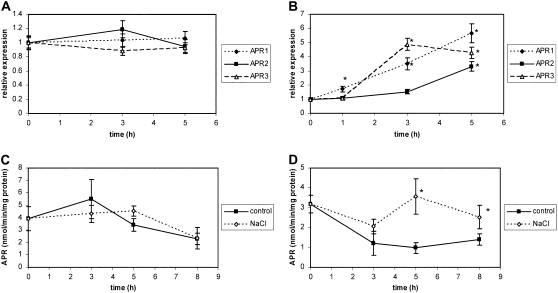
We searched the GENEVESTIGATOR Arabidopsis microarray database to identify abiotic stress conditions that strongly induce APR mRNA levels (Zimmermann et al., 2004). Exposure to 150 mm NaCl appeared to be the most suitable treatment because the mRNA levels of the APR isoforms increased 2- to 3-fold in the leaves and 6- to 10-fold in the roots within 6 h (Supplemental Fig. S1). However, in our experimental system, treatment of hydroponically grown Arabidopsis with 150 mm NaCl did not affect the transcript levels for the three APR isoforms in the leaves within 5 h, whereas they were increased in the roots 3- to 5-fold (Fig. 1, A and B). In leaves, no effect of the salt treatment on APR activity was measured up to 8 h after salt treatment. On the other hand, after 5 h, there was a significant, approximately 3-fold higher APR activity in salt-treated roots compared to controls (Fig. 1, C and D). The activity was still elevated at 8 h, but only by 80%. It has to be noted here that the decrease in APR activity in control plants was observed consistently in all time course experiments performed and can be explained by the well-characterized diurnal rhythm of APR (Kopriva et al., 1999). The increased APR activity in the Columbia-0 (Col-0) ecotype correlated with increased concentration of total GSH after 5 h of salt treatment (Table I). The transcript of the salt-regulated gene RD29A, used as a positive control, was also highly induced in these experimental conditions (Fig. 3E). Because the APR activity was not regulated in the shoots, we focused our attention on the roots only. Because the activity in roots showed a maximal increase 5 h after the beginning of the salt treatment and declined thereafter, most subsequent experiments were performed at this time point only.
Figure 1.
Regulation of APR by salt. Arabidopsis plants (Col-0) grown in hydroculture were treated with 150 mm NaCl. Relative mRNA levels of the three APR isoforms were determined by semiquantitative RT-PCR in shoots (A) and roots (B). Data are presented as ratios of mean transcript levels of four independent RNA preparations from treated versus untreated plants. APR activity was measured in shoots (C) and roots (D). Means ± sds of three independent plants are displayed. Values indicated by asterisks are different from controls at P ≤ 0.05. All experiments were repeated at least twice with similar results.
Table I.
GSH content in Arabidopsis roots treated with salt
GSH was determined in roots of control and salt-treated plants by HPLC. Mean values ± SEM from three to four independent plants are presented. Asterisks mark values significantly (P ≤ 0.05) higher than in control samples.
| Genotype | Total GSH
|
|
|---|---|---|
| Control | Salt | |
| nmol g FW−1 | ||
| Col-0 | 371 ± 44 | 544 ± 12* |
| Ler | 294 ± 167 | 686 ± 79* |
| npr1-2 | 430 ± 57 | 614 ± 32* |
| ein2-1 | 209 ± 30 | 203 ± 36 |
| jar1-1 | 207 ± 13 | 220 ± 35 |
| tir1-1 | 429 ± 25 | 650 ± 54* |
| gai | 310 ± 33 | 895 ± 102* |
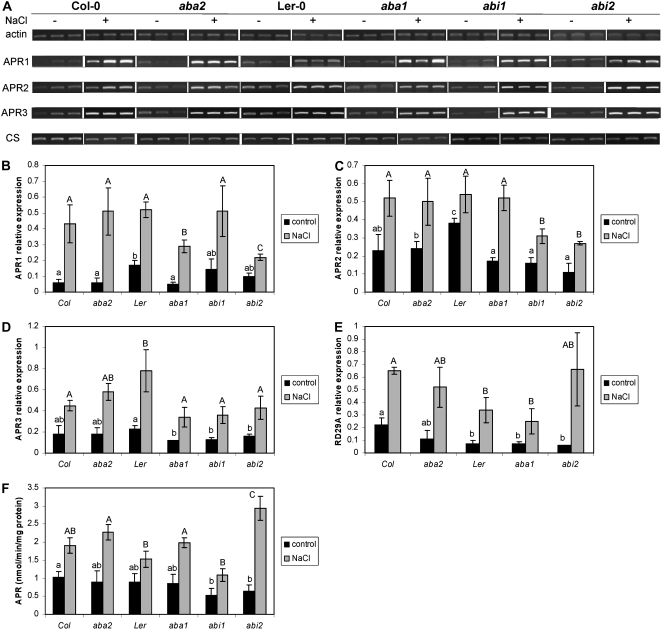
Figure 3.
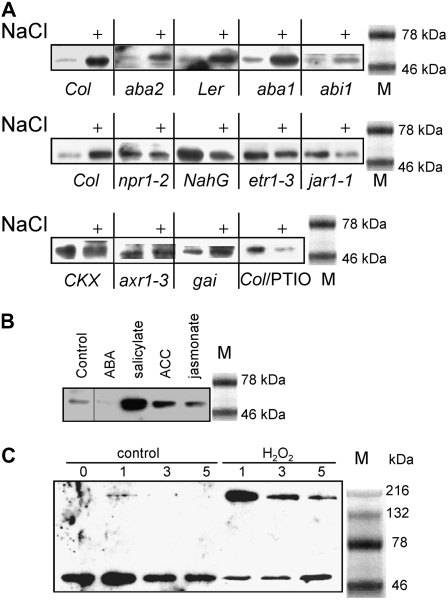
Salt regulation of APR is ABA independent. Arabidopsis mutants deficient in ABA accumulation (aba1 and aba2) or ABA signaling (abi1 and abi2) plus the corresponding wild-type ecotypes Col-0 and Ler were grown in hydroculture and treated with 150 mm NaCl for 5 h. A, Ethidium bromide-stained gels of RT-PCR fragments of actin, APR1, APR2, APR3, and cytosolic OASTL (CS) from a representative experiment are shown. The relative mRNA levels for APR1 (B), APR2 (C), APR3 (D), and RD29A (E) in roots were calculated from the semiquantitative RT-PCR data with the Quantity One software package and standardized with actin 2/7 transcript. F, APR activity was measured in crude root extracts. Data are presented as means ± sds of three independent plants. All treatments were repeated at least twice with similar results. Different indices indicate values significantly different at P ≤ 0.05. All values of salt-treated plants are significantly (P ≤ 0.05) different from nontreated plants.
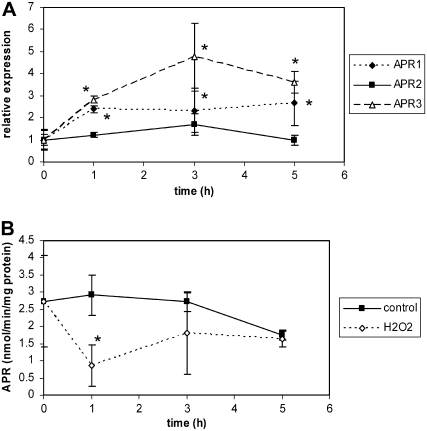
Because salt treatment induces accumulation of ROS, we tested how APR responds to treatment of the roots with hydrogen peroxide (H2O2). Indeed, diaminobenzidine (DAB) staining revealed formation of H2O2 in the salt-treated roots (Supplemental Fig. S2). Exposure to H2O2 rapidly induced mRNA accumulation of APR1 and APR3, but not of APR2 (Fig. 2A). Although an increase in mRNA accumulation occurred after 1 h, the enzyme activity was strongly reduced after 1 h and slowly recovered so that after 5 h it was similar to that in control roots (Fig. 2B). This experiment suggests that the effect of salt on APR may not be primarily caused by ROS.
Figure 2.
Regulation of APR by H2O2. Arabidopsis plants (Col-0) grown in hydroculture were treated with 10 mm H2O2. A, Relative mRNA levels of the three APR isoforms in roots were determined by semiquantitative RT-PCR. Data are presented as ratios of mean transcript levels of four independent RNA preparations from treated versus untreated plants. B, APR activity was measured in crude root extracts. Data are presented as means ± sds of three independent plants. Values indicated by asterisks are different from control at P ≤ 0.05.
Regulation of APR by Salt Is ABA Independent
Response to salt stress is commonly regulated via abscisic acid (ABA) signaling (Zhu, 2002). To test whether ABA is involved in regulation of APR by salt, we compared the induction of APR in Arabidopsis mutants deficient in ABA and ABA signaling (see Table II for a description of the mutants) with their corresponding wild types. After 5 h of salt treatment, the mRNA levels for all three APR isoforms and APR activity were increased in all lines compared to untreated controls (Fig. 3, A–D and F). The increase in APR transcripts is evident in the images of gels resolving reverse transcription (RT)-PCR fragments (Fig. 3A). The mRNA level of control salt and ABA-regulated RD29A gene was also elevated in all genotypes (Fig. 3E). After quantification, the individual levels of induction differed from 2.5- to 8-fold and were statistically significant at a high confidence level (P ≤ 0.01) for all genotypes. On the other hand, the mRNA level of cytosolic OASTL was not affected by the salt treatment (Fig. 3A). Thus, it seems that APR is regulated by salt in an ABA-independent manner.
Table II.
Mutant lines analyzed for regulation of APR by salt stress
| Mutant Allele | Functional Category | Gene | Parental Ecotype | Ref. |
|---|---|---|---|---|
| aba1 | ABA deficient | Zeaxanthin epoxidase | Ler | Koornneef et al. (1982) |
| aba2 | ABA deficient | Short-chain dehydrogenase/reductase | Col-0 | Leon-Kloosterziel et al. (1996) |
| abi1 | ABA insensitive | Protein phosphatase 2C | Ler | Koornneef et al. (1984) |
| abi2 | ABA insensitive | Protein phosphatase 2C | Ler | Koornneef et al. (1984) |
| npr1-2 | Blocked salicylate signaling | Transcription activator | Col-0 | Cao et al. (1994) |
| NahG | Salicylate deficient | Overexpressing bacterial salicylate hydroxylase | Col-0 | Lawton et al. (1995) |
| etr1-3 | Ethylene insensitive | Ethylene receptor | Col-0 | Bleecker et al. (1988) |
| ein2-1 | Ethylene insensitive | NRAMP metal transporter family | Col-0 | Guzmán and Ecker (1990) |
| jar1-1 | Jasmonate resistant | Jasmonate:amino acid synthetase | Col-0 | Staswick et al. (1992) |
| CKX | Cytokinin deficient | Overexpressing cytokinin oxidase | Col-0 | Werner et al. (2003) |
| ahk4 | Cytokinin insensitive | Cytokinin receptor AHK4 | Col-0 | Inoue et al. (2001) |
| axr1-3 | Auxin resistant | Ubiquitin-like activating enzyme | Col-0 | Estelle and Somerville (1987) |
| tir1-1 | Inhibition of auxin transport | Auxin receptor | Col-0 | Ruegger et al. (1998) |
| gai | GA insensitive | DELLA protein | Ler | Koornneef et al. (1985) |
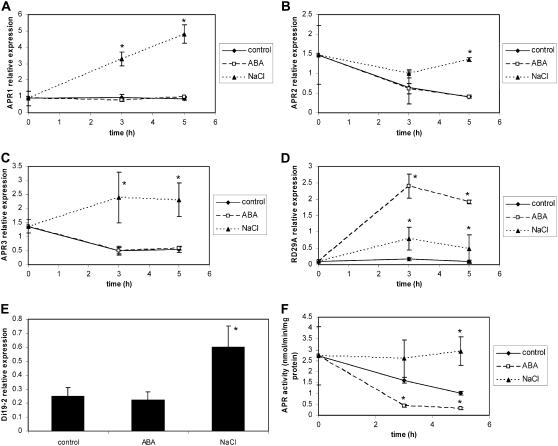
These findings were corroborated by treating Arabidopsis plants with 50 μm ABA. mRNA accumulation of none of the APR isoforms was affected by ABA in the roots after 3 and 5 h, in contrast to treatment with salt (Fig. 4, A–C). In contrast, the RD29A gene was clearly induced by both treatments at both time points (Fig. 4D), whereas the transcript of an ABA-insensitive AtDi19-2 gene (Rodriguez Milla et al., 2006) was induced by salt only (Fig. 4E). Surprisingly, however, ABA treatment resulted in a significant decrease in APR activity at both time points, whereas control salt treatment led to induction of the activity (Fig. 4F). Because the mRNA levels were not affected, it seems that ABA represses APR activity at the posttranscriptional level. These experiments, therefore, indicate that APR is regulated by salt in an ABA-independent manner.
Figure 4.
Regulation of APR by ABA. Wild-type Arabidopsis (Col-0) were grown in hydroculture and treated by addition of ABA or NaCl to the nutrient solution to a final concentration of 50 μm or 150 mm, respectively, for 3 or 5 h. Relative mRNA levels for APR1 (A), APR2 (B), APR3 (C), RD29A (D), and AtDi19-2 (E) were determined in roots by semiquantitative RT-PCR and standardized with actin 2/7 transcript. F, APR activity was determined in crude root extracts. Data are presented as means ± sds of three independent plants. All treatments were repeated at least twice with similar results. Values indicated by asterisks are different from untreated control plants at P ≤ 0.05.
Involvement of Salicylate, Ethylene, Jasmonate, and Nitric Oxide in Regulation of APR by Salt Stress
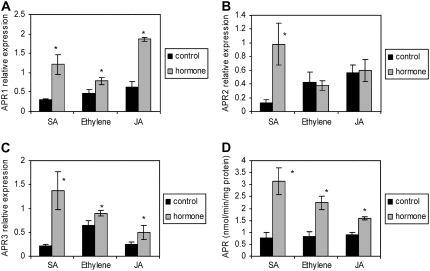
In the search for components of signaling pathways regulating APR, we tested the known stress-signaling molecules salicylate, ethylene, and jasmonate for their ability to affect APR activity and transcript levels in the roots. Addition of 0.1 mm salicylate to the nutrient solution led to an increase of mRNA levels of all three APR isoforms. On the other hand, 0.2 mm 1-aminocyclopropane carboxylic acid (ACC), which stimulates ethylene production, and 45 μm jasmonate increased accumulation of APR1 and APR3 transcripts, but did not affect the APR2 isoform (Fig. 5, A–C). All three compounds significantly induced APR activity (Fig. 5D). It seems that the increase in mRNA for APR1 and APR3 isoforms is sufficient to increase APR activity upon feeding with these phytohormones.
Figure 5.
Regulation of APR by phytohormones. Wild-type Arabidopsis (Col-0) were grown in hydroculture and treated with 100 μm salicylate, 200 μm ACC, or 45 μm jasmonate for 5 h. The relative mRNA levels for APR1 (A), APR2 (B), and APR3 (C) in roots were determined by semiquantitative RT-PCR and standardized with actin 2/7 transcript. D, APR activity was measured in crude root extracts. Data are presented as means ± sds of three independent plants. Values indicated by asterisks are different from untreated plants at P ≤ 0.05. All treatments were repeated at least twice with similar results.
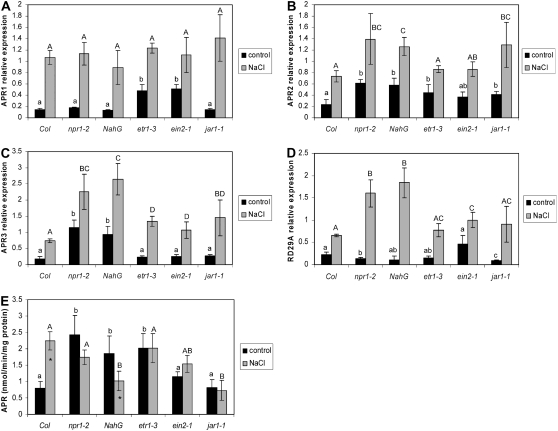
To test whether these hormones are involved in regulation of APR by salt stress, we analyzed plants deficient in the corresponding signaling pathways (Table II). In all these genotypes, mRNA levels of the three APR isoforms and the control RD29A gene were induced by the salt treatment (Fig. 6, A–D). Although some variations in the degree of induction or in steady-state transcript levels in control plants were detected in some genotypes, the general up-regulation of APR transcripts by salt was not affected by modulation of the signaling pathways. Surprisingly, in contrast to wild-type Arabidopsis, APR activity was not affected by salt stress in these plants or was even slightly decreased in the genotypes deficient in salicylate signaling (Fig. 6E). Remarkably, the activity in npr1-2 and NahG was consistently higher than in wild-type Arabidopsis. Thus, the induction of APR activity, but not mRNA, seems to be dependent on correctly functioning stress signaling by salicylate, ethylene, and jasmonate. HPLC analysis revealed no increase in total GSH in ein2 or jar1 following salt treatment, whereas it was increased in npr1 plants to a similar degree as in wild-type plants (Table I).
Figure 6.
Regulation of APR by salt in mutants deficient in stress signaling. Arabidopsis mutants or transgenic lines deficient in salicylate signaling (npr1), salicylate accumulation (NahG), ethylene signaling (etr1 and ein2), or jasmonate signaling (jar1) plus the corresponding wild type (Col-0) were grown in hydroculture and treated with 150 mm NaCl for 5 h. The relative mRNA levels for APR1 (A), APR2 (B), APR3 (C), and RD29A (D) in roots were determined by semiquantitative RT-PCR and standardized with actin 2/7 transcript. E, APR activity was measured in crude root extracts. Data are presented as means ± sds of three independent plants. All treatments were repeated at least twice with similar results. Different indices indicate values significantly different at P ≤ 0.05. All values of salt-treated plants in A to D and those marked with asterisks in E are significantly (P ≤ 0.05) different from nontreated plants.
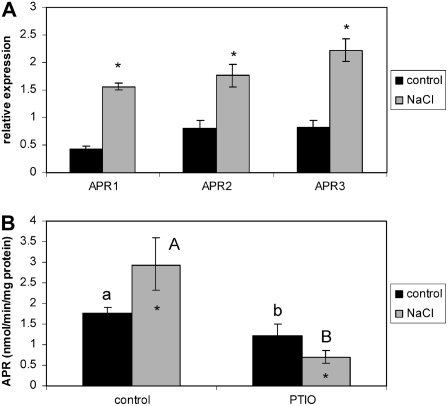
Another molecule associated recently with stress signaling is nitric oxide (NO; Delledonne, 2005). NO signaling can be prevented by the NO scavenger 2-phenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (PTIO). Similar to experiments blocking salicylate, ethylene, and jasmonate signaling, preventing NO signaling by treatment with PTIO only slightly affected the increase in APR mRNA levels by salt treatment, which remained significantly elevated, but prevented the increase in enzyme activity (Fig. 7). NO may thus be another component of the signaling network in regulation of APR by salt stress.
Figure 7.
Involvement of NO in regulation of APR by salt stress. Wild-type Arabidopsis (Col-0) were grown in hydroculture and treated with 0.6 mm PTIO for 30 min before exposure to 150 mm NaCl for 5 h. A, The relative mRNA levels for APR1, APR2, and APR3 in roots were determined by semiquantitative RT-PCR and standardized with actin 2/7 transcript. B, APR activity was measured in crude root extracts. Data are presented as means ± sds of three independent plants. All treatments were repeated at least twice with similar results. Values marked with asterisks are significantly (P ≤ 0.01) different from control values.
GA Is Required for APR Regulation by Salt
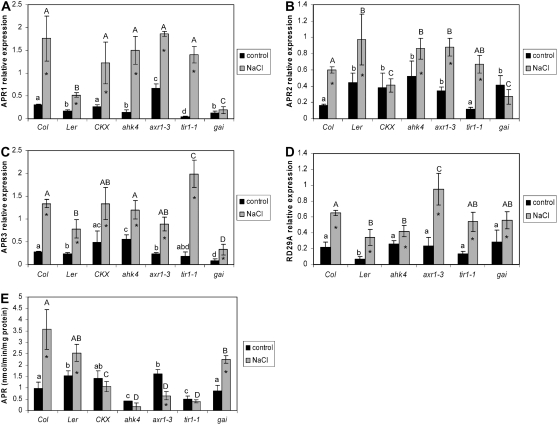
APR has been shown to be regulated by cytokinins (Ohkama et al., 2002); therefore, we tested whether these or two other phytohormones, auxin and GA, are involved in regulation of this enzyme by salt stress. Five further Arabidopsis mutants impaired in phytohormone signaling were used (Table II). These plants were compared to the corresponding wild types for alterations in APR response to salt treatment. In the plants deficient in cytokinin accumulation and signaling and auxin signaling, the mRNA levels of the three APR isoforms were increased upon salt treatment similar to wild type (Fig. 8, A–C), except APR2 in the CKX plants, which was not induced. Enzyme activity measurements revealed that APR activity was not increased by salt stress in cytokinin and auxin-insensitive genotypes (Fig. 8E). On the other hand, the APR1 and APR2 mRNA levels were not significantly affected by salt treatment in the GA-insensitive gai plants (Fig. 8, A–C). APR3 regulation was similar to the corresponding wild type (Landsberg erecta [Ler]) and other genotypes. The induction of RD29A mRNA level has been observed in all genotypes (Fig. 8D). APR activity was, however, increased by salt stress in gai plants despite no effect of the treatment on transcript levels of APR1 and APR2. Because GAs have not yet been implicated in APR regulation, we treated wild-type plants with 50 μm GA3 added to the nutrient solution, which resulted in a significant increase in APR mRNA levels and enzyme activity (Supplemental Fig. S3). Total GSH content in gai and tir1 plants was increased by salt stress in a similar way to corresponding wild types.
Figure 8.
Regulation of APR by salt in mutants deficient in hormone signaling. Arabidopsis mutants or transgenic lines deficient in cytokinin perception (ahk4), cytokinin accumulation (CKX), auxin signaling (axr1), and tir1 or GA-signaling gai plus the corresponding wild types (Col-0 or Ler) were grown in hydroculture and treated with 150 mm NaCl for 5 h. The relative mRNA levels for APR1 (A), APR2 (B), APR3 (C), and RD29A (D) in roots were determined by semiquantitative RT-PCR and standardized with actin 2/7 transcript. E, APR activity was measured in crude root extracts. Data are presented as means ± sds of three independent plants. All treatments were repeated at least twice with similar results. Different indices indicate values significantly different at P ≤ 0.05. Values marked with asterisks are significantly (P ≤ 0.05) different from nontreated plants.
APR Is Regulated at Translational and Posttranslational Levels
To gain further insight into the mechanism of regulation of APR, crude root extracts were analyzed by western blotting using antiserum against recombinant APR2, which cross-reacts with all three APR isoforms (Kopriva et al., 1999; Koprivova et al., 2000). Several genotypes were used, representing different effects of salt treatment on APR transcript levels and activity. In both wild-type ecotypes, in mutants deficient in or insensitive to ABA, and in the GA-insensitive gai plants, APR was more abundant in salt-treated roots (Fig. 9A). Analysis of mutants deficient in salicylate, ethylene, jasmonate, auxin, or cytokinin signaling revealed that, in contrast to wild-type plants, salt treatment of these plants did not induce APR protein levels. The protein accumulation thus correlated well with the APR activity in these genotypes (Fig. 9A; compare with Figs. 3, 4, 6, and 8). The same strict correlation between APR protein level and activity was observed after feeding various phytohormones (Fig. 9B). Thus, ABA reduced APR protein accumulation, whereas salicylate, ACC, and jasmonate induced APR protein levels to a similar degree as enzyme activity. These results reveal a novel mechanism of APR regulation at the posttranscriptional level. This regulation seems to require the correct functioning of multiple signaling pathways, including those involving salicylate, ethylene, jasmonate, auxin, and cytokinins.
Figure 9.
Western-blot analysis. A, APR protein accumulation was compared in various untreated and salt-treated Arabidopsis genotypes by western blotting of the crude root extracts for activity measurement. The analysis was repeated at least twice on independent extracts with similar results. B, Protein extracts from Arabidopsis wild-type (Col-0) roots untreated or treated with ABA, salicylate, ACC, and jasmonate for 5 h were resolved by SDS-PAGE. APR protein accumulation was compared by western blotting with APR2 antiserum. C, Protein extracts from Arabidopsis (Col-0) roots untreated or treated with H2O2 for indicated time were resolved by SDS-PAGE. APR protein accumulation was compared by western blotting with APR2 antiserum. M represents molecular mass marker.
Different results, however, were obtained from analysis of extracts from roots treated with H2O2. The signal corresponding to APR at 52 kD was strongly reduced in these extracts, which again correlated with the decrease in enzyme activity. An additional signal also appeared on the blots corresponding to a high molecular mass protein or a protein adduct (Fig. 9C). The same results were obtained using reducing and nonreducing conditions (data not shown). Such a high molecular mass signal was not observed in extracts from salt-treated plants of any genotype.
DISCUSSION
Exposure to high salinity is connected with ionic stress due to accumulation of Na+ ions, osmotic stress, and ROS production (Hasegawa et al., 2000). Salt stress induces the activities of antioxidative enzymes and accumulation of antioxidants, such as GSH (Ruiz and Blumwald, 2002; Mittova et al., 2003). Therefore, it was not surprising that this appeared to be a suitable treatment for dissecting the regulation of APR by stress. Indeed, in our experimental conditions, GSH content and APR activity were increased by salt treatment (Table I; Fig. 1). The increased APR activity and GSH level in salt-treated roots correlate with previous observations of demand-driven regulation of sulfate assimilation and the key role of APR in control of the pathway (Brunner et al., 1995; Lappartient and Touraine, 1996; Lee and Leustek, 1999; Leustek et al., 2000; Westerman et al., 2001; Vauclare et al., 2002; Kocsy et al., 2004; Kopriva, 2006). In fact, other enzymes of sulfate assimilation are also induced by salt, including ATP sulfurylase, Ser acetyltransferase, and a cytosolic isoform of OASTL (Barroso et al., 1999; Ruiz and Blumwald, 2002).
Salt stress signaling is complex and involves numerous pathways with frequent cross-talk. ABA has a pivotal role among these secondary signals; however, both ABA-dependent and ABA-independent signaling pathways have been described (Zhu, 2002). ABA regulates gene expression directly via ABF/AREB transcription factors (Xiong et al., 2002) or indirectly because ABA also induces ROS production (Guan et al., 2000), which in turn activates ROS signaling, affecting cytosolic Ca2+, NO, ethylene, jasmonate, and salicylate. APR regulation by salt is clearly ABA independent, as demonstrated by the same regulation of APR by salt treatment in aba and abi mutants (Fig. 3). In contrast, the OASTL mRNA was induced after 24 h by salt treatment in an ABA-dependent manner and also by direct ABA treatment (Barroso et al., 1999). In our experiments, the level of cytosolic OASTL mRNA was not increased by salt within the first 5 h of salt exposure (Fig. 3). This suggests that the role of the two genes in the Arabidopsis response to salt stress is different: Whereas OASTL is involved in the acclimation to high salt, APR contributes to the early response. It seems that because of its key role in control of sulfate assimilation, APR has to be increased rapidly to allow a higher rate of Cys synthesis to accommodate the increased demand for GSH.
Because ABA does not seem to be involved in regulation of APR by salt, we addressed the possibility that the regulation is actually triggered by ROS. Indeed, DAB staining revealed that ROS were induced by the salt treatment in our experimental conditions. However, the regulation of APR was different after exposure to salt and to H2O2 (Figs. 1 and 2). Because fumigation with ozone resulted in a posttranslational activation of APR (Bick et al., 2001), the reduction in APR activity in H2O2-treated roots appears surprising. It is, however, supported by our observation that purified recombinant APR2 from Arabidopsis is inactivated by 0.2% H2O2 (S. Kopriva, unpublished data). Presumably, the concentration of H2O2 in the root cells after the treatment was higher than after the ozone fumigation and resulted in enzyme inactivation. It also cannot be excluded that the APR regulation differs in roots and leaves or that it is specific to certain types of ROS, such as superoxide. In salt-treated roots, however, the staining revealed even higher ROS concentration and the activity was still increased. The reason for the discrepancy thus might be the subcellular distribution of the ROS between apoplast, cytosol, and plastids. Nevertheless, it seems that ROS were not the primary elicitors of the APR response to salt stress, or perhaps alternative ROS, such as superoxide (O2−), participate in this regulation.
In the vast majority of previous experiments, a strict correlation between APR mRNA levels, protein accumulation, and enzyme activity was observed leading to the conclusion that APR is regulated primarily on the transcriptional level (Kopriva et al., 1999; Koprivova et al., 2000; Hesse et al., 2003; Vauclare et al., 2002; Kopriva and Koprivova, 2004). Additional means of affecting APR activity via posttranslational redox regulation has been demonstrated in Brassica plants subjected to oxidative stress (Bick et al., 2001) and on recombinant APR2 protein (Kopriva and Koprivova, 2004). In addition, a reduction of APR activity despite an increase in mRNA level was observed after treatment of Brassica juncea with cadmium, although this may have been caused by a direct inhibitory effect of cadmium on the enzyme (Lee and Leustek, 1999). Salt treatment of wild-type Arabidopsis resulted in a coordinated increase in APR mRNA, protein, and activity, which suggested a transcriptional regulation in response to this stimulus as well. From the analysis of mutants disrupted in different signaling pathways, we therefore expected to identify the cascades responsible for the regulation of APR by salt, as described, for example, in Charlton et al. (2005). For most mutants analyzed, the mRNA levels of all three APR isoforms were increased similarly to corresponding wild-type controls. There were small differences in the level of induction in several genotypes; however, the general response was the same. The exception was the gai mutant, revealing that the increase of APR mRNA after salt treatment may be dependent on GA signaling. This is quite surprising because GA has not been described previously as a regulator of APR and because disruption of none of the usual stress signaling pathways (jasmonate, salicylate, ethylene, NO) affected the regulation of APR transcripts.
In addition, the analysis revealed that the regulation of the three APR isoforms is not identical. APR2 mRNA was not induced by H2O2, jasmonate, or ACC, in contrast to APR1 and APR3 (Fig. 5), and also was not induced by salt stress in the cytokinin-deficient plants (Fig. 8). Only little is known about the biochemical or molecular differences between the three APR isoforms. Based on genomic sequence, APR1 and APR3 are more closely related to each other than either of them is to APR2. In previous experiments, the isoforms were all regulated in the same way, but with different time and/or strength of the response (Kopriva et al., 1999; Koprivova et al., 2000, Vauclare et al., 2002). These results indicate that the different APR isoforms have specific functions and are regulated differently, which is likely to allow a more precise fine tuning in the plant stress response.
Interestingly, the GSH content in the signaling mutants did not always correlate with APR activity. In jar1 and ein2, APR activity was not induced by salt stress and correspondingly GSH was also not increased. On the other hand, no increase in APR was detected in tir1 and npr1, but the thiols were increased similar to wild-type plants. This finding shows that, despite its high control over sulfate assimilation, induction of APR activity is not essential for the increase of GSH synthesis after salt stress and that other components of GSH biosynthesis have to be induced by salt as well to enable its accumulation. The other components are probably under the control of jasmonate and ethylene signaling because disruption of these pathways prevented GSH accumulation. The uncoupling of APR regulation from GSH synthesis has been observed before. Loudet et al. (2007) showed that reduction of total APR activity in apr2 T-DNA lines to 20% of the wild-type activity did not affect thiol levels.
Had we stopped our analysis at measuring only transcript levels, which is often the case (Charlton et al., 2005; Ma et al., 2006), this would be the end of a very simple story. However, we also measured the APR enzyme activity because this is more relevant for the physiological response of the plant than mRNA level alone. The activity results were very striking. We could rapidly conclude that APR is indeed regulated by salt stress in an ABA-independent manner (Fig. 3). In all other signaling mutants but gai, APR activity was not increased upon salt treatment despite the increase in mRNA levels. The uncoupling of responses of mRNA and enzyme activity to salt stress in most of the signaling mutants revealed that the regulation is far more complex than we expected and from what is known from literature (Leustek et al., 2000; Kopriva and Koprivova, 2004; Kopriva, 2006). Therefore, we have to postulate that posttranscriptional regulation of APR occurs, requiring the correct functioning of stress signaling. The finding that APR activity was significantly reduced in H2O2-treated plants (Fig. 2) led to a possible explanation for our results. We hypothesized that the disruption of stress signaling pathways resulted in an inefficient detoxification of ROS, elicited by the salt treatment, which inactivated APR. However, because by DAB staining we have not observed any increased ROS production in salt- treated roots of the various signaling mutants (data not shown) and because in extracts from such roots the high molecular mass APR adducts were not detected, this explanation is unlikely. Moreover, in all western-blot experiments the APR protein accumulation strictly correlated with enzyme activity (Fig. 9); therefore, the regulation is likely to be on the level of translation or protein stability.
The lack of induction of APR activity in the signaling mutants could be explained in three ways: the need for a stress-induced activator for the translation of APR; the presence of an inhibitor that is inactivated and/or degraded as a response to stress; or the activation of the APR degradation pathway in the mutants. The results could also possibly be attributed to various pleiotropic effects of the mutations. However, because similar responses were observed in plants where very different signaling pathways have been disrupted and also in plants where the signaling was disrupted by chemical treatment, the pleiotropic effects are probably not the main cause of the observed changes in APR regulation in the mutants. The strict correlation between activity and protein, however, suggests that the posttranslational activation of APR by oxidative stress as described by Bick et al. (2001) does not contribute to the regulation of the enzyme by salt stress.
The increase in APR protein accumulation and activity in gai plants was no less surprising because mRNAs of two of the three APR isoforms were not affected in this genotype. It appears that the small increase in APR3 transcript may be responsible for the increase in enzyme activity. However, the contribution of individual isoforms to total APR activity is not known, and APR3 has never been considered to be the major isoform. Indeed, in APR2 T-DNA lines, the foliar APR activity was reduced to 20% of wild-type levels, pointing to a more significant role of this isoform than APR3 (Loudet et al., 2007). Therefore, posttranscriptional control of APR involving a GA-modulated mechanism is more likely to explain this result.
Such posttranscriptional regulation has not been described for plant APR before, but is similar to regulation of APR in sulfur starvation response mutants of the green alga Chlamydomonas reinhardtii. Specifically, in the sac2 mutant, APR mRNA was induced by sulfate starvation similar to wild-type Chlamydomonas, but a corresponding increase in enzyme activity was prevented by the mutation (Davies et al., 1994; Ravina et al., 2002). This is analogous to our results, and the SAC2 gene thus seems to be equivalent to our postulated translational regulator. Analysis of the sac1 mutant revealed another peculiarity in APR regulation. Transcripts of APR and other genes of sulfate assimilation were not induced by sulfur deficiency in this mutant; nevertheless, the APR activity increased similar to wild type. This was not true for any other enzyme of the pathway; the increases in ATP sulfurylase, Ser acetyltransferase, and OASTL were prevented by the sac1 mutation (Davies et al., 1994; Ravina et al., 2002). Again, this regulation is remarkably similar to the regulation of APR by salt stress in gai plants. The similarity in regulatory mechanisms is very surprising because, until now, discoveries about the regulation of the pathway by sulfur starvation in Chlamydomonas could not be confirmed in higher plants despite the great interest in this biological question.
Interestingly, another gene involved in salt stress response is also regulated at the posttranscriptional level (Hua et al., 2001). The mRNA level of AtP5R (pyrroline-5-carboxylate reductase), which is involved in Pro synthesis, is increased upon salt treatment, but the protein does not accumulate. The translation of the protein is inhibited during salt stress, via a mechanism dependent on the 5′-untranslated region, which is predicted to form a very strong secondary structure (total free energy −59.7 kcal mol−1; Hua et al., 2001). In silico analysis of 5′-untranslated regions of the three APR genes revealed that they also potentially form strong secondary structures with free energies of −5.8, −18.9, and −22.4 kcal mol−1 for APR1, APR2, and APR3, respectively (Supplemental Fig. S4), whereas an average free energy of predicted secondary structures of 5′-untranslated regions is approximately −3 kcal mol−1 (Hua et al., 2001). Whether the 5′-untranslated regions of APR genes are indeed involved in the translational regulation awaits further investigation.
In conclusion, we have demonstrated that regulation of APR by salt cannot be wholly attributed to transcriptional regulation. This has important implications for the analysis of plant responses to different stimuli because many studies up to now have concentrated on transcriptome analysis to deduce the in vivo effects of these stimuli. It is clear that we must look at enzyme activities together with transcriptional information to gain more physiologically relevant insight into plant responses to their environment.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The seeds of the mutants and transgenic plants were kindly provided by Gary Creissen (npr1, NahG, etr1, ein2, jar1), Fred Rook (aba1, aba2, abi1), Nick Harberd (gai), Robert Sablowski (axr1), Jonathan Jones (tir1), Geoff Holroyd (abi1), and Thomas Schmuelling (CKX), or obtained from The Nottingham Arabidopsis Stock Centre (ahk4). The plants were grown in hydroculture in nutrient solution composed of 1.5 mm Ca(NO3)2, 1 mm KNO3, 0.75 mm KH2PO4, 0.75 mm MgSO4, 0.1 mm Fe-EDTA, 10 μm MnCl2, 50 μm H3BO3, 1.75 μm ZnCl2, 0.5 μm CuCl2, 0.8 μm Na2MoO4, 1 μm KI, and 0.1 μm CoCl2 under a 10-h-light/14-h-dark cycle at constant temperature of 22°C, 60% relative humidity, and light intensity of 160 μmol m−2 s−1. The nutrient solution was exchanged weekly. Three weeks after sowing, the plants were transferred into fresh nutrient solution with or without 150 mm NaCl for salt treatment, 10 mm H2O2 or other additives, as indicated, and incubated for 5 h under the same conditions. Roots were collected and immediately frozen in liquid N2.
RNA Extraction and Expression Analysis
Total RNA was isolated from the roots by phenol:chloro-form:isoamylalcohol (25:24:1) extraction and LiCl precipitation. Aliquots of 1 μg were reverse transcribed by SuperScript reverse transcriptase (Invitrogen). For semiquantitative PCR, equivalents of 40 ng of total RNA were amplified by GoTaq Flexi DNA polymerase (Promega) in 20-μL reactions with primers specific for the three APR isoforms APR1 (At4g04610)—APR1f (CTCGTTTCGGTGTTTCATTG) and APR1r (CAATCCCTTGCTCCTAACCA); APR2 (At1g62180)—APR2f (CCACACATCAGCTCCTTCAA) and APR2r (AACGCTGAGTCACATTCACG); and APR3 (At4g21990)—APR3f (TCCAAGCACGTAAACCCTTC) and APR3r (CGGCTTCTCTGAGTTTGTCC). As controls, the cDNA was amplified with primers derived from actin 2/7 (At5g09810)—actf (GGAGCTGAGAGATTCCGTTG) and actr (TGAACAATCGATGGACCTGA); from salt and ABA up-regulated gene RD29A (At5g52310)—RD29Af (GGAGCTGAGCTGGAAAAAGAATTTGATCAGAAG) and RD29Ar (CCAATCTGAAGTTTCTCGGCAACCATATCAG); and an ABA-independent salt-inducible gene AtDi19-2 (At1g02750)— Di19f (ACGCGTCGACATGGAAGACGATATGTGGTGCG) and Di19r (CGCGGATCCGCCTCAGAAGAGTCACATTCATC). The reactions were stopped after 26, 28, and 29 cycles for APR1, APR2, and APR3, respectively, and after 22, 29, and 32 cycles for actin, RD29A, and Di19-2 when the reactions were still in the exponential phase as determined in preliminary experiments (Supplemental Fig. S5). Eighteen microliters of the PCR products were subjected to electrophoresis on ethidium bromide containing 1% agarose gels. The resulting band intensity on a UV transilluminator was calculated with the Quantity One software package (Bio-Rad).
APR Activity Measurement
APR activity was determined as described elsewhere (Kopriva et al., 1999; Koprivova et al., 2000). The roots were homogenized 1:20 (w/v) in 50 mm Na/K phosphate buffer, pH 8, supplemented with 30 mm Na2SO3, 0.5 mm 5′-AMP, and 10 mm dithioerythritol (DTE) and the extract was centrifuged for 30 s at 2,000 rpm to remove cell debris. APR activity was measured in the supernatants as the production of [35S]sulfite, assayed as acid volatile radioactivity formed from [35S]APS and DTE (Brunold and Suter, 1990). The protein concentration in the extracts was determined by Bio-Rad protein assay with bovine serum albumin as a standard.
Western Blotting
APR protein accumulation was assessed by western blotting with polyclonal antisera against recombinant APR2 (Kopriva et al., 1999). Aliquots of the crude extracts from APR activity measurements corresponding to 8 μg of protein were resolved on 12% SDS-PAGE gels and transferred to nitrocellulose membrane by electroblotting. The blots were developed with SuperSignal West Pico system (Pierce).
GSH Measurements
GSH was extracted from the root tissue by grinding 0.1 g of frozen material in 1 mL of 0.1 m HCl. After centrifugation at 20,000g for 10 min, the supernatant was used to measure the content of total GSH after reduction with DTE by HPLC using the monobromobimane derivatization method as described by Creissen et al. (1999).
DAB Staining
H2O2 in the roots of plants treated with salt or H2O2 was detected by staining with DAB according to Thordal-Christensen et al. (1997).
Statistical Analysis
The data were subjected to ANOVA and multiple range tests (lsd). The results from salt treatments were compared with controls by Student's t test at 95% confidence level. Statistical analyses of the data were carried out using SPPS for Windows (release 9.0; SPSS).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. GENEVESTIGATOR microarray data on regulation of APR by salt.
Supplemental Figure S2. Localization of ROS in roots after treatment with NaCl and H2O2.
Supplemental Figure S3. Regulation of APR by GA.
Supplemental Figure S4. Predicted hairpin structures of 5′-untranslated regions of APR mRNA.
Supplemental Figure S5. RT-PCR conditions.
Supplementary Material
Acknowledgments
We would like to thank Gary Creissen, Fred Rook, Nick Harberd, Robert Sablowski (John Innes Centre), Jonathan Jones (Sainsbury Laboratory), Geoff Holroyd (Lancaster University), and Thomas Schmuelling (Free University of Berlin) for seeds of Arabidopsis mutants and transgenic plants.
This work was supported by the Biotechnology and Biological Sciences Research Council of the United Kingdom.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stanislav Kopriva (stanislav.kopriva@bbsrc.ac.uk).
The online version of this article contains Web-only data.
References
- Barroso C, Romero LC, Cejudo FJ, Vega JM, Gotor C (1999) Salt-specific regulation of the cytosolic O-acetylserine(thiol)lyase gene from Arabidopsis thaliana is dependent on abscisic acid. Plant Mol Biol 40 729–736 [DOI] [PubMed] [Google Scholar]
- Bick JA, Seterdahl AT, Knaff DB, Chen YC, Pitcher LH, Zilinskas BA, Leustek T (2001) Regulation of the plant-type 5′-adenylyl sulfate reductase by oxidative stress. Biochemistry 40 9040–9048 [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H (1988) Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241 1086–1089 [DOI] [PubMed] [Google Scholar]
- Brunner M, Kocsy G, Ruegsegger A, Schmutz D, Brunold C (1995) Effect of chilling on assimilatory sulphate reduction and glutathione synthesis in maize. J Plant Physiol 146 743–747 [Google Scholar]
- Brunold C, Suter M (1990) Adenosine 5′-phosphosulphate sulfotransferase. In P Lea, ed, Methods in Plant Biochemistry, Vol 3. Academic Press, London, pp 339–343
- Cao H, Bowling SA, Gordon AS, Dong X (1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlton WL, Matsui K, Johnson B, Graham IA, Ohme-Takagi M, Baker A (2005) Salt-induced expression of peroxisome-associated genes requires components of the ethylene, jasmonate and abscisic acid signalling pathways. Plant Cell Environ 28 513–524 [Google Scholar]
- Creissen G, Firmin J, Fryer M, Kular B, Leyland N, Reynolds H, Pastori G, Wellburn F, Baker N, Wellburn A, et al (1999) Elevated glutathione biosynthetic capacity in the chloroplasts of transgenic tobacco plants paradoxically causes increased oxidative stress. Plant Cell 11 1277–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JP, Yildiz FH, Grossman AR (1994) Mutants of Chlamydomonas with aberrant responses to sulfur deprivation. Plant Cell 6 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delledonne M (2005) NO news is good news for plants. Curr Opin Plant Biol 8 390–396 [DOI] [PubMed] [Google Scholar]
- Estelle MA, Somerville CR (1987) Auxin-resistant mutants of Arabidopsis with an altered morphology. Mol Gen Genet 206 200–206 [Google Scholar]
- Guan LM, Zhao J, Scandalios JG (2000) Cis-elements and trans-factors that regulate expression of the maize Cat1 antioxidant gene in response to ABA and osmotic stress: H2O2 is the likely intermediary signaling molecule for the response. Plant J 22 87–95 [DOI] [PubMed] [Google Scholar]
- Guzmán P, Ecker JR (1990) Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2 513–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann T, Hönicke P, Wirtz M, Hell R, Rennenberg H, Kopriva S (2004) Sulfate assimilation in poplars (Populus tremula x P. alba) overexpressing γ-glutamylcysteine synthetase in the cytosol. J Exp Bot 55 837–845 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51 463–499 [DOI] [PubMed] [Google Scholar]
- Hesse H, Trachsel N, Suter M, Kopriva S, von Ballmoos P, Rennenberg H, Brunold C (2003) Effect of glucose on assimilatory sulfate reduction in roots of Arabidopsis thaliana. J Exp Bot 54 1701–1709 [DOI] [PubMed] [Google Scholar]
- Hua XJ, Van de Cotte B, Van Montagu M, Verbruggen N (2001) The 5′ untranslated region of the At-P5R gene is involved in both transcriptional and post-transcriptional regulation. Plant J 26 157–169 [DOI] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T (2001) Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409 1060–1063 [DOI] [PubMed] [Google Scholar]
- Kocsy G, Szalai G, Galiba G (2004) Effect of osmotic stress on glutathione and hydroxymethylglutathione accumulation in wheat. J Plant Physiol 161 785–794 [DOI] [PubMed] [Google Scholar]
- Kocsy G, von Ballmoos P, Ruegsegger A, Szalai G, Galiba G, Brunold C (2001) Increasing the glutathione content in a chilling-sensitive maize genotype using safeners increased protection against chilling-induced injury. Plant Physiol 127 1147–1156 [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Elgersma A, Hanhart CJ, van Loenen-Martinet EP, van Rijn L, Zeevaart JAD (1985) A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol Plant 65 33–39 [Google Scholar]
- Koornneef M, Jorna ML, Derswan DLCB, Karssen CM (1982) The isolation of abscisic-acid (aba) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L) Heynh. Theor Appl Genet 61 385–393 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Reuling G, Karssen CM (1984) The isolation and characterization of abscisic-acid insensitive mutants of Arabidopsis thaliana. Physiol Plant 61 377–383 [Google Scholar]
- Kopriva S (2006) Regulation of sulfate assimilation in Arabidopsis and beyond. Ann Bot (Lond) 97 479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Koprivova A (2004) Plant adenosine 5′phosphosulfate reductase—the past, the present, and the future. J Exp Bot 55 1775–1783 [DOI] [PubMed] [Google Scholar]
- Kopriva S, Muheim R, Koprivova A, Trachsler N, Catalano C, Suter M, Brunold C (1999) Light regulation of assimilatory sulfate reduction in Arabidopsis thaliana. Plant J 20 37–44 [DOI] [PubMed] [Google Scholar]
- Kopriva S, Rennenberg H (2004) Control of sulphate assimilation and glutathione synthesis: interaction with N and C metabolism. J Exp Bot 55 1831–1842 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Suter M, Op den Camp R, Brunold C, Kopriva S (2000) Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol 122 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappartient AG, Touraine B (1996) Demand-driven control of root ATP sulphurylase activity and SO42− uptake in intact canola. The role of phloem-translocated glutathione. Plant Physiol 111 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappartient AG, Touraine B (1997) Glutathione-mediated regulation of ATP sulphurylase activity, SO42− uptake and oxidative stress responses to intact canola roots. Plant Physiol 114 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawton K, Weymann K, Friedrich L, Vernooij B, Uknes S, Ryals J (1995) Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol Plant Microbe Interact 8 863–870 [DOI] [PubMed] [Google Scholar]
- Lee SM, Leustek T (1999) The affect of cadmium on sulfate assimilation enzymes in Brassica juncea. Plant Sci 141 201–207 [Google Scholar]
- Leon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JA, Koornneef M (1996) Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J 10 655–661 [DOI] [PubMed] [Google Scholar]
- Leustek T, Martin MN, Bick JA, Davies JP (2000) Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu Rev Plant Physiol Plant Mol Biol 51 141–165 [DOI] [PubMed] [Google Scholar]
- Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, North KA, Kopriva S, Daniel-Vedele F (2007) Natural variation for sulfate content in Arabidopsis is highly controlled by adenosine 5′-phosphosulfate reductase. Nat Genet 39 896–900 [DOI] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ (2006) Dissecting salt stress pathways. J Exp Bot 57 1097–1107 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Watanabe-Takahashi A, Inoue E, Yamaya T, Takahashi H (2005) Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J 42 305–314 [DOI] [PubMed] [Google Scholar]
- Mittova V, Theodoulou FL, Kiddle G, Gomez L, Volokita M, Tal M, Foyer CH, Guy M (2003) Coordinate induction of glutathione biosynthesis and glutathione-metabolizing enzymes is correlated with salt tolerance in tomato. FEBS Lett 554 417–421 [DOI] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33 633–650 [DOI] [PubMed] [Google Scholar]
- Noctor G, Arisi A-CM, Jouanin L, Kunert KJ, Rennenberg H, Foyer CH (1998) Glutathione: biosynthesis, metabolism and relationship to stress tolerance explored in transformed plants. J Exp Bot 49 623–647 [Google Scholar]
- Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annu Rev Plant Physiol Plant Mol Biol 49 249–279 [DOI] [PubMed] [Google Scholar]
- Ohkama N, Takei K, Sakakibara H, Hayashi H, Yoneyama T, Fujiwara T (2002) Regulation of sulfur-responsive gene expression by exogenously applied cytokinins in Arabidopsis thaliana. Plant Cell Physiol 43 1493–1501 [DOI] [PubMed] [Google Scholar]
- Ravina CG, Chang CI, Tsakraklides GP, McDermott JP, Vega JM, Leustek T, Gotor C, Davies JP (2002) The sac mutants of Chlamydomonas reinhardtii reveal transcriptional and posttranscriptional control of cysteine biosynthesis. Plant Physiol 130 2076–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Milla MA, Townsend J, Chang IF, Cushman JC (2006) The Arabidopsis AtDi19 gene family encodes a novel type of Cys2/His2 zinc-finger protein implicated in ABA-independent dehydration, high-salinity stress and light signaling pathways. Plant Mol Biol 61 13–30 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev 12 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JM, Blumwald E (2002) Salinity-induced glutathione synthesis in Brassica napus. Planta 214 965–969 [DOI] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH (1992) Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc Natl Acad Sci USA 89 6837–6840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohm M, Jouanin L, Kunert KJ, Pruvost C, Polle A, Foyer CH, Rennenberg H (1995) Regulation of glutathione synthesis in leaves of transgenic poplar (Populus tremula X P. alba) overexpressing glutathione synthetase. Plant J 7 141–145 [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11 1187–1194 [Google Scholar]
- Vauclare P, Kopriva S, Fell D, Suter M, Sticher L, von Ballmoos O, Krähenbuhl U, Op den Camp R, Brunold C (2002) Flux control of sulphate assimilation in Arabidopsis thaliana: adenosine 5′-phosphosulphate reductase is more susceptible to negative control by thiols than ATP sulphurylase. Plant J 31 729–740 [DOI] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15 2532–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerman S, Stulen I, Suter M, Brunold C, De Kok LJ (2001) Atmospheric H2S as sulphur source for Brassica oleracea: consequences for the activity of the enzymes of the assimilatory sulphate reduction pathway. Plant Physiol Biochem 39 425–432 [Google Scholar]
- Xiong L, Schumaker KS, Zhu JK (2002) Cell signaling during cold, drought, and salt stress. Plant Cell 14 S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.